Abstract
Objective
The objective with this study was to determine and follow antimicrobial resistance in faecal bacteria over time in hospital wastewater pipe sediment. A further aim was to determine bacterial growth rates of sensitive, intermediate and resistant intestinal enterococci in different ciprofloxacin concentrations as a measure of bacterial fitness.
Methods
A system enabling the collection of settled particles over time was installed at Kalmar County Hospital. Samples were collected bi-monthly for a 14-month period. Coliform bacteria and enterococci were isolated from the sediment with standard methods and investigated for resistance to ciprofloxacin (CIP), imipenem (IMI), trimetroprim-sulfamethoxazole (TS), ampicillin (AMP) and vancomycin (VAN) by the disc diffusion method. Resistant isolates were further typed with the PhenePlateTM system. Growth assessments were performed with an automated spectrophotometer.
Results
The rate of intestinal enterococci resistance was <0.6, 1.3, 1.9 and 13% to VAN, IMI, AMP and CIP respectively. Coliform resistance frequencies were 1.1, 2.2 and 2.2% to CIP, IMI and TS respectively. At two sampling occasions, significantly higher rates of ciprofloxacin resistant enterococci were found and the establishment of a resistant clone in the sewer was indicated by the PhP-analysis. Ciprofloxacin resistant intestinal enterococci had a significantly longer lag-phase time than sensitive isolates, but from 500 µg ml−1 (half MIC) resistant isolates had a competitive advantage in terms of significantly faster generation time.
Discussion
Despite high concentration of antimicrobials in the sediment, resistance frequencies were generally low. This can depend on limited growth possibilities for faecal bacteria. However, the establishment of a resistant clone shows that hospital sewers can serve as a reservoir for antibiotic resistant bacteria.
Keywords: hospital wastewater, sediments, antimicrobial resistance, enterococci, coliform bacteria
After ingestion, oral antimicrobials are excreted mainly into wastewater. Some are excreted completely unchanged, whereas others are more or less modified by chelate bindings, degradation or inactivation by metabolic systems inside or outside the host (1). Antimicrobial excretion in combination with high microbial biomass and an abundance of nutrients makes wastewater a potential habitat for horizontal gene transfer and selection of antimicrobial resistant bacteria. Although due to dilution in wastewater, concentrations of antimicrobials in general are too low to inhibit growth, susceptible bacteria can be affected, leading to selection of resistant bacteria (2, 3). Some studies have detected an increase in resistance in wastewater, for example over treatment (4–7), while others have detected a decrease (2, 5, 8, 9). However, sewage pipes also contain settled particles. This sediment of settled particles forms a dynamic ecosystem in which hydrophobic chemical substances are enriched. In a study by Petersen et al. (10), antimicrobial resistance increased in Acinetobacter and enterococci in the sediment-water interface in integrated fish farm ponds receiving poultry and pig manure as a fertiliser. The manure originated from livestock routinely fed antimicrobials as growth promoters. The increase in resistance was explained by either the introduction of resistant bacteria or the selective pressure of antimicrobials in the pond environment. In another fish farm study, oxytetracycline resistance initially increased in sediment bacteria before declining again between 33 and 73 days after the end of antimicrobial treatment (11). If the sediment is constantly exposed to antibiotics, like for example in hospital wastewater systems, the risk of resistance development is likely to increase since enrichment of antimicrobials in sewer sediments can form microniches for a selective pressure on bacterial populations. A study of the degradation and genotoxicity of antimicrobials showed that none of the compounds tested (ciprofloxacin, ofloxacin and metronidazole) were biodegraded (3). Hospital sewers may thus act as an environmental reservoir for antibiotic resistant bacteria (12, 13).
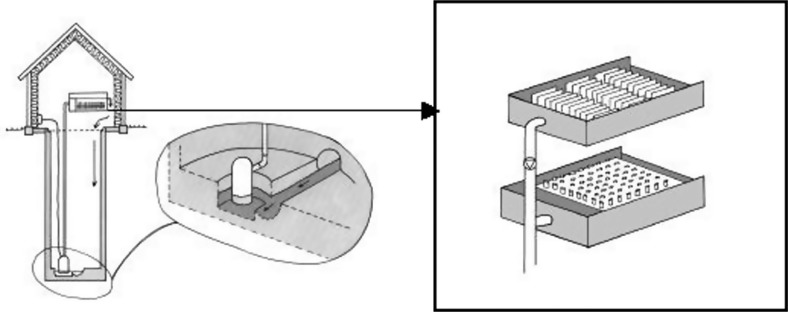
Therefore a system was developed to enable studies of the dynamics of resistance development and antimicrobial concentrations in hospital wastewater (Fig. 1). The specific aim of the reported study was to determine resistance of faecal indicator bacteria to some of the commonly used antimicrobials at the hospital and to follow resistance development over a period of 60 weeks. A further study determined the generation and lag-phase times of bacterial isolates classified as resistant, sensitive and intermediate.
Fig. 1.
Sampling arrangement in a sewage line at Kalmar county hospital. The sampling equipment was installed in a manhole. Wastewater was pumped from a sump placed with the bottom lower than the main furrow in the manhole to ensure the continuous presence of water in the sump. In the tank, settled particles were collected in tubes.
Materials and methods
In the main sewage pipe from a newly established section of Kalmar County Hospital, containing 127 beds (72 general and urology surgery, 33 gynaecology and 22 paediatric), a device that allowed chemical and microbiological sampling of biofilms and trapped sediment from wastewater at chosen intervals was installed in a 4-m deep manhole (14). Wastewater was pumped from a sump placed laterally to the main pipe to a test tank, where the content of solids settled into sampling tubes (Fig. 1). Water supply data was continuously monitored and the amount of antibiotics used at each ward registered by the hospital pharmacy. Results from these studies have been published by Jarnheimer et al. (14).
Sampling and isolation of bacteria
Bi-monthly sampling was carried out over a 14-month period. Three tubes were collected and transported to the laboratory. Ten grams settled sediment was mixed with 90 ml phosphate buffered saline (PBS) 0.5% Tween 80 in a Stomacher as described by Långmark et al. (15). The extract was serially diluted and colonies of intestinal enterococci and coliform bacteria were isolated by standard methods (16, 17). Briefly, 0.1 ml from a suitable dilution was spread onto m-Endo agar, LES (Difco; Becton, Dickinson and Company; Franklin Lakes, New Jersey) and incubated for 21±3 h at 35 °C. Typical metallic colonies were further tested for oxidase production and oxidase negative colonies confirmed as being coliform bacteria. To isolate intestinal enterococci, 0.1 ml was spread onto m-Enterococcus agar (Difco) and incubated for 44±4 h at 35 °C. Typical dark red colonies were verified as intestinal enterococci by their ability to ferment esculine at 44 °C.
Antimicrobial resistance determination
Twelve to fifty-eight colonies from every sampling date were tested for resistance by the disc diffusion method on paper disc media (PDM) agar (AB Biodisk; Solna, Sweden) following the recommendations of the Swedish Reference Group for Antibiotics (18). The coliform bacteria were tested for resistance to ciprofloxacin (CIP), imipenem (IMI), and trimetroprim-sulphamethoxasol (TS). Intestinal enterococci were tested for resistance to CIP, ampicillin (AMP), vancomycin (VAN) and IMI. The following disc concentrations were used: CIP, 5 µg; AMP, 10 µg; VAN, 5 µg; IMI, 10 µg and TS, 25 (23.2+1.8) µg. Inhibition zone diameters were measured after 24±3 h incubation at 37 °C and isolates classified according to the respective breakpoints as shown in Table 1.
Table 1.
Breakpoints for sensitive (S), intermediate (I) and resistant (R) categorisation by the paper disc method and minimum inhibitory concentration (MIC) breakpoints for resistance categorisation by the spread plate method
| Zone diameter breakpoints (mm) | MIC breakpoints (mg l−1 agar) | ||||
|---|---|---|---|---|---|
| Antimicrobial | Paper disc concentration (µg) | Coliforms | Enterococci | Coliforms | Enterococci |
| Ampicillin | 10 | – | 20/16 | – | 8 |
| Imipenem | 10 | 23/16 | 20/16 | 8 | 8 |
| Vancomycin | 5 | – | 11/9 | – | 4 |
| Ciprofloxacin | 5 | 24/17 | 32/12 | 1 | 2 |
| Trimetroprim–Sulphamethoxasol | 25 | 17/13 | – | 32 | – |
Zone breakpoints 23/16, i.e. clear zone with a diameter: S≥23 mm, R≤16 mm and 16<I<23. MIC breakpoint 8: R can grow in antimicrobial concentration>8 mg l−1 (18).
In parallel, the frequency of resistant presumptive enterococci and coliform bacteria was assessed by plating sediment samples on selective media, with and without antimicrobials in concentrations corresponding to the MIC of the respective organism (Table 1), as described above. The resistance frequency was expressed as the ratio of CFU ml−1 on agar with antimicrobials to CFU ml−1 on agar without antimicrobials.
Typing of resistant isolates
Intestinal enterococci isolates that were classified as resistant with the disc diffusion method were typed using the PhenePlateTM PhP-RF system (PhPlate; Stockholm, Sweden) (19). Resistant coliform bacteria were typed with PhenePlateTM PhP-RS system (PhPlate) (20). These systems give each isolate a biochemical fingerprint based on the fermentation kinetics of 11 carbohydrates. Isolates were given an identity by comparing the PhP profile with reference strains ran at the same time.
Growth assessment
Nine intestinal enterococci isolates, three each from the groups CIP resistant (R), CIP intermediate (I) and CIP sensitive (S) were chosen for growth assessments. Using an automated spectrophotometer, Bioscreen C (Lab Systems; Helsinki, Finland), the lag-phase and generation time during the log-phase at 37 °C were measured in Luria Bertoni broth (Difco) at OD 600 nm (21). The generation times were calculated in relation to a standard curve validated with plate count.
Data analysis
The Simpson's diversity index between resistant isolates was calculated with the PhP system's software and similarities were illustrated with a dendrogram produced by the same software (PhPlate). Statistical analyses were performed in SigmaStat 3.0 (SPSS Inc.; Chicago, Illinois) using the z-test for frequencies and ANOVA, all pairwise multiple comparison procedures (Holm-Sidak method).
Results
With the disc diffusion method, nine resistant coliform isolates out of 273 tested were resistant and the frequencies were 1.1, 2.2 and 2.2% to CIP, IMI and TS respectively (Table 2). None of the isolates were resistant to two or more substances. The five TS resistant isolates from January 2002 were of the same phenotype, but no close relationship between isolates could otherwise be verified by the PhP-typing (Simpson's diversity index=0.972; data not shown). The screening method gave resistance frequencies from <0.1 to 5.4% (Table 2).
Table 2.
Resistant coliform bacterial isolates (/total isolates) by the disc diffusion method on PDM agar and percentage resistant coliform bacteria as determined by the ratio CFU ml−1 on LES endoagar with and without antimicrobials (%) in concentrations corresponding to Table 1
| Ciprofloxacin | Imipenem | Trimetroprim–Sulphamethoxazole | ||||
|---|---|---|---|---|---|---|
| Antimicrobial Medium | PDM | LES | PDM | LES | PDM | LES |
| Sampling date | ||||||
| 13 December 2000 | 0/44 | 1.4 | 0/44 | <0.1 | 0/44 | 0.7 |
| 15 February 2001 | 2/43 | <0.1 | 1/43 | <0.1 | 0/43 | 0.2 |
| 17 April 2001 | 0/58 | 1.1 | 3/58 | <0.1 | 0/58 | 3.4 |
| 07 June 2001 | 0/16 | ND | 0/16 | ND | 0/16 | ND |
| 30 August 2001 | 0/46 | ND | 0/46 | ND | 0/46 | ND |
| 24 October 2001 | 0/34 | <0.1 | 1/29 | <0.1 | 1/29 | 5.4 |
| 24 January 2002 | 1/37 | <0.1 | 1/37 | 0.1 | 5/37 | 3.8 |
| 3/273 | 6/273 | 6/273 | ||||
Results from the disc diffusion method on intestinal enterococci are presented in Table 3. No VAN resistant enterococci were detected (N=158). The highest frequency measured was to CIP with 21 of 158 (13%) isolates being resistant. On two sampling occasions (30 August, 10 October 2001) significantly higher frequencies of resistant isolates were found (P<0.05). Also with the screening method significantly higher CIP resistance frequencies were detected at two occasions (P<0.05), however at different sample dates (Table 3).
Table 3.
Resistant intestinal enterococci isolates (/total isolates) by the disc diffusion method on PDM agar and percentage resistant enterococci as determined by the ratio CFU ml−1 on ES agar with and without antimicrobials (%) in concentrations corresponding to Table 1
| Ciprofloxacin | Imipenem | Ampicillin | Vancomycin | |||||
|---|---|---|---|---|---|---|---|---|
| Antimicrobial Medium | PDM | ES | PDM | ES | PDM | ES | PDM | ES |
| Sampling date | ||||||||
| 13 December 2000 | 2/12 | 0.2 | 2/12 | 0.2 | 2/12 | 0.1 | 2/12 | <0.1 |
| 15 February 2001 | 0/24 | 3.5a | 0/24 | <0.1 | 0/24 | <0.1 | 0/24 | <0.1 |
| 17 April 2001 | 2/24 | 4.0a | 0/24 | 1.0 | 1/24 | <0.1 | 0/24 | <0.1 |
| 07 June 2001 | 0/20 | ND | 0/20 | ND | 0/20 | ND | 0/20 | ND |
| 30 August 2001 | 9/24a | ND | 0/24 | ND | 0/20 | ND | 0/20 | ND |
| 24 October 2001 | 8/30a | 0.4 | 0/30 | 0.3 | 0/30 | <0.1 | 0/30 | <0.1 |
| 24 January 2002 | 0/24 | <0.1 | 0/24 | <0.1 | 0/24 | <0.1 | 0/24 | <0.1 |
|
| ||||||||
| 21/158 | 2/158 | 3/158 | 0/158 | |||||
a(P<0.05).
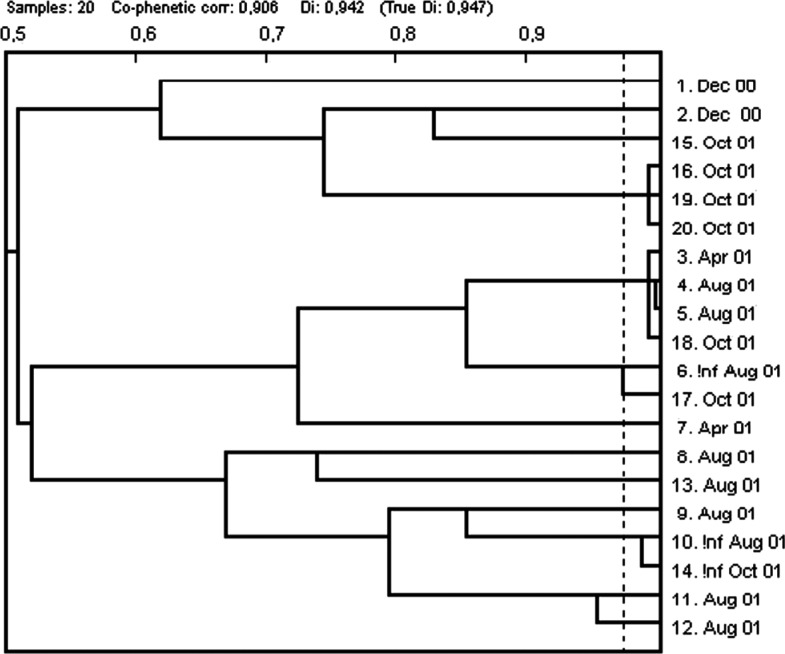
Phenotyping of CIP resistant isolates with the PhP system for enterococci (19) produced a diversity index of 0.947 (Fig. 2). However, two clusters could be identified. One of these contained isolates from the same sampling occasion whereas the other cluster contained isolates from three different samplings, April, Aug and Oct (Fig. 2).
Fig. 2.
Dendrogram from PhP-typing of ciprofloxacin-resistant faecal enterococci at sample dates.
Three intestinal enterococci isolates each from the CIP resistance classes R, I and S (based on clear zone diameter, Table 1) were further assessed for growth in different CIP concentrations. Sensitive isolates had significantly shorter lag-phase in medium without antibiotics and up to CIP concentration 1000 µg l−1. In 500 µg l−1 S grew significantly slower (P<0.01) than R and I respectively. At 1000 µg l−1 R had the fastest growth rate and at 4000 µg l−1 only resistant isolates could grow (Table 4).
Table 4.
Generation and lag-phase times (±standard deviation) (min) for nine independent intestinal enterococci isolates from three CIP resistance classes in different ciprofloxacin concentrations (µg l−1)
| 0 (µg l−1) | 100 (µg l−1) | 500 (µg l−1) | 1000 (µg l−1) | 4000 (µg l−1) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CIP resistance class | Generation time (min) | Lag-phase time (min) | Generation time (min) | Lag-phase time (min) | Generation time (min) | Lag-phase time (min) | Generation time (min) | Lag-phase time (min) | Generation time (min) | Lag-phase time (min) |
| Resistant | 39.8±0.2 | 127±12 | 39.8±0.1 | 127±12 | 40.2±0.4b’ | 130±17 | 46.6±3.9b,c’ | 138±19 | 52.8±6.6 | 150±17 |
| Intermediate | 40.4±11 | 133±21 | 40.4±0.1 | 133±21 | 43.9±0.3b’ | 133±7.6 | 109±18 | 120±20 | – | – |
| Sensitive | 40.4±11 | 97±6a | 42.1±14 | 97±6a | 61.2±9.6 | 90±10b | 175±21 | 90±10a’ | – | – |
aStatistically shorter time (P<0.05).
a’Statistically shorter time than the longer of the other two (P<0.05).
bStatistically shorter time (P<0.01).
b'Statistically shorter time than the longer of the other two (P<0.01).
c’Statistically shorter time than the longer of the other two (P<0.001).
Discussion
Enterococci and coliform bacteria are commonly assessed for various water quality analysis and both groups include important opportunistic pathogens (22, 23). Resistance to the antimicrobials tested among enterococci and coliform bacteria was low and no evidence of resistance development was detected. Resistance against the tested antimicrobial substances varied between 1.1 and 2.2% of the coliform isolates (N=273) (Table 2). The same figures for enterococci varied from less than 0.6 (VAN) to 13% (CIP) (Table 3). These frequencies are lower than what many other studies have reported. Enterococci faecalis and E. faecicum isolated in Portuguese wastewater had a resistance rate of 23 and 33% against CIP respectively and a selection for resistance over treatment was confirmed (7). From Polish wastewater, Luczkiewicz et al. (24) reported 29% CIP, 3.2% VAN, 7% AMP. On the other hand no carbapenem (IMI and Meropenem) resistant E. coli were found (N=153) but TS resistance was 11% compared to 2.2% in the present study.
Quinolones are not normally used for treating infections caused by enterococci due to their low affinity to enterococcal gyrase. Enterococci are resistant to many antimicrobials and have the ability to require new resistance traits (25). In the present study intestinal enterococci were used as model organisms to study bacterial growth rate in different antibiotic concentrations. These growth assessments showed that CIP resistance in intestinal enterococci is associated with a fitness cost in terms of a significantly longer lag-phase in sub-MIC concentrations. However at 500 µg l−1 equal to half the reported MIC value of 1 mg l−1 (18), sensitive isolates had a significantly slower growth rate than resistant and intermediate isolates (Table 4).
In the hospital wastewater pipes where the present study took place, the highest CIP concentration measured was 100 µg l−1 in the water phase (26). However, in the sediment phase, from which the bacteria were actually isolated, the CIP concentration was 151.4 µg g−1 (∼150 mg l−1) (14). Why a consistent development of increased resistance could not be demonstrated despite these high concentrations of ciprofloxacin present may have several reasons: (1) Reduced antimicrobial effect due to CIP binding strongly to particulate matter (1); (2) bacteria associated with biofilm being less susceptible to antimicrobials (27); or (3) limited growth possibilities for faecal bacteria within the studied system. The importance of bacterial multiplication in the selection for resistant bacteria has been shown, for example for oxytetracycline resistance (28). It would appear that the growth possibilities for faecal bacteria were not as good in this system as has been shown in other wastewater installations and sediments (29, 30). The Acinetobacter spp. used by Guardabassi et al. (31–33), might have been a better target bacterium for the purposes of this study. However, most bacteria of clinical importance transmitted via wastewater are gastrointestinal pathogens. Therefore faecal indicators were considered to be the most suitable organisms to study.
Twice, higher frequencies of CIP resistant enterococci were isolated. This sudden rise may be explained by the large temporal variation of antimicrobial concentrations in the studied system (26), reflecting daily activities. Though sludge should provide a more continuous environment than water, it cannot be excluded that isolates emanated from a single patient. However, evidence for establishment of a CIP resistant clone of Enterococcus faecium was obtained. Isolates from this clone were detected on sampling occasions from April–October 2001. Similarly, a VAN resistant Enterococcus faecalis strain was isolated on several occasions from Swedish sewage, indicating persistence in the system or leakage from a reservoir, e.g. a hospital (12). Further the finding of a clone of TS resistant E. coli in one sample can be interpreted as an indication that growth of resistant bacteria may have taken place within the system.
The rate of antimicrobial resistance in the studied system was lower compared to what has been reported in several other European studies (7, 34, 35), reflecting the general lower use and resistance to antimicrobials in the Swedish society compared to these countries (36). Further investigations quantified genes mediating resistance to aminoglycosides, β-lactams and tetracyclines from DNA extracted from the sediment in the studied system. A higher concentration of all genes was found in the hospital sewer compared to other studied environments (soil and municipal wastewater) (37), indicating the hospitals role as a resistome within the human society. Disinfection of hospital wastewater, e.g. ozonation, could be considered as a means to limit the further transmission of resistant bacteria and active antimicrobials (38) in the environment.
Acknowledgements
The authors thank Stefan Nilsson, Daniel Glatz, Bert Helmersson and Sten-Uno Frisk for their expert technical assistance.
Conflict of interest and funding
There are no conflicts of interest. This study was supported by the Swedish Federation of County Councils, the Swedish Strategic Programme for the Rational Use of Antimicrobial Agents and Surveillance of Resistance (STRAMA) and the Research Council of South Eastern Sweden (FORSS).
References
- 1.Edlund C, Lindqvist L, Nord CE. Norfloxacin binds to human fecal material. Antimicrob Agents Chemother. 1988;32:1869–74. doi: 10.1128/aac.32.12.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hassani L, Imziln B, Boussaid A, Gauthier M. Seasonal incidence of and antibiotic resistance among Aeromonas species isolated from domestic watewater before and after treatment. Microbial Ecol. 1992;23:227–37. doi: 10.1007/BF00164098. [DOI] [PubMed] [Google Scholar]
- 3.Kummerer K, Al-Ahmad A, Mersch-Sundermann V. Biodegradability of some antibiotics, elimination of the genotoxicity and affection of wastewater bacteria in a simple test. Chemosphere. 2000;40:701–10. doi: 10.1016/s0045-6535(99)00439-7. [DOI] [PubMed] [Google Scholar]
- 4.Meckes MC. Effect of UV light disinfection on antibiotic-resistant coliforms in wastewater effluents. Appl Environ Microbiol. 1982;43:371–7. doi: 10.1128/aem.43.2.371-377.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell JB, Elliott GE, Smith DW. Influence of sewage treatment and urbanization on selection of multiple resistance in fecal coliform populations. Appl Environ Microbiol. 1983;46:227–32. doi: 10.1128/aem.46.1.227-232.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mezrioui N, Echab K. Drug-resistance in salmonella strains isolated from domestic waste-water before and after treatment in stabilization ponds in an Arid Region (Marrakech, Morocco) World J Microbiol Biotechnol. 1995;11:287–90. doi: 10.1007/BF00367100. [DOI] [PubMed] [Google Scholar]
- 7.Ferreira da Silva M, Tiago I, Verissimo A, Boaventura RA, Nunes OC, Manaia CM. Antibiotic resistance of enterococci and related bacteria in an urban wastewater treatment plant. FEMS Microbiol Ecol. 2006;55:322–9. doi: 10.1111/j.1574-6941.2005.00032.x. [DOI] [PubMed] [Google Scholar]
- 8.Bell RB. Antibiotic resistance patterns of fecal coliforms isolated from domestic sewage before and after treatment in an aerobic lagoon. Can J Microbiol. 1978;24:886–8. doi: 10.1139/m78-147. [DOI] [PubMed] [Google Scholar]
- 9.Guardabassi L, Lo Fo Wong D, Dalsgaard A. The effects of tertiary wastewater treatment on the prevalence of antimicrobial resistant bacteria. Water Res. 2002;36:1955–64. doi: 10.1016/s0043-1354(01)00429-8. [DOI] [PubMed] [Google Scholar]
- 10.Petersen A, Andersen JS, Kaewmak T, Somsiri T, Dalsgaard A. Impact of integrated fish farming on antimicrobial resistance in a pond environment. Appl Environ Microbiol. 2002;68:6036–42. doi: 10.1128/AEM.68.12.6036-6042.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerry J, Hiney M, Coyne R, Cazabon D, Nicgabhainn S, Smith P. Frequency and distribution of resistance to oxytetracycline in microorganisms isolated from marine fish farm sediments following therapeutic use of oxytetracycline. Aquaculture. 1994;123:43–54. [Google Scholar]
- 12.Iversen A, Kuhn I, Franklin A, Mollby R. High prevalence of vancomycin-resistant enterococci in Swedish sewage. Appl Environ Microbiol. 2002;68:2838–42. doi: 10.1128/AEM.68.6.2838-2842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guillaume G, Verbrugge D, Chasseur-Libotte M, Moens W, Collard J. PCR typing of tetracycline resistance determinants (Tet A-E) in Salmonella enterica serotype Hadar and in the microbial community of activated sludges from hospital and urban wastewater treatment facilities in Belgium. FEMS Microbiol Ecol. 2000;32:77–85. doi: 10.1111/j.1574-6941.2000.tb00701.x. [DOI] [PubMed] [Google Scholar]
- 14.Jarnheimer PÅ, Ottoson J, Lindberg R, Stenström TA, Johansson M, Tysklind M, et al. Fluoroquinolone antibiotics in a hospital sewige line; occurrence, distribution and impact on bacterial resistance. Scand J Inf Dis. 2004;36:752–5. doi: 10.1080/00365540410021027. [DOI] [PubMed] [Google Scholar]
- 15.Långmark J, Ashbolt NJ, Szewzyk U, Stenström TA. Adequacy of in situ glass slides and direct sand extractions to assess the microbiota within sand columns used for drinking water treatment. Can J Microbiol. 2001;47:601–7. [PubMed] [Google Scholar]
- 16.ISO. Geneva: International Standardisation Organisation; 2000. Water quality – detection and enumeration of intestinal enterococci – part 2: membrane filtration method. Report No. 7899-2. [Google Scholar]
- 17.ISO. Geneva: International Standardisation Organisation; 2000. Water quality – detection and enumeration of coliform organisms, thermotolerant coliform organisms and presumptive Escherichia coli – part 1: membrane filtration method. Report No. 9308-1. [Google Scholar]
- 18.Olsson-Liljequist B, Larsson P, Walder M, Miorner H. Antimicrobial susceptibility testing in Sweden. III. Methodology for susceptibility testing. Scand J Infect Dis. 1997;105(Suppl):13–23. [PubMed] [Google Scholar]
- 19.Kühn I, Burman LG, Haeggman S, Tullus K, Murray BE. Biochemical fingerprinting compared with ribotyping and pulse-field gel electrophoreses of DNA for epidemiological typing of enterococci. J Clin Microbiol. 1995;33:2812–17. doi: 10.1128/jcm.33.11.2812-2817.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kühn I, Allestam G, Stenström TA, Möllby R. Biochemical fingerprinting of water coliform bacteria, a new method for measuring phenotypic diversity and for comparing different bacterial populations. Appl Environ Microbiol. 1991;57:3171–7. doi: 10.1128/aem.57.11.3171-3177.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nilsson AI, Zorzet A, Kanth A, Dahlstrom S, Berg OG, Andersson DI. Reducing the fitness cost of antibiotic resistance by amplification of initiator tRNA genes. PNAS. 2006;103:6976–81. doi: 10.1073/pnas.0602171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linden PK, Miller CB. Vancomycin-resistant enterococci: the clinical effect of a common nosocomial pathogen. Diagn Microbiol Infect Dis. 1999;33:113–20. doi: 10.1016/s0732-8893(98)00148-5. [DOI] [PubMed] [Google Scholar]
- 23.Raymond J, Bergeret M, Gendrel D. Impact of bacterial resistance on severe infections. Arch Pediatr. 2001;8(Suppl 4):697s–704. doi: 10.1016/s0929-693x(01)80184-6. [DOI] [PubMed] [Google Scholar]
- 24.Luczkiewicz A, Jankowska K, Fudala-Ksiazek S, Olanczuk-Neyman K. Antimicrobial resistance of fecal indicators in municipal wastewater treatment plant. Water Res. 2010;44:5089–97. doi: 10.1016/j.watres.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 25.Tankovic J, Mahjoubi F, Courvalin P, Duval J, Leclerco R. Development of fluoroquinolone resistance in Enterococcus faecalis and role of mutations in the DNA gyrase gyrA gene. Antimicrob Agents Chemother. 1996;40:2558–61. doi: 10.1128/aac.40.11.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindberg R, Jarnheimer PA, Olsen B, Johansson M, Tysklind M. Determination of antibiotic substances in hospital sewage water using solid phase extraction and liquid chromatography/mass spectrometry and group analogue internal standards. Chemosphere. 2004;57:1479–88. doi: 10.1016/j.chemosphere.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 27.Stewart PS. Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol. 2002;292:107–13. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- 28.Kerry J, Slattery M, Vaughan S, Smith P. The importance of bacterial multiplication in the selection, by oxytetracycline-HCl, of oxytetracycline-resistant bacteria in marine sediment microcosms. Aquaculture. 1996;146:103–19. [Google Scholar]
- 29.Ottoson J, Stenström TA. Faecal contamination of greywater and associated microbial risks. Water Res. 2003;37:645–55. doi: 10.1016/S0043-1354(02)00352-4. [DOI] [PubMed] [Google Scholar]
- 30.Ottosson J, Stenstrom TA. Growth and reduction of microorganisms in sediments collected from a greywater treatment system. Lett Appl Microbiol. 2003;36:168–72. doi: 10.1046/j.1472-765x.2003.01286.x. [DOI] [PubMed] [Google Scholar]
- 31.Guardabassi L, Dalsgaard A, Olsen JE. Phenotypic characterization and antibiotic resistance of Acinetobacter spp. isolated from aquatic sources. J Appl Microbiol. 1999;87:659–67. doi: 10.1046/j.1365-2672.1999.00905.x. [DOI] [PubMed] [Google Scholar]
- 32.Guardabassi L, Dalsgaard A, Raffatellu M, Olsen JE. Increase in the prevalence of oxolinic acid resistant Acinetobacter spp. observed in a stream receiving the effluent from a freshwater trout farm following the treatment with oxolinic acid-mediated feed. Aquaculture. 2000;188:205–18. [Google Scholar]
- 33.Guardabassi L, Petersen A, Olsen JE, Dalsgaard A. Antibiotic resistance in Acinetobacter spp. isolated from sewers receiving waste effluent from a hospital and a pharmaceutical plant. Appl Environ Microbiol. 1998;64:3499–502. doi: 10.1128/aem.64.9.3499-3502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caplin JL, Hanlon GW, Taylor HD. Presence of vancomycin and ampicillin-resistant Enterococcus faecium of epidemic clonal complex-17 in wastewaters from the south coast of England. Environ Microbiol. 2008;10:885–92. doi: 10.1111/j.1462-2920.2007.01507.x. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Aljaro C, Moreno E, Andreu A, Prats G, Blanch AR. Phylogroups, virulence determinants and antimicrobial resistance in stx(2) gene-carrying Escherichia coli isolated from aquatic environments. Res Microbiol. 2009;160:585–91. doi: 10.1016/j.resmic.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Cars O. Annual reports of antibiotic use and resistance – for whom? Neth J Med. 2004;62:405–6. [PubMed] [Google Scholar]
- 37.Borjesson S, Dienues O, Jarnheimer PA, Olsen B, Matussek A, Lindgren PE. Quantification of genes encoding resistance to aminoglycosides, beta-lactams and tetracyclines in wastewater environments by real-time PCR. Int J Environ Health Res. 2009;19:219–30. doi: 10.1080/09603120802449593. [DOI] [PubMed] [Google Scholar]
- 38.Sharma VK. Oxidative transformations of environmental pharmaceuticals by Cl, ClO, O, and Fe(VI): kinetics assessment. Chemosphere. 2008;73:1379–86. doi: 10.1016/j.chemosphere.2008.08.033. [DOI] [PubMed] [Google Scholar]