Abstract
TAR DNA binding protein 43 (TDP-43) is a nucleic acid binding protein that is associated with the pathology of cystic fibrosis and neurodegenerative diseases such as amyotrophic lateral sclerosis and frontotemporal lobar dementia. We have developed a robust, quantitative, nonradiometric high-throughput assay measuring oligonucleotide binding to TDP-43 using AlphaScreen® technology. Biotinylated single-stranded TAR DNA (bt-TAR-32) and 6 TG repeats (bt-TG6) bound with high affinity to TDP-43, with KD values of 0.75 nM and 0.63 nM, respectively. Both oligonucleotides exhibited slow dissociation rates, with half-lives of 750 min for bt-TAR-32 and 150 min for bt-TG6. The affinities of unlabeled oligonucleotides, as determined by displacement of either bt-TAR-32 or bt-TG6, were consistent with previous reports of nucleic acid interactions with TDP-43, where increasing TG or UG repeats yield greater affinity. A diversity library of 7360 compounds was screened for inhibition of TDP-43 binding to bt-TAR-32, and a series of compounds was discovered with nascent SAR and IC50 values ranging from 100 nM to 10 μM. These compounds may prove to be useful biochemical tools to elucidate the function of TDP-43 and may lead to novel therapeutics for indications where the TDP-43 nucleic acid interaction is causal to the associated pathology.
Keywords: TDP-43, AlphaScreen, TAR DNA, ALS, cystic fibrosis
INTRODUCTION
Trans-activating response (TAR) DNA binding protein-43 (TDP-43) is a nucleic acid binding protein that is expressed primarily in the nucleus, where it functions as a regulator of gene transcription and RNA splicing.1 TDP-43 was initially discovered as a protein that binds to TAR DNA sequences and functions as a repressor of HIV-1 gene expression.2 Later studies demonstrated that TDP-43 is integral to pathological exon 9 skipping in the cystic fibrosis transmembrane conductance regulator (CFTR) gene in cystic fibrosis.3–5 Using siRNA to decrease TDP-43 levels, it has been shown that TDP-43 regulates expression of human cyelin-dependent kinase 6 (Cdk6), resulting in changes in the cell cycle with subsequent apoptosis.6 The structure of TDP-43 is typical of heterogeneous nuclear ribonucleoproteins (hnRNPs) with 2 RNA recognition motifs, designated RRM1 and RRM2, along with a glycine-rich C-terminal domain.1,7 The RRM domains bind TAR DNA and TG/UG-rich DNA and RNA oligonucleotides, whereas the glycine-rich domain is known to interact with other hnRNPs and functions as a regulator of gene splicing.2,7,8
Recently, TDP-43 has been implicated in certain neurodegenerative diseases such as frontotemporal lobar degeneration (FTLD) and amyotrophic lateral sclerosis (ALS).1,9–13 Immunocytochemical studies have shown that both TDP-43 and ubiquitinylated TDP-43 are present in neuronal inclusions in FTLD and ALS patients and that this aggregation is accompanied by a decrease of TDP-43 in the nucleus.13–16 This cytoplasmic aggregation of TDP-43 is often associated with point mutations in both familial and sporadic ALS patients.17 Accumulation of TDP-43 or mutant TDP-43 (TDP-43A315T) in the cytoplasm appears to be toxic to primary neurons, where TDP-43A315T is more toxic than wild-type.18 This cytotoxicity may be the result of TDP-43-mediated sequestration of cytoplasmic RNAs such as neurofilaments.18 Conversely, studies characterizing TDP-43 Drosophila knockouts have shown a lack of TDP-43 results in ALS-like symptoms.19 Therefore, additional studies are needed to further understand the role of both wild-type and mutant TDP-43 in ALS and FTLD.
The discovery of small molecules that disrupt nucleotide binding to TDP-43 would be useful probe molecules to further understand the role of TDP-43 in neurodegenerative diseases such as FTLD and ALS, HIV infection, and cystic fibrosis and may eventually serve as the basis for therapeutics. Previous studies have characterized oligonucleotide binding to TDP-43 using electromobility shift assays.7,8 However, although these assays are useful in providing qualitative estimates of affinity and rank order, they are unable to provide quantitative estimates of affinity and binding kinetic rate constants in a rapid and convenient format required for modern drug discovery efforts. We have developed a robust assay suitable for high-throughput screening (HTS) that measures inhibition of oligonucleotide binding to TDP-43 using Amplified Luminescence Proximity Homogenous Assay (AlphaScreen®) technology. We then characterized the binding of oligonucleotides to TDP-43 and have generated estimates of affinity and association and dissociation rate constants using this assay. Screening of a diversity set of more than 7000 compounds identified a class of related compounds with sub-micromolar affinities that disrupt oligonucleotide binding to TDP-43.
MATERIALS AND METHODS
Materials
TDP-43 with a glutathione S-transferase (GST) tag on the C-terminus was purchased from Abnova (Tapei City, Taiwan). Single-stranded DNA and RNA oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA), where TAR-32 and TG6 were biotinylated (bt) at the 5′ end. The TAR-32 sequence is (5′-CTG CTT TTT GCC TGT ACT GGG TCT CTG TGG TT-3′) and corresponds to the first 32 nucleotides from the sequence identified by Ou et al.2 to bind to TDP-43. AlphaScreen® GST detection kit and AlphaScreen® TruHTS assay kit were purchased from PerkinElmer Life Sciences (Waltham, MA).
Our chemical library was assembled by culling from large libraries to represent a variety of heterocyclic chemotypes compatible with drug suitability and CNS penetration. The molecular weight range was 200 to 400, which allows great flexibility for hit-to-lead chemistry. Twenty percent of the library comes directly from a lead generation set of compounds. The following core heterocycles were included: azaindole, quinolone, pyrimidine, benzopyrimidine, isoxazole, oxadiazole, oxazole, purine, thiazole, and triazole. Filters for removal included 2 or more rule-of-5 violators; anionically charged compounds such as carboxylates, phosphates, phosphonates, and sulfates; N-oxides; nitro compounds, hydrazines, hydrazides; thiocarbonyls, α- and β-unsaturated carbonyls; Michael acceptors; and sulfhydryl groups.
bt-TAR-32 and bt-TG6 binding to TDP-43
Assay mixtures contained a series of concentrations of TDP-43 (0.47–30 ng/mL), a series of concentrations of bt-TAR-32 or bt-TG6 (10–320 pM), and 10 μg/mL of AlphaScreen® streptavidin donor beads and anti-GST acceptor beads in a total volume of 40 μL assay buffer (25 mM Tris [pH 7.4], 0.1% bovine serum albumin [BSA], 0.1% Triton X-100) in 384-well plates. AlphaScreen® beads were handled in a dark room under green light to avoid photo-depleting the AlphaScreen® beads. Nonspecific binding was determined in the absence of TDP-43 and was less than 10% of total binding. Assays were incubated in the dark at room temperature for 3 h to ensure the binding reaction was at equilibrium, and then the AlphaScreen® signal was measured on a Synergy 2 plate reader (BioTek, Winooski, VT).
Saturation binding of bt-TAR-32 and bt-TG6 to TDP-43
TDP-43 (5 μg/mL, 40 pM) was preincubated with 10 μg/mL anti-GST acceptor beads in assay buffer for 30 min at room temperature and then added to assays containing a series of concentrations of bt-TAR-32 or bt-TG6 ranging from 4.2 pM to 10 nM that had also been preincubated with 10 μg/mL streptavidin donor beads for 30 min at room temperature. Nonspecific binding was determined in the presence of 10 μM TG4. After incubation in the dark at room temperature for 3 h to ensure the binding reaction was at equilibrium, the AlphaScreen® signal was measured on a Synergy 2 plate reader. The equilibrium dissociation constants (KD) for TAR-32 and TG6 binding to TDP-43 were determined from nonlinear regression fits of the data to a 1-site binding model in GraphPad Prism®, version 5.0 (GraphPad Software, San Diego, CA).
Association and dissociation rates of bt-TAR-32 and bt-TG6 binding to TDP-43
A series of concentrations of bt-TAR-32 or bt-TG6 ranging from 0.25 to 6.0 nM were preincubated with 10 μg/mL streptavidin donor beads for 30 min at room temperature. Assays were initiated at various times by the addition of TDP-43 (5 ng/mL, 40 pM) with 10 μg/mL anti-GST donor beads that were preincubated as described above, and the AlphaScreen® signal was measured at the end of the time course. The observed association rate constant (kobs) was determined from nonlinear regression fits of the data for each ligand concentration to a 1 -site exponential association model in GraphPad Prism®, version 5.0. A plot of the resulting kobs values versus ligand concentration for both TAR-32 and TG6 yielded a straight line with the slope equal to the association rate constant (kon) and the Y-intercept equal to the dissociation rate constant (koff). The KD value determined by koff/kon was compared against the KD value determined by saturation binding using an unpaired, 2-tailed r-test using GraphPad Prism®.
Inhibition of bt-TAR-32 and bt-TG6 binding to TDP43 by unlabeled oligonucleotides and test compounds
Test compounds or oligonucleotides were diluted in assay buffer at 4× the desired final concentration. DMSO concentrations were less than 1% because this was the level of DMSO tolerance in the assay. Dilutions of test compound were preincubated with TDP-43 (5 ng/mL, 40 pM) prebound to AlphaScreen® anti-GST acceptor beads (10 μg/mL) for 30 min at room temperature. Assays were initiated by the addition of bt-TAR-32 or bt-TG6 (0.1 nM) prebound to AlphaScreen® strepavidin donor beads (10 μg/mL). After incubation in the dark at room temperature for 3 h to ensure the binding reaction was at equilibrium, the AlphaScreen® signal was measured. IC50 values were determined from nonlinear regression fits of the data to a 1-site competition model in GraphPad Prism®. For unlabeled oligonucleotides, KD values were determined from Cheng-Prusoff corrections of the IC50 values. Z′ values were determined for each plate to assess assay robustness.20 Initial hits with inhibition values greater than 75% from screening ~7400 compounds were then titrated in bt-TAR-32 binding to TDP-43 to confirm activity and determine IC50 values. Confirmed actives were then tested in the AlphaScreen® TruHTS assay according to the manufacturer’s directions to rule out nonspecific interference with the AlphaScreen® signal by compounds that have an inner filter effect, are singlet oxygen quenchers, or are biotin mimetics.
RESULTS
Characterization of bt-TAR-32 and bt-TG6 binding to TDP-43
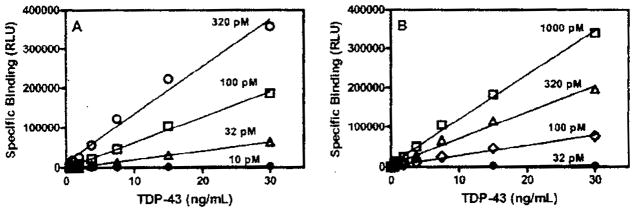
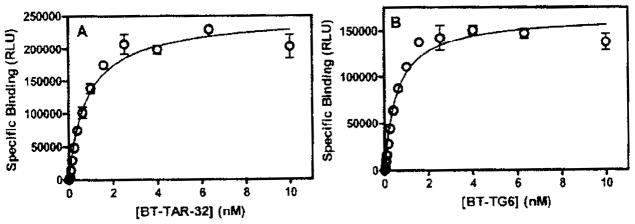
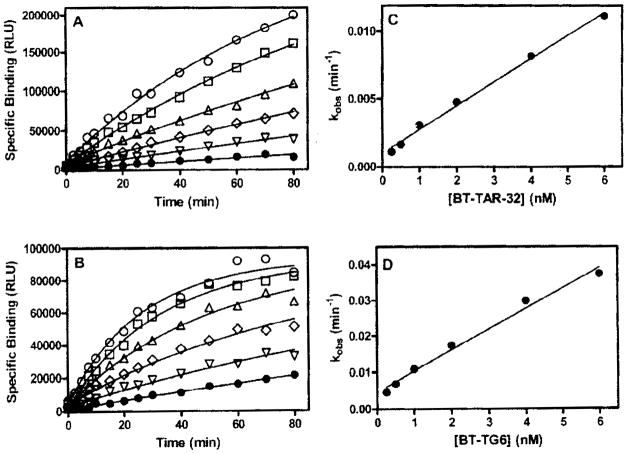
Binding of bt-TAR-32 and bt-TG6 was linear with TDP-43 concentration as well as dependent on the concentration of labeled oligonucleotide consistent with a specific binding interaction (Fig. 1). Saturation binding experiments showed that binding of both bt-TAR-32 and bt-TG6 to TDP-43 was saturable and with high affinity, with KD values of 0.75 nM (95% confidence interval [CI), 0.29–1.9 nM) and 0.63 nM (95% CI, 0.54–0.74 nM), respectively (Fig. 2). Time course experiments showed that binding of bt-TAR-32 and bt-TG6 was both time and concentration dependent. The association rates for both bt-TAR-32 and bt-TG6 to TDP-43 were similar, with Kon values of 2.4 ± 0.93 μmol−1 min−1 and 4.5 ± 1.3 μmol−1 min−1, respectively (Fig. 3 and Table 1). The observed association rate constants (kobs) for both bt-TAR-32 and bt-TG6 binding to TDP-43 were linearly proportional to ligand concentration, which is consistent with binding to a single population of noninteracting sites (Fig. 3).21 Both labeled oligonucleotides exhibited slow dissociation rates, with koff values of 8.8 × 10−4 min−1 (t1/2 = 750 min) for bt-TAR-32 and 4.5 × 10−3 min−1 (t1/2 = 150 min) for bt-TG6 (Fig. 3 and Table 1). The KD values obtained by saturation experiments were not statistically different from the KD values calculated by the ratio of koff/kon, as determined by an unpaired, 2-tailed t-test (p = 0.12), consistent with the binding reactions reaching equilibrium at the 3-h time point.
FIG. 1.
Concentration dependence of bt-TAR-32 and bt-TG6 binding to TDP-43. (A) Specific binding of bt-TAR-32 DNA to TDP-43. (B) Specific binding of bt-TG6 DNA to TDP-43. Data are from a single experiment that was repeated 3 times with similar results.
FIG. 2.
Saturation isotherms of bt-TAR-32 and bt-TG6 DNA binding to TDP43. TDP-43 (5 ng/mL) was incubated for 3 h with a series of concentrations of either (A) bt-TAR-32 or (B) bt-TG6 as described in the Methods section. Data shown are the means ± SEM of triplicate values from a single experiment that was repeated 3 times with similar results.
FIG. 3.
Association rate of bt-TAR-32 and bt-TG6 binding to TDP43. TDP43 (5 ng/mL) was incubated with a series of concentrations of either bt-TAR-32 or bt-TG6 as described in the Methods section. Specific binding over time of (A) bt-TAR-32 and (B) bt-TG6 at 6.0 nM (○), 4.0 nM (□), 2.0 nM (△), 1.0 nM (◇), 0.50 nM (▽), and 0.25 nM (■), Observed association rate constants (kobs) at different concentrations of either (C) bt-TAR-32 or (D) bt-TG6. Data are means of duplicate determinations from a single experiment that was repeated 3 times with similar results.
Table 1.
Association and Dissociation Rate Constants of bt-TAR-32 and bt-TG6 Binding to TDP-43
| Oligonucleotide | kon (μmol−1 min−1) | koff (10−3 min−1) | KD (nM)a | KD (nM)b |
|---|---|---|---|---|
| TAR-32 | 2.4 ± 0.93 | 0.88 ± 0.19 | 0.40 (0.090–1.8) | 0.75 (0.29–1.9) |
| TG6 | 4.5 ± 1.3 | 4.5 ± 0.49 | 1.1 (0.33–3.7) | 0.63 (0.54–0.74) |
Association and dissociation rates were determined from linear regression fit of kobs as a function of ligand concentration where the slope is equal to kon and the Y-intercept is equal to koff. Rate constants expressed as the arithmetic mean ± SEM, whereas the KD values are expressed as the geometric mean and 95% confidence intervals of 3 determinations.
Determined by the quotient of koff/kon, as described in the Methods section.
Determined by saturation binding, as described in the Methods section.
Pharmacology of bt-TAR-32 and bt-TG6 binding to TDP-43
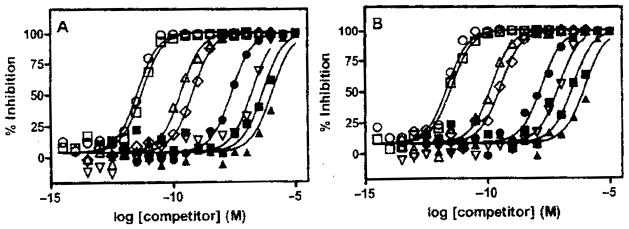
A series of unlabeled single-stranded DNA and RNA oligonucleotides were tested for their ability to inhibit either bt-TAR-32 or bt-TG6 binding to TDP-43 (Fig. 4 and Table 2). In general, the affinities for all unlabeled oligonucleotides were largely independent of the competing ligand, either bt-TAR-32 or bt-TG6. In addition, DNA oligonucleotides bound with higher affinity than RNA oligonucleotides. Binding to TDP-43 was highly sensitive to the number of TG repeats in the DNA oligonucleotides and UG repeats in the RNA nucleotides, with rank order affinities of TG12 > TG8 ≫ TG6 ≫ TG4 and UG12 > UG8 > UG6. There was no detectable binding of TDP-43 to the TG or AC dinucleotide. Both DNA and RNA AC repeats exhibited low affinity for TDP-43, with KD values 3.3 μM and 12 μM in bt-TAR-32 binding, respectively. This rank order potency is consistent with previous findings using electromobility shift assays to measure binding to TDP-43.8 In addition, the RNA sequence of 3 UCUU repeats exhibited low affinity at TDP-43, which is also consistent with previous studies.8 The affinity of unlabeled TAR-32 was similar to TG6. The KD values for both TAR-32 and TG6 were slightly greater than their respective KD values, suggesting that the biotinylation and/or binding to the streptavidin donor beads cause a modest decrease in affinity.
FIG. 4.
Inhibition of bt-TAR-32 and bt-TG6 binding to TDP-43. TDP-43 (5 ng/mL) and 0.1 nM of (A) bt-TAR-32 or (B) bt-TG6 were incubated for 2 h with a series of concentrations of unlabeled oligonucleotides: TG12 (○), TG8 (□), TAR-32 (△), TG6 (◇), TG4 (●), UG12(▽), UG8 (■), and UG6 (▲). Data are single determinations from a single experiment that was repeated at least 5 times with similar results.
Table 2.
Affinities of ssDNA and ssRNA Oligonucleotides Determined by Displacement of Either bt-TAR-32 or bt-TG6 Binding to TDP-43
| KD (95% CI), nM
|
||
|---|---|---|
| TAR-32 | TG6 | |
| ssDNA oligonucleotide | ||
| TG12 | 0.0018 (0.00084–0.0038) | 0.0013 (0.00047–0.0036) |
| TG8 | 0.0058 (0.0033–0.010) | 0.0064 (0.002–0.015) |
| TAR-32 | 0.14 (0.11–0.19) | 0.12 (0.074–0.21) |
| TG6 | 0.41 (0.26–0.66) | 0.29 (0.11–0.75) |
| TG4 | 12 (5.3–26) | 13 (7.8–20) |
| dAC12 | 3300 (1500–7200) | NA |
| TG | > 100,000 | > 100,000 |
| ssRNA oligonucleotide | ||
| UG12 | 92 (47–180) | 130(86–190) |
| UG8 | 370 (160–370) | 450 (270–750) |
| UG6 | 750 (380–1500) | 1300 (990–1700) |
| UCUU3 | 14,000 (7400–27,000) | 9800 (7600–13,000) |
| rAC12 | 12,000 (4200–33,000) | NA |
Oligonucleotides were titrated in bt-TAR-32 and bt-TG6 binding to TDP-43 and the KD values determined as described in the Methods section. Values shown are the geometric mean and 95% confidence intervals for Between 5 and 7 determinations. NA, AC repeats hybridize with the bt-TG6 and therefore prevent the determination of affinity at TDP-43.
Screening for novel inhibitors of bt-TAR-32 binding to TDP-43
Given the similar pharmacology of bt-TAR-32 and bt-TG6 binding to TDP-43, we chose to use the bt-TAR-32 sequence for compound screening because the signal-to-noise ratio was slightly higher for bt-TAR-32. To evaluate the performance of the assay in screening mode, a 384-well plate was tested with 4 concentrations of TG4 dispensed to the 4 quadrants on the assay plate. There was no overlap in inhibition values between the 5-nM and 16-nM concentrations, and the 50-nM and 160-nM concentrations were greater than 75%, indicating the assay was robust and reliable. Furthermore, the Z′ value in this experiment was 0.55, as calculated using the 5-nM quadrant as the negative control and the 50-nM quadrant as the positive control. From these data, with a screening concentration of 10 μM and a threshold of 75%, the assay can reliably identify novel compounds with IC50 values 2 μM or less.
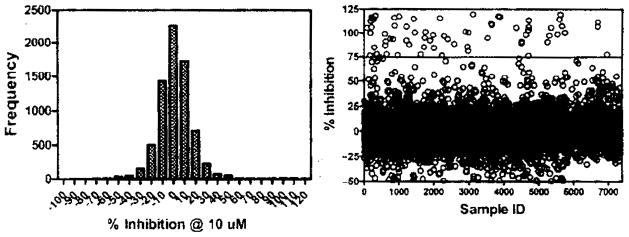
A set of 7360 chemically diverse, drug-like compounds was screened at 10 μM against bt-TAR-32 binding to TDP-43. The mean inhibition value was 2.2% with a standard deviation of 19% (Fig. 5). Z′ values ranged from 0.4 to 0.7, and the mean coefficient of variation (CV) value for plates was 8%. There were 69 compounds with inhibition values greater than 75%, yielding a hit rate of 0.94%. The initial hits were then retested and the activity of 27 compounds repeated, yielding a confirmation rate of 39% and overall hit rate of 0.3%. The confirmed actives were then tested for nonspecific interference in the assay using the AlphaScreen® TruHTS assay kit. Two of the 27 compounds inhibited AlphaScreen® TruHTS signal by greater than 50% at 10 μM, indicating that there is significant nonspecific interference with these compounds. Within the remaining compounds, a nascent structure-activity relationship (SAR) in a series of compounds was observed, with KD values ranging from 100 nM to 10 μM (Table 3).
FIG. 5.
Frequency histogram and scatter plot of inhibition values from 7360 compounds screened for inhibition of bt-TAR-32 binding to TDP43. TDP-43 (5 ng/mL) and 0.1 nM bt-TAR-32 were incubated with 10 μM test compound. The mean inhibition value for the compounds tested was 2.2% with a standard deviation of 19%. Data represent single determinations across twenty-four 384-well plates tested on 4 different days.
Table 3.
Affinities of Screening Hits as Determined by Displacement of bt-TAR-32 Binding to TDP-43
| Compound | IC50 (95% CI), μM |
|---|---|
| 1 | 0.12 (0.067–0.21) |
| 2 | 0.15 (0.065–0.33) |
| 3 | 0.28 (0.19–0.42) |
| 4 | 0.47 (0.29–0.76) |
| 5 | 0.50 (0.25–1.0) |
| 6 | 0.71 (0.49–1.0) |
| 7 | 1.1 (0.65–1.2) |
| 8 | 1.2 (0.5–2.9) |
| 9 | 1.3 (1.2–1.5) |
| 10 | 3.9 (2.8–5.5) |
| 11 | >10 |
Values shown are the geometric means with 95% confidence intervals of 3 determinations.
DISCUSSION
In this present study, we have developed and validated a novel, quantitative, nonradiometric assay for nucleic acid binding to TDP-43. We used this assay to characterize the binding of bt-TAR-32 and bt-TG6 and are the first to report association and dissociation rate constants for these interactions. The binding of both bt-TAR-32 and bt-TG6 to TDP-43 was saturable and of high affinity, with sub-nanomolar KD values. The association rates for both bt-TAR-32 and bt-TG6 binding to TDP-43 were similar, whereas the dissociation rate of bt-TAR-32 was slower than that of bt-TG6. The observed association rates of both bt-TAR-32 and bt-TG6 increased linearly with ligand concentration, which is consistent with a lack of cooperativity between binding sites.21 Furthermore, the equilibrium dissociation constants as determined by the ratio of the dissociation and association rate constants were the same as that obtained from equilibrium binding experiments. These results suggest that the stoichiometry of binding of DNA oligonucleotides to TDP-43 is 1:1, with no apparent cooperativity, and that binding is consistent with the law of mass action.
The rank order affinities of unlabeled DNA and RNA oligonucleotides obtained in this study were consistent with pharmacology reported by other investigators for TDP-43.8 Similar to previous studies, the greater number of TG repeats resulted in increased affinity for TDP-43. It is important to note that the apparent affinities of both TG8 and TG12 were in the picomolar range and therefore may be an underestimation of their actual affinities because of significant ligand depletion, given the concentration of TDP-43 (40 pM) in these experiments. It is also interesting that the affinity of TG8 was more than 100-fold greater than TG6, and the affinity of TG6 was 30-fold greater than TG4, thereby indicating that TDP-43 is highly sensitive to the number of TG repeats in an oligonucleotide sequence. Although the affinities of RNA oligonucleotides were much less than those of DNA oligonucleotides, the rank order affinities were consistent with TDP-43 pharmacology.8 Although it has been shown that TDP-43 does not bind TAR RNA,2 it is capable of binding well to both TG DNA repeats and UG RNA repeats in electromobility shift assays.7,8 In our assay, we found that UG repeats were at least 3 orders of magnitude less potent than corresponding TG repeats. It should be noted that although there is an overall shift in potency between RNA and DNA, the weaker interactions with RNA are sub-micromolar and would yield signals in the UV-crossing electromobility shift assays used previously.8 Interestingly, investigators have reported that TDP-43 binding to DNA oligonucleotides resulted in multiple species of complexes, whereas those for RNA formed single complexes.8 It may therefore be that the potency shift for DNA relative to RNA in our quantitative assay is due to multiple noncooperative binding interactions between DNA and TDP-43 that do not occur with RNA.
The dissociation rates of both bt-TAR-32 and bt-TG6 were extremely slow, with half-lives of 750 min and 150 min, respectively, which are consistent with their high affinity for TDP-43. In addition, the affinities of TG12 and TG8 for TDP-43 would suggest that the dissociation rates of these oligonucleotides may be even slower than that of bt-TAR-32. This characteristic “tight-binding” property of these oligonucleotides is interesting in light of the putative role of TDP-43 as a transcription repressor. Theoretically, once TDP-43 is bound to these DNA sequences, the interaction is “pseudo-irreversible,” and a conformational change in the protein is necessary for dissociation and subsequent transcription to occur. Such a conformational change could occur through phosphorylation and/or allosteric binding of another factor to TDP-43. Investigators have shown that the glycine-rich C-terminal tail of TDP-43 is necessary for recruitment of other splicing inhibitory proteins in exon 9 skipping of the CFTR mRNA.1,8,22 Although a direct negative allosteric interaction with TDP-43 has not yet been described, our quantitative assay could be useful in identifying such interactions.
The assay we have developed using AlphaScreen® technology is suitable for HTS. The assay performance was robust, as indicated by Z′ values greater than 0.5 and variability within acceptable limits.20 As a result of screening a small library of 7360 chemically diverse and drug-like compounds, we successfully identified a series of structurally similar compounds that disrupt bt-TAR-32 binding to TDP-43 with affinities ranging from 100 nM to 10 μM. These compounds had no effect in the TruHTS® counterscreen assay, thereby indicating that the observed inhibition of these compounds is specific for the DNA oligonucleotide binding to TDP-43. However, it is important to note that this assay will identify not only compounds that inhibit the interaction by binding to TDP–43 but also compounds that bind to bt-TAR-32. Future studies will determine the mechanism by which these compounds disrupt the TDP-43–nucleic acid interaction and their utility in understanding TDP-43’s normal and pathological functions.
Inhibitors of TDP-43’s binding to nucleic acids have potential utility as biochemical tools and possibly novel therapeutics. Small-molecule inhibitors of TDP-43 nucleic acid binding may improve respiratory function in cystic fibrosis by restoring the production of functional CFTR protein because investigators have shown that depletion of TDP-43 results in increased inclusion of exon 9 in transcription of the CFTR gene3–5 As biochemical tools, TDP-43 inhibitors could be useful for further understanding the role of TDP-43 in neurodegenerative diseases such as ALS. For example, although TDP-43 knockout mice are embryonic lethal,23 a pharmacological inhibitor of TDP-43 might be more tolerated and produce ALS-like symptoms in mice such as those seen in Drosophila.19 In addition, fluorescently labeled TDP-43 binding ligands could be used to develop high-content screening assays to identify novel compounds that inhibit the cytoplasmic aggregation and restore TDP-43 to the nucleus.
In summary, we have characterized DNA oligonucleotide binding to TDP-43 using a robust, high-throughput assay based on AlphaScreen® technology, which is easily scalable to 1536-well plates for screening of large libraries. We have reported association and dissociation rates for TAR-32 and TG6 oligonucleotides as well as equilibrium dissociation constants and demonstrated that these oligonucleotides bind to a single population of noninteracting sites. From screening our diverse, small-member chemical library, we have discovered a set of structurally similar compounds with nascent SAR and sub-micromolar inhibition of the TDP-43–nucleic acid interaction. Finally, we have discussed the potential utility of such compounds as biochemical tools and possibly as novel therapeutics.
References
- 1.Buratti E, Baralle FE. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci. 2008;13:867–878. doi: 10.2741/2727. [DOI] [PubMed] [Google Scholar]
- 2.Ou SH, Wu F, Harrich D, Garcia-Martinez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buratti E, Brindisi A, Pagani F, Baralle FE. Nuclear factor TDP-43 binds to the polymorphic TG repeats in CFTR intron 8 and causes skipping of exon 9: a functional link with disease penetrance. Am J Hum Genet. 2004;74:1322–1325. doi: 10.1086/420978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayala YM, Pagani F, Baralle FE. TDP43 depletion rescues aberrant CFTR exon 9 skipping. FEBS Let. 2006;580:1339–1344. doi: 10.1016/j.febslet.2006.01.052. [DOI] [PubMed] [Google Scholar]
- 6.Ayala YM, Misteli T, Baralle FE. TDP-43 regulates retinoblastoma protein phosphorylation through the repression of cyclin-dependent kinase 6 expression. Proc Natl Acad Sci U S A. 2008;105:3785–3789. doi: 10.1073/pnas.0800546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuo PH, Doudeva LG, Wang YT, Shen CK, Yuan HS. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 2009;37:1799–1808. doi: 10.1093/nar/gkp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- 9.Pawlyk AC, CJ, Reitz AB. Current nervous system related drug targets for the treatment of amyotrophic lateral sclerosis. Curr Pharm Des. 2010;16:2053–2073. doi: 10.2174/138161210791293024. [DOI] [PubMed] [Google Scholar]
- 10.Buratti E, Baralle FE. The molecular links between TDP-43 dysfunction and neurodegeneration. Adv Genet. 2009;66:1–34. doi: 10.1016/S0065-2660(09)66001-6. [DOI] [PubMed] [Google Scholar]
- 11.Geser F, Martinez-Lage M, Kwong LK, Lee VM, Trojanowski JQ. Amyotrophic lateral sclerosis, frontotemporal dementia and beyond: the TDP-43 diseases. J Neurol. 2009;256:1205–1214. doi: 10.1007/s00415-009-5069-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strong MJ. The syndromes of frontotemporal dysfunction in amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2008;9:323–338. doi: 10.1080/17482960802372371. [DOI] [PubMed] [Google Scholar]
- 13.Kwong LK, Neumann M, Sampathu DM, Lee VM, Trojanowski JQ. TDP-43 proteinopathy: the neuropathology underlying major forms of sporadic and familial frontotemporal lobar degeneration and motor neuron disease. Acta Neuropathol. 2007;114:63–70. doi: 10.1007/s00401-007-0226-5. [DOI] [PubMed] [Google Scholar]
- 14.Banks GT, Kuta A, Isaacs AM, Fisher EM. TDP-43 is a culprit in human neurodegeneration, and not just an innocent bystander. Mamm Genome. 2008;19:299–305. doi: 10.1007/s00335-008-9117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagier-Tourenne C, Cleveland DW. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136:1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liscic RM, Grinberg LT, Zidar J, Gitcho MA, Caims NJ. ALS and FTLD. two faces of TDP-43 proteinopathy. Eur J Neurol. 2008;15:772–780. doi: 10.1111/j.1468-1331.2008.02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pesiridis GS, Lee VM, Trojanowski JQ. Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18:R156–R162. doi: 10.1093/hmg/ddp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barmada SJ, Skibinski G, Korb E, Rao EJ, Wu JY, Finkbemer S. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J Neurosci. 2010;30:639–649. doi: 10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feiguin F, Godena VK, Romano G, D’Ambrogio A, Klima R, Baralle FE. Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett. 2009;583:1586–1592. doi: 10.1016/j.febslet.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 20.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 21.Limbird L. Cell Surface Receptors: A Short Course on Theory and Methods. Norwell, MA: Kluwer Academic; 1996. [Google Scholar]
- 22.Ayala YM, Pantano S, D’Ambrogio A, Buratti E, Brindisi A, Marchetti C, et al. Human, Drosophila, and C. elegans TDP43: nucleic acid binding properties and splicing regulatory function. J Mol Biol. 2005;348:575–588. doi: 10.1016/j.jmb.2005.02.038. [DOI] [PubMed] [Google Scholar]
- 23.Sephton CF, Good SK, Atkin S, Dewey CM, Mayer P, III, Herz J, et al. TDP-43 is a developmentally regulated protein essential for early embryonic development. J Biol Chem. 2010;285:6826–6834. doi: 10.1074/jbc.M109.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]