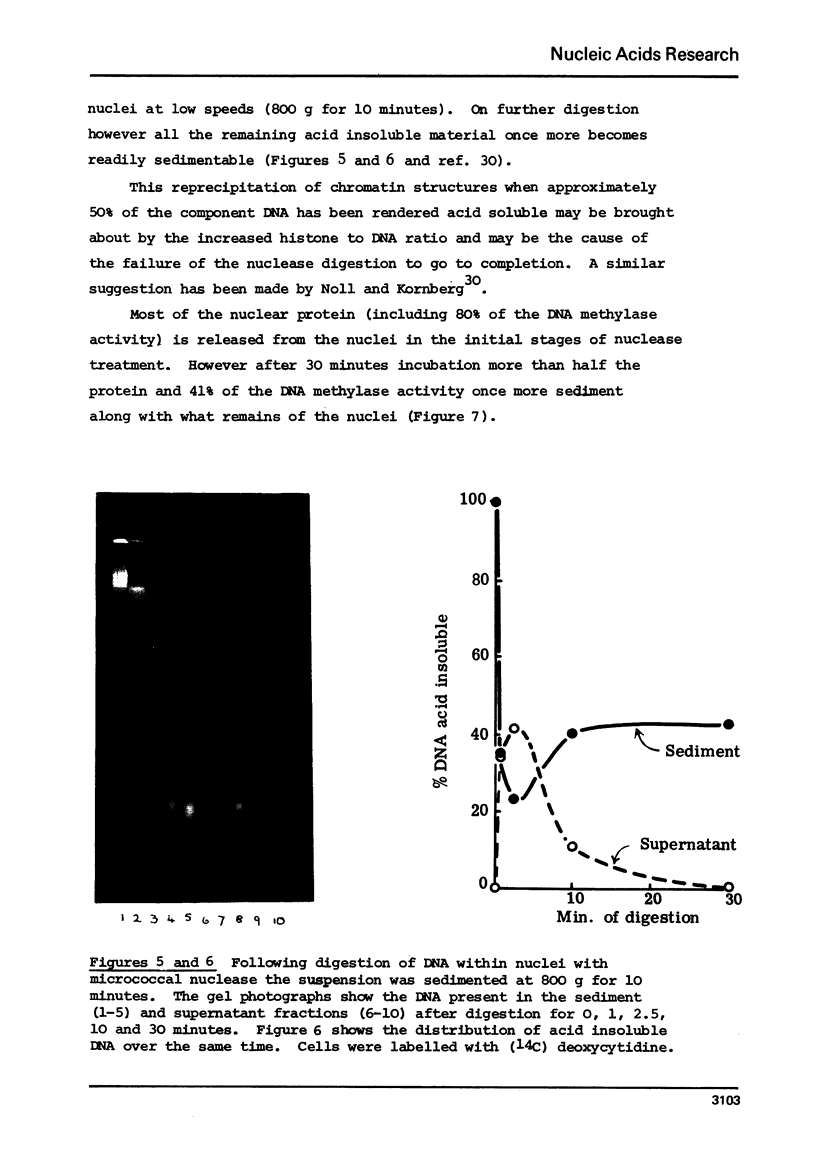
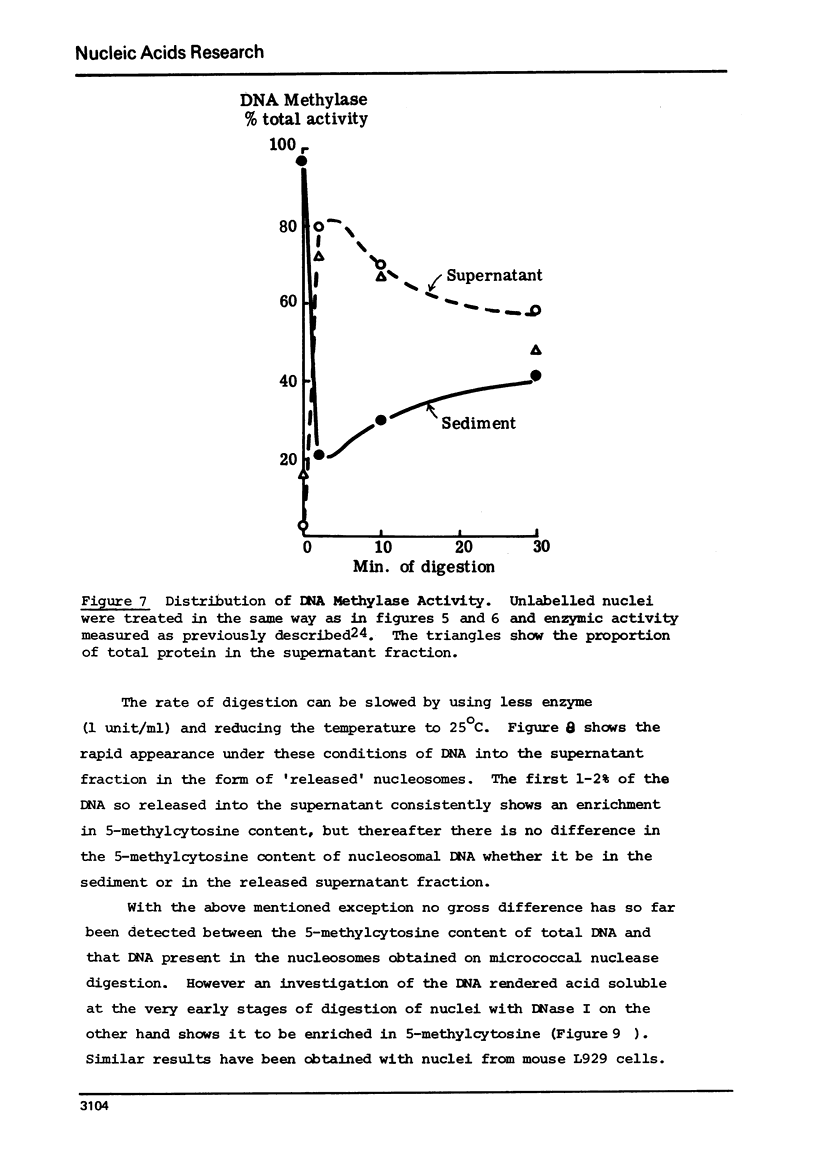
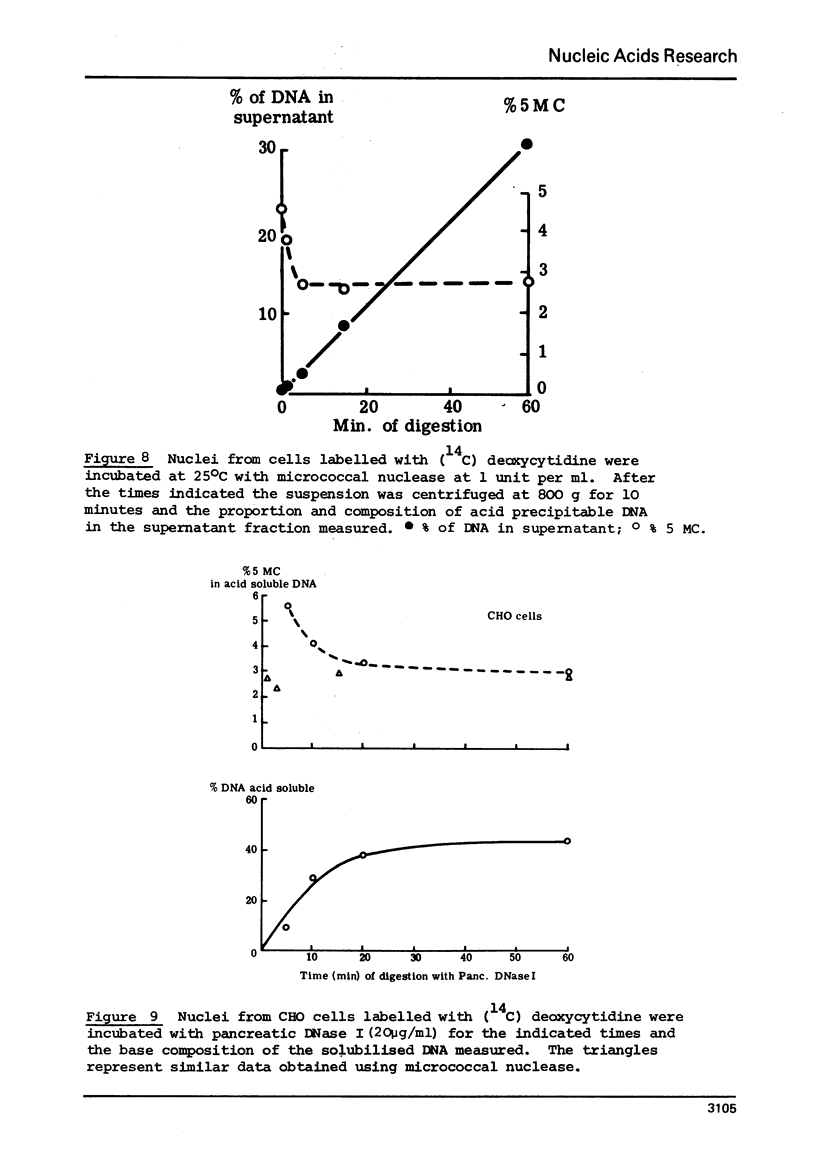
Abstract
The proportion of cytosines methylated in the DNA of nucleosome oligomers and of core particles appears indistinguishable from that of total nuclear DNA from CHO cells. However the DNA in nucleoprotein which is initially released from nuclei by treatment with very low levels of micrococcal nuclease and the first 10% of material rendered acid soluble by treatment of nuclei with DNase I are enriched 2 fold in their content of 5 methylcytosine. (Cessation of hydrolysis by nuclease occurs concomitantly with precipitation of nucleosomal core particles).
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axel R., Cedar H., Felsenfeld G. Chromatin template activity and chromatin structure. Cold Spring Harb Symp Quant Biol. 1974;38:773–783. doi: 10.1101/sqb.1974.038.01.082. [DOI] [PubMed] [Google Scholar]
- Axel R. Cleavage of DNA in nuclei and chromatin with staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2921–2925. doi: 10.1021/bi00684a020. [DOI] [PubMed] [Google Scholar]
- Axel R., Melchior W., Jr, Sollner-Webb B., Felsenfeld G. Specific sites of interaction between histones and DNA in chromatin. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4101–4105. doi: 10.1073/pnas.71.10.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne M. J., Burdon R. H. The sequence specificity of vertebrate DNA methylation. Nucleic Acids Res. 1977 Apr;4(4):1025–1037. doi: 10.1093/nar/4.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne M. J., Turnbull J. F., McKay E. L., Adams R. L., Burdon R. H. The sequence specificity of a mammalian DNA methylase. Nucleic Acids Res. 1977 Apr;4(4):1039–1045. doi: 10.1093/nar/4.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor C. R. A possible explanation for the nuclease limit digestion pattern of chromatin. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3391–3393. doi: 10.1073/pnas.73.10.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comings D. E. Methylation of euchromatic and heterochromatic DNA. Exp Cell Res. 1972 Oct;74(2):383–390. doi: 10.1016/0014-4827(72)90391-6. [DOI] [PubMed] [Google Scholar]
- Compton J. L., Bellard M., Chambon P. Biochemical evidence of variability in the DNA repeat length in the chromatin of higher eukaryotes. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4382–4386. doi: 10.1073/pnas.73.12.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton J. L., Hancock R., Oudet P., Chambon P. Biochemical and electron-microscopic evidence that the subunit structure of Chinese-hamster-ovary interphase chromatin is conserved in mitotic chromosomes. Eur J Biochem. 1976 Nov 15;70(2):555–568. doi: 10.1111/j.1432-1033.1976.tb11047.x. [DOI] [PubMed] [Google Scholar]
- Duerksen J. D., Paul I. J. Satellite DNA sequence content of polylysine-titratable and nuclease-resistant fractions of mouse liver hepatoma chromatin. Nucleic Acids Res. 1976 Sep;3(9):2277–2291. doi: 10.1093/nar/3.9.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J. M., Murphy R. F., Bonner J. Structure of transcriptionally active chromatin. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4404–4408. doi: 10.1073/pnas.72.11.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Kappler J. W. The 5-methylcytosine content of DNA: tissue specificity. J Cell Physiol. 1971 Aug;78(1):33–36. doi: 10.1002/jcp.1040780106. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Thomas J. O. Chromatin structure; oligomers of the histones. Science. 1974 May 24;184(4139):865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- Lacy E., Axel R. Analysis of DNA of isolated chromatin subunits. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3978–3982. doi: 10.1073/pnas.72.10.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. J., Nathans D. Deletion mutants of simian virus 40 generated by enzymatic excision of DNA segments from the viral genome. J Mol Biol. 1974 Oct 15;89(1):179–193. doi: 10.1016/0022-2836(74)90169-7. [DOI] [PubMed] [Google Scholar]
- Levy B., Dixon G. H. Renaturation kinetics of cDNA complementary to cytoplamic polyadenylated RNA from rainbow trout testis. Accessibility of transcribed genes to pancreatic DNase. Nucleic Acids Res. 1977 Apr;4(4):883–898. doi: 10.1093/nar/4.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A., Cohn K., Scheinman M. M. Right atrial versus left atrial echo zones: a proposed new criterion for determining the atrial site of retrograde preexcitation. J Electrocardiol. 1976;9(4):357–363. doi: 10.1016/s0022-0736(76)80029-5. [DOI] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Pfeiffer W., Horz W., Igo-Kemenes T., Zachau H. G. Restriction nucleases as probes of chromatin structure. Nature. 1975 Dec 4;258(5534):450–452. doi: 10.1038/258450a0. [DOI] [PubMed] [Google Scholar]
- Rill R. L., Nelson D. A., Oosterhof D. K., Hozier J. C. Structural repeat units of Chinese hamster ovary chromatin. Evidence for variations in repeat unit DNA size in higher eukaryotes. Nucleic Acids Res. 1977 Apr;4(4):771–789. doi: 10.1093/nar/4.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M., Streeck R. E., Zachau H. G. Nucleosome formation abolishes base-specific binding of histones. Nature. 1975 Dec 4;258(5534):447–450. doi: 10.1038/258447a0. [DOI] [PubMed] [Google Scholar]
- Tegtmeyer P., Macasaet F. Simian virus 40 deoxyribonucleic acid synthesis: analysis by gel electrophoresis. J Virol. 1972 Oct;10(4):599–604. doi: 10.1128/jvi.10.4.599-604.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull J. F., Adams R. L. DNA methylase: purification from ascites cells and the effect of various DNA substrates on its activity. Nucleic Acids Res. 1976 Mar;3(3):677–695. doi: 10.1093/nar/3.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Winocour E., Kaye A. M., Stollar V. Synthesis and transmethylation of DNA in polyoma-infected cultures. Virology. 1965 Oct;27(2):156–169. doi: 10.1016/0042-6822(65)90155-8. [DOI] [PubMed] [Google Scholar]