Abstract
Background
Facing critically low return per dollar invested on clinical research and clinical care, the American biomedical enterprise is in need of a significant transformation. A confluence of high-throughput “omic” technologies and increasing adoption of the electronic health record has fueled excitement for a new paradigm for biomedical research and practice. The ability to simultaneously measure thousands of molecular variables and assess their relationships with clinical data collected during the course of care could enable reclassification of disease not only by gross phenotypic observation but according to underlying molecular mechanism and influence of social determinants.In turn, this reclassification could enable development of targeted therapeutic interventions as well as disease prevention strategies at the individual and population levels.
Methods/Design
The MURDOCK Study consists of distinct project “horizons” or stages. Horizon 1 entailed the generation and analysis of molecular data for existing large,clinically well-annotated cohorts in four disease areas. Horizon 1.5 involves creating and maintaining a 50,000-person,community volunteer registry for biomarker signature validation and prospective studies, including integration of environmental and social data. Horizon 2 leverages and prospectively recruits Horizon 1.5 volunteers, and extends the study to additional disease areas of interest. Horizon 3 will expand the study through regional, national,and international partnerships.
Discussion
The MURDOCK Study embodies a new model of team science investigation and represents a significant resource for translational research. The study team invites inquiries to form new collaborations to exploit the rich resources provided by these biospecimens and associated study data.
Keywords: Stratified medicine, personalized medicine, biomarkers, disease reclassification, community registry, biorepository
Introduction
The American biomedical enterprise is in need of a fundamental transformation. In 2010, the United States (US) spent $2.6 trillion on health care [1], and unless significant changes are made, health care expenditures are projected to continue to increase at a rate that exceeds that of the Gross Domestic Product (GDP) [2]. Despite this substantial investment in health care, the US system was ranked 37th best in the world by the World Health Organization [3]. This poor health system performance is thought to be driven by sub-optimal allocation of expensive resources at both the individual and the population level [4]. In addition to health care spending, investment in research and development for new drugs is also increasing rapidly. Between 2004 and 2010, investment in pharmaceutical research and development increased approximately 6% per year on average, exceeding $60 billion in 2010, while the number of new molecular entities approved each year remained flat or even decreased [5,6].
Strategic application of recent and future advances in technology and informatics can fuel the transformation needed by enabling new approaches to both drug development and management of human health and disease. Technologies for high-throughput molecular assays, increased computational power and digital storage capacity, and novel methods for analysis of large data sets will enhance understanding of molecular mechanisms of health and disease. Searchable electronic health records (EHR) coupled with biospecimen repositories, and the integration of social and geospatially plotted environmental data, will further elucidate complex interactions between genes, gene expression, and environmental factors. Increased mechanistic understanding of human health and disease is expected to enable the development of targeted approaches to disease prevention, early detection and tailored treatment. Variably referred to as personalized, stratified, genomic, precision, or “P4” medicine [7-10], this concept fundamentally focuses on ensuring that the right counseling, lifestyle modification, or treatment is given to the right patient, at the right time. Importantly, this targeted approach can also enable more rational and effective population health management, thus facilitating the fundamental transformation that is required.
Rewriting the textbook of medicine
Nearly a decade into the genomic era, new technologies and analytical approaches enable the vision for personalized medicine [11]. However, the actualization of genomic medicine continues to be hindered by a pre-genomic-era medical framework. Disease classification taxonomies widely in use today (e.g., International Classification of Disease-9), were developed similarly to Linnaeus’ taxonomy of living organisms, i.e., based primarily on macroscopic symptoms and observations. Today, we anticipate that through the use of technology platforms that generate molecular level data, many diseases previously considered a single entity at a macroscopic level may be revealed to encompass multiple distinct underlying conditions. Molecular profiling may also reveal commonalities among diseases previously considered to be mechanistically distinct.
Significant advances in mechanistic understanding and reclassification of disease will require an innovative approach to research. It will be essential to collect baseline and longitudinal clinical data, biospecimens, environmental exposure data, and information about use of medical and lifestyle support resources from large numbers of participants. Success will also require a coordinated, end-to-end informatics infrastructure to facilitate data management and analysis across molecular, clinical, and environmental data and among samples, subjects, and populations. Finally, the translation of information to actual application in clinical care will require a team-science approach involving collaborations not only among interdisciplinary academic researchers but also with industry.
Here we present the Measurement to Understand the Reclassification of Disease of Cabarrus/Kannapolis (MURDOCK) Study (www.murdock-study.org), a large-scale research initiative designed to foster the transition to predictive, preventive, personalized medicine through biomarker discovery, elucidation of environmental health factors, and the molecular reclassification of disease. Centered in Kannapolis, North Carolina, the MURDOCK Study was named after its benefactor, David H. Murdock, and represents a rich resource for collaborative, genomeera biomedical research.
Methods and design
The MURDOCK Study is organized in stages, called “horizons” (Figure 1). Horizon 1 consists of initial exploratory studies performed using data and samples from existing cohorts. Horizon 1.5, known as the MURDOCK Study Community Registry and Biorepository and launched concurrently with Horizon 1, entails the creation of a 50,000-participant volunteer registry in Cabarrus County and the city of Kannapolis. Horizon 2 leverages the Horizon 1.5 Registry for validation and prospective studies, and to recruit targeted population cohorts. Horizon 3 will expand the study with regional, national, and international partnerships, enabling meta-analyses and cross-population studies.
Figure 1.
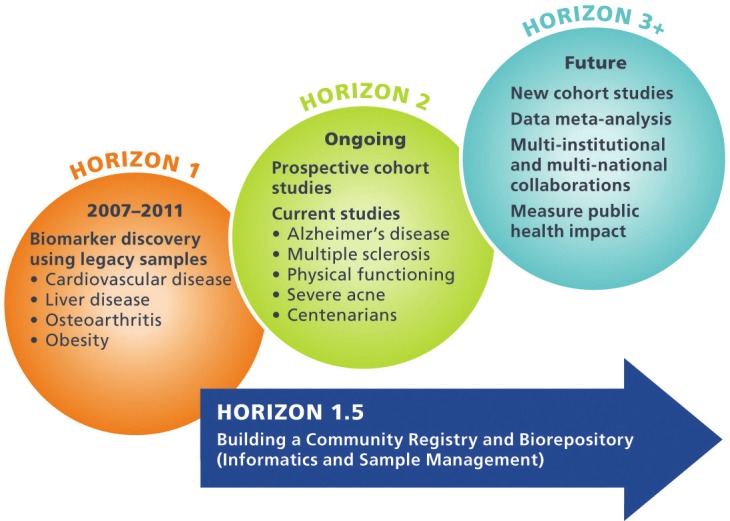
Distinct horizons of the MURDOCK Study. Horizon 1 entailed biomarker discovery in four existing cohorts. Horizon 1.5, launched concurrently with Horizon 1, creates a community registry of 50,000 subjects. Horizon 2 leverages the Horizon 1.5 Registry for validation studies, prospective studies, and to recruit targeted populations. Horizon 3 will expand the study with regional and national partnerships.
Horizon 1
The goal of Horizon 1 was to enable biomarker discovery in the short term, using existing cohorts in specific disease areas. Horizon 1 encompasses four previously existing, independent cohorts studied for cardiovascular disease, obesity, liver disease, or osteoarthritis. Within each cohort, biospecimens had been collected and stored for future research use along with documented clinical outcomes and other characterizing information. Each Horizon 1 investigative team applied one or more high-throughput assays (genomic, proteomic, and/or metabolomic) to existing biospecimens to address project-specific, clinically relevant hypotheses. The osteoarthritis study additionally used specialized image analysis. Details of each Horizon 1 project are provided in Table 1 [12-15].
Table 1.
Overview of Horizon 1 studies
| H1 Study | CV | Hep C | OA | WLM |
|---|---|---|---|---|
| Principal Investigator | L Kristin Newby | John McHutchison | Virginia Kraus | Laura Svetkey |
| Therapeutic area | Cardiovascular disease | Gastrointestinal/Infectious disease | Osteoarthritis | Obesity/Weight loss maintenance |
| Source of samples and clinical data | Biological samples: Duke CATHeterization GENetics biorepository (www.cathgen.duhs.duke.edu/index.htm); Clinical data from the Duke Databank for Cardiovascular Disease [12] | Duke Hepatology Clinical Research | Predict Osteoarthritis Progression study, described in [25,26] and the Genetics of Generalized Osteoarthritis (GOGO) study [27] | The weight loss maintenance randomized controlled trial [28] |
| Clinical questions | 1. What baseline clinical features best stratify risk for coronary disease events? 2. What -omic profiles best stratify risk for coronary disease events and how do they contribute incrementally to clinical characteristics? | 1. What protein biomarkers (host or viral) predict response to interferon therapy? 2. Are there protein biomarkers (host or viral) associated with different Hep C genotypes? | 1. What imaging biomarkers predict OA progression? 2. What molecular biomarkers predict OA progression? | 1. What are metabolomic biomarkers to stratify who is able to lose weight and keep the weight off? 2. What are metabolomic biomarkers associated with metabolic consequences of obesity? To what extent are these consequences are affected by weight loss? |
| N | 6447 clinical modeling 2023 molecular cohort | 55 discovery 41 validation | 2047 | 1000 |
| Biospecimens | Plasma, PaxGene RNA, DNA | Serum | Synovial fluid, serum, urine | Serum, plasma at 3 time points |
| Platforms | ||||
| Targeted metabolomics | 2023 plasma | _ | _ | 500 plasma, serum at baseline |
| Discovery proteomics | 60 plasma (30 + 30) | 96 serum | 48 urine, 14 synovial fluids | _ |
| Targeted proteomics | 500 plasma (250 + 250) | 96 | Pending 108 sera | _ |
| GWAS | 2023 | 96* | 1258 | _ |
| Gene expression | 500 PaxGene RNA (250 + 250) | _ | _ | _ |
| Imaging | Angiography | _ | 138 individuals (248 non-replaced knees) | _ |
| Planned follow-on studies | NHLBI GO Grant (Go Get It) to add GWAS and complete RNA profiling in the 2023 molecular cohort | |||
| References | [12] | [13,16,29-32] | [15] |
HCV genome-wide association data were generated through external funding.
Standard and advanced statistical methods were used to analyze the clinical and molecular data generated from the Horizon 1 studies. Mapping common clinical data elements among disparate disease cohorts, coupled with the use of a given molecular platform to study more than one clinical disease (e.g., proteomics in both cardiovascular and liver disease), may allow identification of molecular variation that is common across seemingly disparate clinical disease states. In addition, Horizon 1 studies (except the obesity study) used multiple molecular platforms to enable a systems biology approach, i.e., exploration of molecular pathways of disease and analysis of molecular interactions in relation to disease state or response to treatment. For example, analysis of proteomic data coupled with genotypic data in hepatitis C patients enabled mechanistic inquiry into the relationship between treatment response and genotypic variation in the IL28B gene [16].
Horizon 1.5
The goal of Horizon 1.5 is to develop the infrastructure necessary to conduct efficient, large-scale studies for biomarker discovery and population stratification. This infrastructure has been realized through the creation of a participant registry and biorepository from which volunteers for prospective studies can be drawn. The details of Horizon 1.5 and initial recruitment are to be described elsewhere (manuscript in preparation). Briefly, Horizon 1.5 was launched in parallel with Horizon 1 investigations with the goal of enrolling 50,000 participants, roughly one-third of the adult population of the catchment area: approximately 35,000 recruited through open enrollment methods, and a population random sample of 15,000 individuals. Enrollment began in February, 2009, and through May, 2012, >8000 subjects were enrolled via open enrollment. At this point, the MURDOCK cohort was 65% female, the median age was 54.2 years, and the racial composition was primarily Caucasian (78.4%), with 13.5% African American (vs. 15.3% in the Cabarrus County population) and 8.4% Hispanic (vs. 9.4% in the Cabarrus County population).
Registry volunteers consent to provide blood (for DNA, RNA, plasma and serum), urine, and self-reported clinical and demographic information, as well as undergo a brief physical exam. At the time of enrollment and annually thereafter, the participant provides information regarding medical history, general health and wellbeing, mental health, socioeconomic status, environment, and lifestyle. Unless a subject chooses to withdraw from participating, he or she will be contacted for annual updates indefinitely, as long as the MURDOCK Study is funded. Biospecimens are stored in a biobank in Kannapolis that has capacity for over 10 million samples, operated by LabCorp, Inc. Participants also consent to be contacted annually for follow-up and up to four times per year to consider participation in future studies for which they might qualify. A key component of registry participation is consent for access to EHR over time. Additionally, address information enables geospatial mapping of a participant’s residence, allowing environmental information to be incorporated, e.g., proximity to recreational spaces, health care services, or grocery stores. This component enhances the study’s interdisciplinary collaboration with environmental and public health researchers.
The MURDOCK Study registry operations are based in Kannapolis, the location of the North Carolina Research Campus, a public-private venture between David H. Murdock, the State of North Carolina, and eight North Carolina universities. Community outreach is a cornerstone of the Horizon 1.5 efforts. Study personnel have developed relationships with health care providers, community leaders, and local organizations, and participate in networking and recruiting events through venues such as local churches, schools, fairs, and farmers’ markets. Advertising is done through local print and radio media in addition to social networking and other electronic sources. A network of sites has been developed for conducting enrollment, and mass enrollment events are regularly held on-site at local organizations within the community, e.g., at businesses, schools, churches, and fire stations.
Horizon 2
The goal of Horizon 2 is to leverage the Horizon 1.5 infrastructure for prospective biomarker discovery in an optimized translational research environment. This phase builds upon Horizons 1 and 1.5 with biomarker validation and focused recruitment in specific disease areas. New areas of focus include multiple sclerosis, mild cognitive impairment, physical functioning, aging, and severe acne. In some cases, these studies have identified participants from Horizon 1.5 based on patient-reported clinical data. Others are recruiting new subjects to the MURDOCK Horizon 1.5 Registry and Biorepository through targeted recruitment at specialized clinics or at the time of enrollment into the main registry.
Horizon 3
The goal of Horizon 3 is to expand the scope of the study to include diverse populations around the world. This phase will involve the formation of collaborative partnerships regionally, nationally, and globally, which will enable meta-analyses, cross-population studies, and validation of findings in geographically, environmentally, ethnically, and racially diverse populations. Specifically, with a medical school in Singapore [17], a set of major partnerships in India in which a cohort study is being launched this year [18], and a university in Shanghai [19], the opportunity arises for combined molecular epidemiology studies involving diverse genetic pools and health care delivery systems.
Informatics as a bridging discipline
Central to the success of each study horizon, and key to bridging the gaps between them, is informatics, including the collection, storage, integration, and retrieval of data. The informatics cornerstone for the MURDOCK Study is the MURDOCK Integrated Data Repository (MIDR). The initial phase of the MIDR houses data from MURDOCK Horizon I studies. As each horizon matures, the MIDR is extended to accommodate new data and new uses of data, and will continue to be developed over time. Near-term developments include employing a metadata-driven approach to augment study data with study descriptions, e.g., using OCRe (Ontology of Clinical Research) [20], as well as sample status information over time. The key short-term use case is to enable any authenticated user to identify a potential collaboration based on mutual interest or complementary data and contact MURDOCK Study personnel to follow up, thus enabling data mining, analysis, and discovery with appropriate data access permission.
The next major release of the MIDR will draw upon data from a number of sources. Some data will be extracted into a central location, while other data will remain in their original location, with the metadata pulled into the central data repository as needed to support key use cases. For example, raw “-omic” and imaging files, which tend to have a large data footprint, will be stored in their original location. Initial MIDR use cases focus on queries that enable a would-be collaborator to understand what studies, samples, and data sets are available, and whom to contact for more information. Future releases of the MIDR will incorporate row-level, processed -omic data as well, to enable molecular data mining and analysis with appropriate governance and approval. Examples of initial and future use cases are given in Box 1.
Box 1.
Different query types for short-term and medium-term use cases.
| Initial use case queries |
| How many samples do we have from women over 50 who were later diagnosed with hypertension and who have a family history of cardiovascular disease? |
| How many male subjects do we have under the age of 50 who have been diagnosed with prostate cancer, for whom we have GWAS data? |
|
|
| Initial use case queries |
| How many samples do we have from women over 50 who were later diagnosed with hypertension and who have a family history of cardiovascular disease? |
| How many male subjects do we have under the age of 50 who have been diagnosed with prostate cancer, for whom we have GWAS data? |
The first challenge in creation of the MIDR was to compare, align, and integrate Horizon 1 clinical data into a canonical set of common data elements. Data from the four independent case report forms were categorized and standardized, with each original data element mapped to a standards-based data element in the MIDR. Importantly, the mapping rules themselves were stored in the MIDR as metadata to archive exactly how mapped data were derived. As a simple example, the data value White from the osteoarthritis study mapped to the value Caucasian in the MIDR. For each Horizon 1 study, a subset of the full data set was extracted from the original source database, transformed, and loaded into the MIDR. Protocol metadata were also imported into the MIDR, primarily as free text. In future versions of the MIDR, study metadata will be represented in structured form using terms from OCRe [20].
Version 2.0 of the MIDR is currently under development. In addition to Horizon 1 clinical, demographic, and protocol data included in the first release, MIDR 2.0 will incorporate Horizon 1.5 Registry data, including biospecimen information, -omics and imaging metadata, consent data, and data from EHR (see Figure 2). Incorporation of EHR data is a key facet of the Horizon 1.5 Registry. When the Health Information Technology for Economic and Clinical Health (HITECH) Act was passed as part of the American Recovery and Reinvestment Act of 2009, it was estimated that only approximately 17% of doctors and 10% of hospitals in the country used some form of EHR [21]. Cabarrus County is an outlier to the relatively low national rate for health Information Techonology adoption, which led to its selection as an ONC (Office of the National Coordinator) Beacon Community [22]. As described in the Beacon Community proposal, three major health care providers, each of which has one or more operating EHR systems, deliver health care for a substantial majority of the Cabarrus population. Regular automated imports of EHR data for registry participants are currently under development. This approach will supplement the annual patient self-report questionnaire and enable a cost-efficient method to track participants longitudinally. It will also provide a valuable resource for research on data quality and secondary use, with the ability to compare and contrast patient-reported information with data collected through clinical care.
Figure 2.
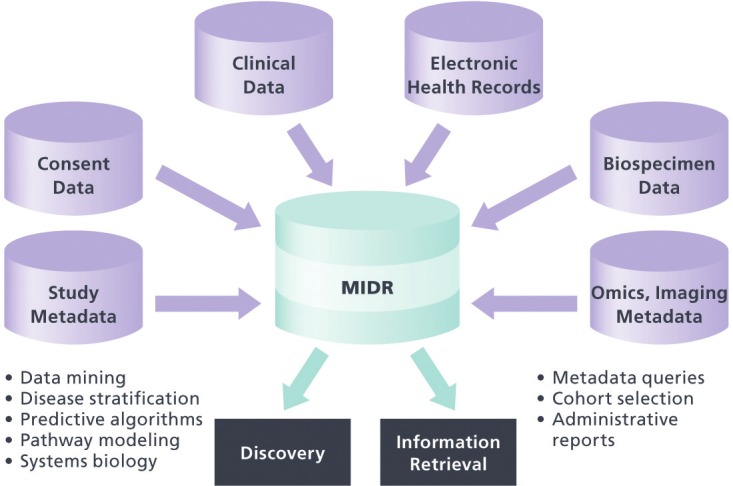
The MURDOCK Integrated Data Repository (MIDR). In addition to the clinical data stored in the current version of the MIDR, the next release will include subject consent data and structured metadata for study protocol, biospecimens, -omic, and imaging data. The resource will enable both administrative information retrieval and scientific discovery through hypothesis generation.
Discussion
To achieve the vision for stratified medicine, a new medical paradigm is required. One crucial first step will be the creation of a new, molecularly based taxonomy of disease and the application of this new taxonomy to clinical studies and clinical trials, enabling its incorporation into guidelines. To this end, an ad hoc committee of the National Research Council (NRC) was convened in 2011 at the request of the Office of the Director of the National Institutes of Health to explore the feasibility and need for “creating a ‘New Taxonomy’ of human diseases based on molecular biology” [23]. Among the conclusions presented in the NRC’s report was the need for new models for population-based research that would incorporate EHRs, enabling the conduct of research at the point of care [23]. In addition, the report asserted the need for “a creative period of bottom-up research activity, organized through pilot projects of increasing scope and scale,” emphasizing cooperation among these different pilot approaches [23]. The MURDOCK Study embodies just such a project. The study is designed and being conducted to facilitate data sharing, both technically through a standards-based approach to data management and logistically through a clear governance structure and documentation of data access policies.
While initial efforts have been productive, many challenges remain in fulfilling the vision of personalized medicine. Key challenges are summarized in Table 2, along with descriptions of how the MURDOCK Study will contribute to addressing them. This effort provides complementary information to preceding large-scale biorepositories and epidemiological studies. The MURDOCK Study adds a population with more ethnic diversity than the Framingham Study [33], and provides a broader age range than the UK Biobank [34]. It also provides a venue for exploration into leveraging recent developments in health information technology, i.e., EHR for longitudinal follow-up. The Electronic Medical Records and Genomics (eMERGE) consortium uses biobanks linked with electronic medical record systems to perform large-scale genome-wide studies, but these analyses are limited to genotypic variation and are done on deidentified data, precluding follow-up with individual research participants [35].
Table 2.
Translational barriers and how they are being addressed by the MURDOCK Study
| Translational Challenge | How addressed by MURDOCK |
|---|---|
| Large scale required for sufficient statistical power in high dimensional analyses | Recruitment of 50,000 participants Extensive community engagement and support |
| Interdisciplinary expertise required | Team science approach that includes clinicians, platform experts, statisticians, informaticists, ethicists, biobanking experts, and business development and public health experts |
| Ability to identify predictive biomarkers prior to disease onset | Collection of biospecimens at baseline in healthy participants, or in participants who have not yet developed a given disease |
| Novel informatics methodologies required | MIDR, informaticists as part of the core team, pilot funding for methods development, solicitation of external collaborations |
| Longitudinal data collection | Annual follow-up, leverage EHR, partner with regional health information exchange |
| Access to high-throughput technology instrumentation and expertise | Partner with David H. Murdock Research Institute and Duke Core Facilities |
| Environmental effects | Geospatial data for participant residence, along with geographical information regarding health care services and community resources |
| Commercialization of novel biomarkers | Partnership with LabCorp for clinical test development, creation of BioMarker Factory |
| Lower paylines in the current funding environment | Emphasis on data reuse and partnerships with industry |
| Data sharing and data access | Formal governance structure and documentation |
Another key aspect of the MURDOCK Study not found in other large-scale epidemiological initiatives is a broad academia-industry partnership. The Biomarker Factory, LLC (www.biomarkerfactory.com/) is a collaboration between Duke University and LabCorp providing a fully integrated biomarker development and commercialization outlet for the project and its collaborators, and serving as a bridge across a marked gap in funding between discovery and therapeutic application to bring novel biomarker tests to market [24]. This collaboration builds from and leverages the work of the MURDOCK Study to translate potential insights generated by this effort into technologies with actual clinical impact.
Finally, the MURDOCK Study uses informatics standards that facilitate linkage with ongoing epidemiological studies. Of particular interest are studies being initiated at Medanta, in Gurgaon, India and efforts being initiated at the Duke-NUS Medical School in Singapore and the Duke-Kunshan University in Kunshan, China (http://today.duke.edu/2010/01/kunshan.html) [17,18]).
Conclusion
We have created an array of rich data sets that can be used both for predictive biomarker discovery and for a mechanistic, systems biology approach to the study of disease. Our ability to analyze biomarkers not just within a given disease cohort but across groups with differing diagnoses-within the MURDOCK Registry and in combination with other populations-will enable the breakdown of superficial disease distinctions toward reclassification of disease. Finally, MURDOCK’s standards-based informatics approach combined with Duke’s regional, national, and global partnerships enable expansion of this approach to translational research beyond a single county, state, or region.
Utility of the tools and data assets generated through each horizon of the MURDOCK Study will be measured in terms of uptake by the re- search community and generation of currency ranging from impact in peer-reviewed publication to products launched from commercial sector collaboration. The authors invite interested investigators to take full advantage of this rich and innovative resource by contacting the MURDOCK Study team (murdock-study@duke.edu) regarding potential collaborations.
Acknowledgments
The authors wish to thank the population and health care providers of Kannapolis and Cabarrus County for their enthusiasm and participation. Thanks also to Peter Hoffmann for meticulous, timely, and courteous copyediting. Jeanette McCarthy co-led the Horizon 1 Hepatitis C Study and contributed to study governance. Planning for this work was enabled through funding from Duke Clinical and Translational Science Award UL1RR024128. The MURDOCK Study was made possible by a gift from David H Murdock.
Abbreviations
- EHR
Electronic Health Record
- eMERGE
Electronic Medical Records and Genomics
- GDP
Gross Domestic Product
- HITECH
Health Information Technology for Economic and Clinical Health
- MIDR
MURDOCK Integrated Data Repository
- MURDOCK
Measurement to Understand the Reclassification of Disease of Cabarrus/Kannapolis
- NCRC
North Carolina Research Campus
- NRC
National Research Council
- OCRe
Ontology of Clinical Research
- ONC
Office of the National Coordinator [for Health Information Technology]
- US
United States
Authors’ contributions
JDT drafted the manuscript and coordinates the informatics aspects of the MURDOCK Study. VC collaborated with JDT to draft the manuscript and review and revise it for important intellectual content, participated in the design of the MURDOCK Study, and is the Executive Manager for the MURDOCK study. GSG reviewed the manuscript and revised it for critical intellectual content, participated in the design of the MURDOCK Study, and serves as a member of the Leadership committee. RMC reviewed and revised the manuscript for critical intellectual content, secured funding for and led the design of the MURDOCK Study and serves as the Principal Investigator of the MURDOCK Study. VBK reviewed and revised the manuscript for critical intellectual content, participated as a Horizon 1 investigator, and contributed to overall MURDOCK study design. LKN reviewed and revised the manuscript for critical intellectual content, participated as Horizon 1 investigator, is co-Principal Investigator of the Horizon 1.5 Community Registry and Biorepository, and participated in the overall MURDOCK study design, JGM reviewed and revised the manuscript for critical intellectual content, participated as a Horizon 1 investigator, and participated in the overall MURDOCK Study design. LPS reviewed and revised the manuscript for critical intellectual content and served as a Horizon 1 investigator. RJD reviewed and revised the manuscript for critical intellectual content and serves as the co-Principal Investigator for the Horizon 1.5 Community Registry and Biorepository. AAD reviewed and revised the manuscript for critical intellectual content and is the Project Manager for the Horizon 1.5 Community Registry and Biorepository. KU reviewed and revised the manuscript for critical intellectual content and participated in the design of the MURDOCK Study. MLN reviewed and revised the manuscript for critical intellectual content and participated in the design of the data infrastructure for the MURDOCK Study. MC reviewed and revised the manuscript for critical intellectual content and participated in the design of the MURDOCK Study, the Horizon 1.5 Community Registry and Biorepository, and the development of its community engagement plan and internal and external collaborations. All authors approved the final version of the manuscript prior to submission.
References
- 1.CMS. Historical National Health Expenditure Data.2011. 2012. [Google Scholar]
- 2.CBO. The Long Term Outlook for Health Care Spending.2007. 2012. [Google Scholar]
- 3.WHO. The world health report 2000 - health systems: improving performance. 2000. [PubMed] [Google Scholar]
- 4.OECD. Health at a Glance 2011: OECD Indicator. 2011. [Google Scholar]
- 5.Bunnage ME. Getting pharmaceutical R&D back on target. Nat Chem Biol. 2011;7:335–339. doi: 10.1038/nchembio.581. [DOI] [PubMed] [Google Scholar]
- 6.PhRMA. PhRMA 2011 Profile.2011. 2012. [Google Scholar]
- 7.Hood L, Friend SH. Predictive, personalized, preventive, participatory (P4) cancer medicine. Nat Rev Clin Oncol. 2011;8:184–187. doi: 10.1038/nrclinonc.2010.227. [DOI] [PubMed] [Google Scholar]
- 8.Ginsburg GS, Willard HF. Genomic and personalized medicine: foundations and applications. Transl Res. 2009;154:277–287. doi: 10.1016/j.trsl.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Carlson B. P4 could transform healthcare, but payers and physicians are not yet convinced. Biotechnol Healthc. 2010;7:7–8. [PMC free article] [PubMed] [Google Scholar]
- 10.Moving toward precision medicine. Lancet. 2011;378:1678. doi: 10.1016/S0140-6736(11)61725-X. [DOI] [PubMed] [Google Scholar]
- 11.Guttmacher AE, Collins FS. Welcome to the genomic era. N Engl J Med. 2003;349:996–998. doi: 10.1056/NEJMe038132. [DOI] [PubMed] [Google Scholar]
- 12.Shah SH, Granger CB, Hauser ER, Kraus WE, Sun JL, Pieper K, Nelson CL, Delong ER, Califf RM, Newby LK. Reclassification of cardiovascular risk using integrated clinical and molecular biosignatures: design of and rationale for the Measurement to Understand the Reclassification of Disease of Cabarrus and Kannapolis (MURDOCK) Horizon 1 Cardiovascular Disease Study. Am Heart J. 2010;160:371–379e2. doi: 10.1016/j.ahj.2010.06.051. [DOI] [PubMed] [Google Scholar]
- 13.Patel K, Lucas JE, Thompson JW, Dubois LG, Tillmann HL, Thompson AJ, Uzarski D, Califf RM, Moseley MA, Ginsburg GS, McHutchison JG, McCarthy JJ. High predictive accuracy of an unbiased proteomic profile for sustained virologic response in chronic hepatitis C patients. Hepatology. 2011;53:1809–1818. doi: 10.1002/hep.24284. [DOI] [PubMed] [Google Scholar]
- 14.Svetkey LP, Ard JD, Stevens VJ, Loria CM, Young DY, Hollis JF, Appel LJ, Brantley PJ, Kennedy BM, Kumanyika SK, Batch BC, Corsino L, Lien LF, Vollmer WM for the Weight Loss Maintenance Collaborative Research Group. Predictors of long-Term Weight Loss in Adults With Modest Initial Weight Loss, by Sex and Race. Obesity (Silver Spring) 2011 doi: 10.1038/oby.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kraus VB, Feng S, Wang S, White S, Ainslie M, Brett A, Holmes A, Charles HC. Trabecular morphometry by fractal signature analysis is a novel marker of osteoarthritis progression. Arthritis Rheum. 2009;60:3711–3722. doi: 10.1002/art.25012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cyr DD, Lucas JE, Thompson JW, Patel K, Clark PJ, Thompson A, Tillmann HL, McHutchison JG, Moseley MA, McCarthy JJ. Characterization of serum proteins associated with IL28B genotype among patients with chronic hepatitis C. PLoS One. 2011;6:e21854. doi: 10.1371/journal.pone.0021854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams RS, Casey PJ, Kamei RK, Buckley EG, Soo KC, Merson MH, Krishnan RK, Dzau VJ. A global partnership in medical education between Duke University and the National University of Singapore. Acad Med. 2008;83:122–127. doi: 10.1097/ACM.0b013e318160b8bc. [DOI] [PubMed] [Google Scholar]
- 18.Ackerly DC, Udayakumar K, Taber R, Merson MH, Dzau VJ. Perspective: global medicine: opportunities and challenges for academic health science systems. Acad Med. 2011;86:1093–1099. doi: 10.1097/ACM.0b013e318226b455. [DOI] [PubMed] [Google Scholar]
- 19. http://today.duke.edu/2010/01/kunshan.html. [Google Scholar]
- 20.Sim I, Carini S, Tu S, Wynden R, Pollock BH, Mollah SA, Gabriel D, Hagler HK, Scheuermann RH, Lehmann HP, Wittkowski KM, Nahm M, Bakken S. The human studies database project: federating human studies design data using the ontology of clinical research. AMIA Summits Transl Sci Proc. 2010;2010:51–55. [PMC free article] [PubMed] [Google Scholar]
- 21.Blumenthal D. Stimulating the adoption of health information technology. N Engl J Med. 2009;360:1477–1479. doi: 10.1056/NEJMp0901592. [DOI] [PubMed] [Google Scholar]
- 22.HHS. HealthIT. hhs.gov: Southern Piedmont Beacon Community. 2012. [Google Scholar]
- 23.NRC. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. 2011. [PubMed] [Google Scholar]
- 24.Butler D. Translational research: crossing the valley of death. Nature. 2008;453:840–842. doi: 10.1038/453840a. [DOI] [PubMed] [Google Scholar]
- 25.Kraus VB, McDaniel G, Worrell TW, Feng S, Vail TP, Varju G, Coleman RE. Association of bone scintigraphic abnormalities with knee malalignment and pain. Ann Rheum Dis. 2009;68:1673–1679. doi: 10.1136/ard.2008.094722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Addison S, Coleman RE, Feng S, McDaniel G, Kraus VB. Whole-body bone scintigraphy provides a measure of the total-body burden of osteoarthritis for the purpose of systemic biomarker validation. Arthritis Rheum. 2009;60:3366–3373. doi: 10.1002/art.24856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraus VB, Jordan JM, Doherty M, Wilson AG, Moskowitz R, Hochberg M, Loeser R, Hooper M, Renner JB, Crane MM, Hastie P, Sundseth S, Atif U. The Genetics of Generalized Osteoarthritis (GOGO) study: study design and evaluation of osteoarthritis phenotypes. Osteoarthritis Cartilage. 2007;15:120–127. doi: 10.1016/j.joca.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Svetkey LP, Stevens VJ, Brantley PJ, Appel LJ, Hollis JF, Loria CM, Vollmer WM, Gullion CM, Funk K, Smith P, Samuel-Hodge C, Myers V, Lien LF, Laferriere D, Kennedy B, Jerome GJ, Heinith F, Harsha DW, Evans P, Erlinger TP, Dalcin AT, Coughlin J, Charleston J, Champagne CM, Bauck A, Ard JD, Aicher K. Comparison of strategies for sustaining weight loss: the weight loss maintenance randomized controlled trial. JAMA. 2008;299:1139–1148. doi: 10.1001/jama.299.10.1139. [DOI] [PubMed] [Google Scholar]
- 29.Tillmann HL, Patel K, Muir AJ, Guy CD, Li JH, Lao XQ, Thompson A, Clark PJ, Gardner SD, McHutchison JG, McCarthy JJ. Beneficial IL28B genotype associated with lower frequency of hepatic steatosis in patients with chronic hepatitis C. J Hepatol. 2011;55:1195–1200. doi: 10.1016/j.jhep.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rowell J, Thompson AJ, Guyton JR, Lao XQ, McHutchison JG, McCarthy JJ, Patel K. Serum apolipoprotein C-III is independently associated with chronic hepatitis C infection and advanced fibrosis. Hepatol Int. 2011 doi: 10.1007/s12072-011-9291-x. [DOI] [PubMed] [Google Scholar]
- 31.McCarthy JJ, Li JH, Thompson A, Suchindran S, Lao XQ, Patel K, Tillmann HL, Muir AJ, McHutchison JG. Replicated association between an IL28B gene variant and a sustained response to pegylated interferon and ribavirin. Gastroenterology. 2010;138:2307–2314. doi: 10.1053/j.gastro.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li JH, Lao XQ, Tillmann HL, Rowell J, Patel K, Thompson A, Suchindran S, Muir AJ, Guyton JR, Gardner SD, McHutchison JG, McCarthy JJ. Interferon-lambda genotype and low serum low-density lipoprotein cholesterol levels in patients with chronic hepatitis C infection. Hepatology. 2010;51:1904–1911. doi: 10.1002/hep.23592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dawber TR, Meadors GF, Moore FE Jr. Epidemiological approaches to heart disease: the Framingham Study. Am J Public Health Nations Health. 1951;41:279–281. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmer LJ. UK Biobank: bank on it. Lancet. 2007;369:1980–1982. doi: 10.1016/S0140-6736(07)60924-6. [DOI] [PubMed] [Google Scholar]
- 35.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, Li R, Masys DR, Ritchie MD, Roden DM, Struewing JP, Wolf WA. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]


