Abstract
MicroRNAs (miRs) are small, noncoding RNAs that are emerging as crucial regulators of cardiac remodeling in left ventricular hypertrophy (LVH) and failure (LVF). However, there are no data on their role in right ventricular hypertrophy (RVH) and failure (RVF). This is a critical question given that the RV is uniquely at risk in patients with congenital right-sided obstructive lesions and in those with systemic RVs. We have developed a murine model of RVH and RVF using pulmonary artery constriction (PAC). miR microarray analysis of RV from PAC vs. control demonstrates altered miR expression with gene targets associated with cardiomyocyte survival and growth during hypertrophy (miR 199a-3p) and reactivation of the fetal gene program during heart failure (miR-208b). The transition from hypertrophy to heart failure is characterized by apoptosis and fibrosis (miRs-34, 21, 1). Most are similar to LVH/LVF. However, there are several key differences between RV and LV: four miRs (34a, 28, 148a, and 93) were upregulated in RVH/RVF that are downregulated or unchanged in LVH/LVF. Furthermore, there is a corresponding downregulation of their putative target genes involving cell survival, proliferation, metabolism, extracellular matrix turnover, and impaired proteosomal function. The current study demonstrates, for the first time, alterations in miRs during the process of RV remodeling and the gene regulatory pathways leading to RVH and RVF. Many of these alterations are similar to those in the afterload-stressed LV. miRs differentially regulated between the RV and LV may contribute to the RVs increased susceptibility to heart failure.
Keywords: heart failure, right ventricle
cardiac remodeling in response to stress results from activation and repression of genes encoding proteins that regulate cardiac contractility and structure. As novel mechanisms of remodeling are uncovered to understand the basic mechanisms of disease, an additional layer of regulation mediated by stress-responsive microRNAs (miRs) has emerged. miRs are small, noncoding RNAs of 18–25 nucleotides that regulate gene expression by degradation or translational suppression of mRNA. miRs have retained sequence conservation across species, indicating a strong evolutionary role in fundamental biologic processes. Since the discovery of the first miR in 1993, their role in cell differentiation, proliferation, apoptosis, and stress responses has been elucidated. Their role in cardiovascular development, left ventricular hypertrophy (LVH), and heart failure is only now being understood. (8, 13).
Van Rooij et al. (56) elucidated the miR expression profile in LVH in a model of thoracic aortic constriction. Many of the induced and repressed miRs were regulated in the same direction in fetal, hypertrophic, and failing human hearts, consistent with the known reactivation of the fetal gene program with stress (18, 51). Also noted were unique subsets of miRs dysregulated with each disease state: hypertrophy, ischemic heart disease, and dilated cardiomyopathy. Gain and loss of function studies of aberrantly expressed miRs in hypertrophied hearts have revealed specific miRs to be key regulators of cardiac hypertrophy and fibrosis such as miR-208, miR-21, miR-1, and miR-133 (7, 10, 22). As the functional role of miRs is elucidated, they are developing as tools for both the understanding of basic cardiovascular regulatory processes and as potential therapeutics for cardiovascular disease by the ability to manipulate their expression with miR mimics and inhibitors. Since miR expression is dynamic during a disease state, it is also conceivable that they can also be utilized as biomarkers for diagnosis and prognosis (47).
Despite extensive data in the left ventricle (LV), there is currently no information on miR expression in the afterload-stressed right ventricle (RV), either during the period of compensated hypertrophy or during the transition to heart failure. This is a critical issue for patients with congenital heart disease, where the RV is uniquely at risk of failure, e.g., in patients with complex congenital heart disease involving right-sided obstructive lesions (tetralogy of Fallot, pulmonary atresia), in patients with systemic RV (L-transposition, hypoplastic left heart syndrome), and in patients with pulmonary hypertension (23, 60). Given evidence suggesting that the stress response of the RV and LV may be marked by key differences (29, 53) and the failure of many common heart failure therapies to work in patients with failing RVs (24, 46), we sought to develop a better understanding of the role of miRs in the mechanisms of RV remodeling and failure. Using a murine model of RV afterload stress that recapitulates many of the clinical features of right ventricle hypertrophy-right ventricle failure (RVH/RVF) in humans, we sought to identify the role of miRs in the adaptation of the RV to hypertrophy and failure, to examine their predicted target genes for altered regulation and to compare miRs altered in RVH with those altered in LVH.
METHODS
Model of RV Afterload Stress
We have developed a murine model of RVH and RVF using pulmonary artery constriction (PAC) providing a unique platform for studying the physiological and molecular events of RV remodeling. (53) We used male FVB mice aged 12 wk for all studies. Following tracheal intubation, anesthesia was maintained with 1.5% isoflurane for the duration of the procedure. Via a right thoracotomy, a 7-0 silk suture was tied around the main pulmonary artery over a 27 G needle to create PAC with a reproducible band gradient of >35 mmHg. PAC mice were compared with sham-operated controls that underwent identical surgical procedures with the exception of placement of the constricting suture. Sham controls were not failed band placements, but a separate group of purposeful sham experiments. We have previously reported on this model, which demonstrates many key components that are observed in clinical RV disease: right bundle branch block by electrocardiogram, elevated RV end-diastolic pressure by invasive hemodynamics, and progression to RV contractile failure, manifested by decreased RV shortening fraction and RV free wall velocity. By 10 days, these mice develop overt signs of “clinical” RVF with liver enlargement, ascites, and high mortality (53). Hearts were harvested at 2, 4, and 10 days after PAC and total RNA isolated.
Model of LV Afterload Stress
LVH and left ventricular failure (LVF) were induced via transverse aortic constriction (TAC). This has been previously described by our group and others (60). Anesthetic procedures were similar between PAC and TAC. A 7-0 silk suture is placed between the innominate and carotid arteries over a 27 G needle to produce a gradient of >35 mmHg across the band similar to that in PAC. A similar group of sham controls was used. Hearts were harvested at 2, 4, and 10 days after TAC and total RNA isolated.
Echocardiography
Transthoracic echocardiograms were performed using a GE Vivid 7 ultrasound platform with a 13 MHz probe. PAC mice and sham-operated controls underwent echocardiograms at 2, 4, and 10 days after surgery to quantify the severity of RV outflow tract obstruction and its effect on RV morphology and function. As previously described (53), mice with a PAC peak pressure gradient >35 mmHg, RV dilation, interventricular septal (IVS) shift with encroachment on the LV cavity, and tricuspid insufficiency were characterized as having severe pulmonary stenosis (PS) and were used for this study. Following echocardiograms at each time point, the hearts were excised, and the RV free wall was separated from the IVS and LV and weighed.
miR and Gene Expression Profiling
Mice with severe PS were evaluated at three time points after surgery: early hypertrophy (2 days), decompensated hypertrophy (4 days), and heart failure (10 days) (n = 4/group/time point). Total RNA was isolated from the RV free wall of PS and sham-operated mice (miRNeasy Mini Kit, Qiagen). We used 10 ng of total RNA from each sample to synthesize cDNA, following which cDNA labeled with cyanine-3 was synthesized, amplified, and purified. This was hybridized to Agilent mouse whole genome oligonucleotide microarrays representing ∼41,000 probes; scanned and probe features were extracted using Agilent feature extraction software (Agilent One-Color Microarray Low Input Quick Amp Labeling Protocol). Total RNA (100 ng) from the same samples was labeled with cyanine-3 to generate fluorescent miR, purified, and hybridized to Agilent mouse miR microarrays representing 567 distinct probes. The slides were scanned, and data extracted using Agilent feature extraction software for miR expression.
Microarray Analysis
miR and gene expression analyses were performed using GeneSpring GX 11.5 software. Normalized data between the 20th and 100th percentiles with detected probes were used for further analysis. Quality control was performed, following which unpaired t-test with Benjamini Hochberg multiple testing correction was applied to the data. Significantly altered miRs and gene transcripts with a corrected p value of <0.05 and with a fold change ≥2 up- or downregulated were considered for further analysis. Putative target genes were identified using the TargetScan algorithm with sequence specificity, binding site accessibility, and evolutionary conservation of binding sites. The gene database containing statistically significant microarray data from sham vs. PAC was queried for the above-identified targets. The target gene subset enriched in PAC was then compared with statistically significant gene microarray datasets from mice undergoing TAC previously reported by our lab (60). Gene Ontology (GO) and pathway analyses were performed to identify important biologic processes, nodal points, and pathways unique and common to compensated and decompensated hypertrophy and heart failure.
Reverse Transcriptase Polymerase Chain Reaction
Expression of key target genes was confirmed in a one-step qRT-PCR using SYBR green technology (Qiagen). In brief, 200 ng of total RNA was reverse transcribed to cDNA and amplified over 40 cycles using the ABI 7900 Thermocycler. Primers were designed using the Primer 3 Output program. The expression of a subset of dysregulated miRs was confirmed by Taqman two-step qRT-PCR using 50 ng of starting template, reverse transcribing to cDNA followed by amplification (Applied Biosystems). Ambion mirVana qRT-PCR Primer Sets were used. Fold change in expression was compared between PAC mice and sham-operated controls and between PAC and TAC.
Western Blotting
Western blotting of selected miR targets was performed to assess if miR-induced transcriptional changes led to translational changes. Proteins were separated by gel electrophoresis, transferred onto a nitrocellulose membrane, and detected using the following antibodies: CaMKII, JNK1, p38 (Santa Cruz Biotechnology, sc-571, sc-6187, sc-5306, respectively), Akt, and GSK3b (Cell signaling, #4691 and #9315). GAPDH was used as the housekeeping protein.
Isolation of RV Cardiomyocytes and Nonmyocytes
We isolated calcium-tolerant adult RV myocytes and RV nonmyocytes from sham and PAC-operated mice. In brief, the heart was retrograde perfused and enzymatically digested in a calcium-free solution, following which CaCl2 was added for a final concentration of 50 mM and further perfused. The RV was separated and digested followed by step-wise calcium readministration to render calcium-tolerant cells. The cell suspension was centrifuged, and the pellet resuspended and centrifuged to separate the myocyte and nonmyocyte fractions (19).
Statistical Analysis
Data other than microarray datasets are represented as means ± SD. Unpaired Student t-test for two-group and ANOVA for three or more group comparison was performed on continuous, normally distributed data with a P value of ≤0.05 being considered significant.
Animal Care
All procedures were performed in accordance with National Institutes of Health standards and were approved by the Administrative Panel on Laboratory Animal Care at Stanford University.
RESULTS
Model of RVH and RVF
Echocardiography.
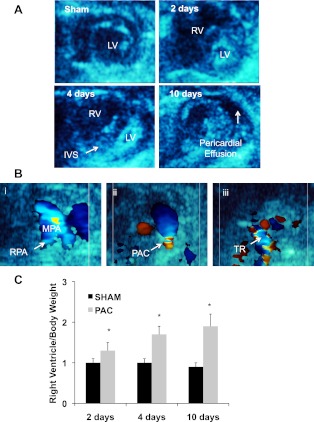
As early as 2 days after PAC, there was RV dilation with flattening of the IVS (Fig. 1A, top right). At 4 days after PAC, the RV continued to dilate with further flattening of the IVS (Fig. 1A, bottom left) and the development of tricuspid regurgitation (Fig. 1B, right). By 10 days, there was posterior bowing of the IVS into the LV with evidence of pericardial effusion (Fig. 1A, bottom right). We have previously reported on the abnormalities of RV function developing over the first 10 days after PAC including a decrease in the RV outflow tract fractional shortening and RV free wall velocity (53).
Fig. 1.
Parasternal short-axis 2D echocardiograms demonstrating right ventricle (RV) dilation and RV failure after pulmonary artery constriction (PAC). A: sham-operated control: normal circular left ventricle (LV) and crescent-shaped RV cavities; PAC at 2 days with RV dilation; PAC at 4 days with RV dilation and pancaking of the LV cavity due to interventricular septal (IVS) shift; PAC at 10 days with the development of pericardial effusion. B: color Doppler images demonstrating sham-operated control: normal main pulmonary artery and branch pulmonary arteries (i), flow turbulence at the site of the pulmonary artery band (ii), tricuspid regurgitation (TR, iii). C: RV mass increased after PAC vs. sham as early as 2 days after banding, with a further increase at 4 and 10 days postoperatively (*P < 0.05). MPA, main pulmonary artery; RPA, right pulmonary artery.
RVH.
RV free wall weight was significantly increased as early as 2 days after banding and increased further at 4 and 10 days compared with sham-operated controls (Fig. 1C). We have previously reported complete morphometric analysis of hearts from these mice (53).
miR Expression in the Normal and Stressed RV vs. LV
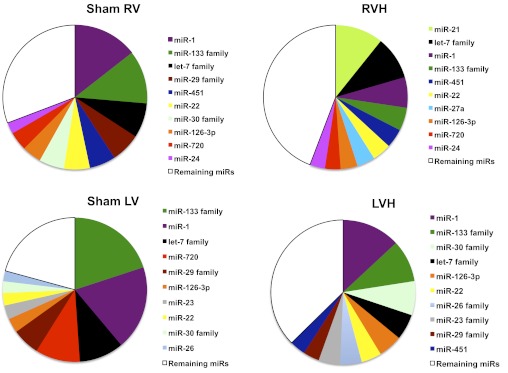
miRs most abundantly expressed in the RV of PAC and sham-operated mice at 10 days are shown in Fig. 2. This was compared with miRs most abundantly expressed in the normal LV [GSM357026, National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) database] and in serum response factor (SRF)-induced LVH (GSE23044, NCBI GEO database) where raw array data were available for analysis (33, 58). Prior studies on TAC did not provide the data to allow the expression of the most abundant miRs to be expressed as a percentage of total miRs. However, since key upregulated miRs are similar between TAC and SRF-induced LVH, we used SRF data for this comparison. Much of the expression profile was similar between the sham RV and LV, with miR-1, 133, and let-7 families being the most abundantly expressed miRs in both ventricles. These miRs were, however, much more abundant in the LV than in the RV. We next compared the abundance of miR expression in RVH after PAC and in SRF-induced LVH. miRs that were noted to be the most abundant in the normal RV and LV are also the most abundant miRs in RVH and LVH. In addition, while miR-21 is more abundant in RVH vs. LVH, miR families-23, 26, 29, and 30 are more abundant in LVH.
Fig. 2.
The 10 most abundantly expressed miRs in the RV of sham and PAC animals were compared with the 10 most abundant miRs in the LV of sham and SRF-induced hypertrophy (58). Data are expressed as median signal intensity. LV data were obtained from GEO datasets. microRNA (miR) distribution is similar in both the RV and LV with miR-1, 133, and let-7 family being the most abundant. See text for details of data selection.
Dynamic miR Changes from RVH to RVF
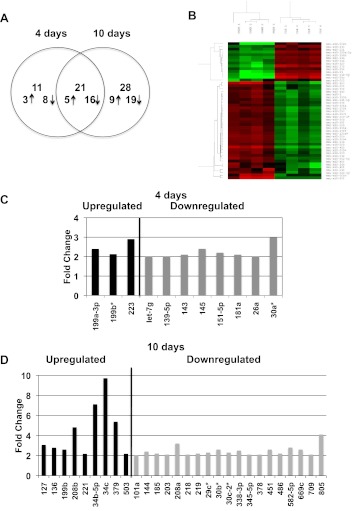
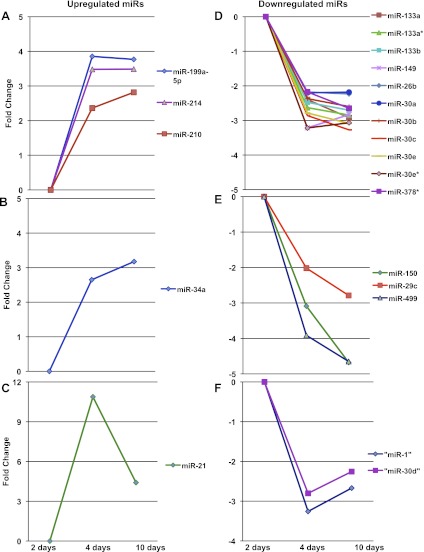
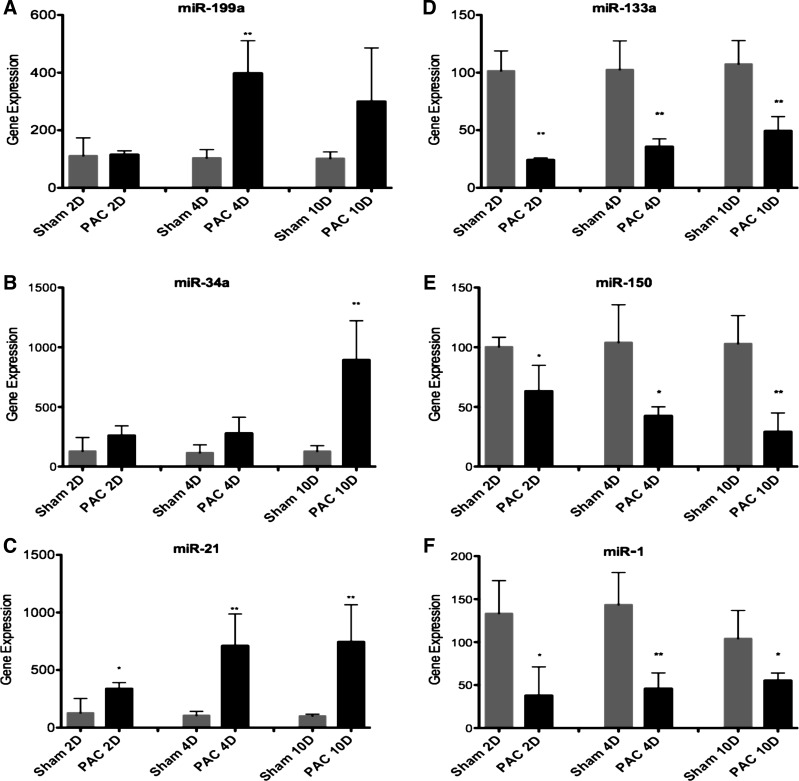
At 2 days, which is the stage of early compensated hypertrophy, there were no differentially expressed miRs. At 4 days, which is the stage of early decompensated hypertrophy without overt heart failure, 32 miRs were differentially expressed: eight miRs were upregulated and 24 were downregulated. At 10 days, which is the stage of overt heart failure, 49 were differentially expressed: 14 miRs were upregulated and 35 were downregulated (Fig. 3A). Of these, 21 differentially expressed miRs were common to both four and 10 days: five were upregulated and 16 were downregulated. Figure 3B demonstrates the heat map of significantly altered miRs at 10 days. Figure 3, C and D, identifies miRs differentially expressed at only 4 or 10 days after PAC with their fold changes. Many of these miRs parallel those reported in LVH and LVF. Interestingly, these miRs followed three patterns of expression over time (Fig. 4). Of the five miRs upregulated at 4 days, three continued to be upregulated with no further increase in expression at 10 days (Fig. 4A), while one (miR-34a) continued to further increase (Fig. 4B) and one (miR-21) decreased in expression (Fig. 4C). Downregulated miRs followed a similar pattern, with the majority (n = 11/16) showing a plateau in expression from 4 to 10 days (Fig. 4D), while others continued to further decrease (n = 3/16) (Fig. 4E) or began to increase (n = 2/16) (Fig. 4F). qRT-PCR was performed on the first miR listed in each of the six subgroups in Fig. 4 to validate the array data. In addition, each miR was tested at three time points: 2, 4 and 10 days. The following miRs were tested: miRs 199a-5p, 34a, 21, 133a, 150, and 1. There was excellent correlation with the microarray data for all miRs at the 4- and 10-day time points. However, while the array data indicated no statistical difference between Sham and PAC at 2 days, four of the six RT-PCRs demonstrated a statistically significant change at 2 days. miR-21 was upregulated at 2 days but to a lesser degree than at 4 and 10 days. miRs 133a, 150, and 1 were downregulated at 2 days similar to the 4-day time point (Fig. 5).
Fig. 3.
miRs altered with hypertrophy and heart failure. A: at 2 days (phase of early RV hypertrophy) no miRs were differentially expressed between PAC and sham, at 4 days (phase of compensated RV hypertrophy) 32 miRs were differentially expressed, and at 10 days (phase of RV failure) 49 were differentially expressed >2-fold. Corrected P value < 0.05. Twenty-one miRs were common to both hypertrophy and heart failure. B: heat map of significantly altered miRs at 10 days between sham and PAC. The color intensity in red represents gene expression being increased and green represents gene expression being decreased. C and D demonstrate miRs dysregulated at only the 4- and 10-day time points with their fold changes.
Fig. 4.
Patterns of miR expression during the progression from early to late hypertrophy and heart failure for differentially expressed miRs common to both 4 and 10 days. (Fold change >2, corrected p value <0.05.) Upregulated miRs follow 3 patterns of expression: increase at 4 days followed by a plateau at 10 days (A), increase at 4 days and further increase at 10 days (B), and increase at 4 days and decrease at 10 days (C). Downregulated miRs follow a similar pattern (D, E, F).
Fig. 5.
qRT-PCR validation of miR microarray data. Six representative up- and downregulated miRs, from each of the 6 groups in Fig. 4, were validated by qRT-PCR. Each miR was evaluated at 2, 4, and 10 days after PAC and compared with age-matched sham-operated controls. There was excellent correlation with the miR microarray data at the 4- and 10-day time points. At the 2-day time point, miR-21 was upregulated and miRs-133a, 150, and 1 were significantly downregulated. These were not statistically significant in the miR microarray data. *P < 0.05, **P < 0.01.
miR-Gene Correlation in RVH and RVF
Significant alterations in miR expression at 10 days were then correlated with significant alterations in gene expression at 10 days from Agilent whole genome oligonucleotide microarrays. We further narrowed our examination to those genes that met the following criteria: 1) genes predicted by both TargetScan and DIANA microT to be regulated by any of the altered miRs; 2) predicted targets that were appropriately up- or downregulated, i.e., decreased if their regulatory miRs were upregulated or increased if their regulatory miRs were downregulated; and 3) predicted targets that were also literature-validated, when possible. Pathway analysis of target genes identified in the PAC gene microarray database revealed nodal points involving Nfκb1, c-jun, ErbB2, Ep300, Raf1, and VEGF networks.
Phase of hypertrophy: target genes increase cardiomyocyte size and fibrosis.
Gene expression data were not available for this phase. Hence only literature-validated target genes are reported. Of the three miRs upregulated at 4 days, miR-199a-3p downregulates mTOR and c-met, miR-199b* downregulates Dyrk1a (NFAT pathway), and miR-223 downregulates glucose uptake via Glut-4. The miR 143/145 cluster, which is downregulated at 4 days, upregulates Krüppel-like factor-4 (Klf4). Downregulation of miR-181a and Let-7 family upregulate their target gene Bcl-2, while Let-7g upregulates Nfκb1. Downregulation of the miR-30 family upregulates its target gene connective tissue growth factor (Ctgf).
Phase of heart failure: enhanced apoptosis and reactivation of the fetal gene program.
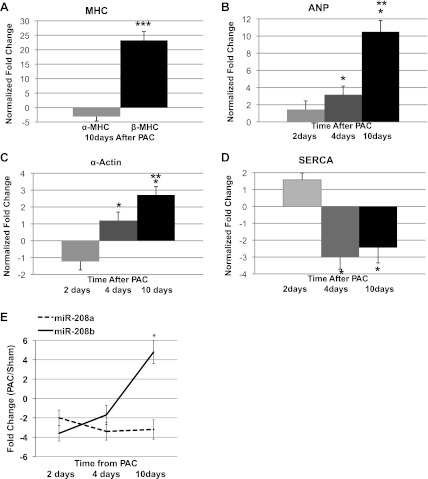
miR-199 and miR-30 families continued to be up- and downregulated, respectively, during this phase, although the isoforms differed compared with hypertrophy. miRs upregulated only during the phase of heart failure such as the miRs-34b and c were associated with downregulation of cell cycle genes. miR-379 and 503 were upregulated by five- and twofold, respectively, and were associated with downregulation of Ccne1 and cdc25A. This phase was also characterized by reactivation of the fetal gene program with a significant increase in the expression of fetal β-cardiac myosin heavy chain gene (β-MHC) and a concomitant decrease in the expression of adult α-cardiac myosin heavy chain gene (α-MHC). miR-208b is encoded within an intron of β-MHC gene, while miR-208a is encoded within an intron of α-MHC. The expression of these miRs paralleled the expression of these genes and was confirmed by RT-PCR (Fig. 6). Downregulation of the miR-144/451 cluster was associated with upregulation of Cugbp2. Expression of the hypertrophy/heart failure program was also assessed during this stage using RT-PCR. There was a progressive increase in atrial natriuretic peptide (ANP) and α-actin expression from 2 days to 10 days with a decrease in SERCA expression at 4 and 10 days.
Fig. 6.
Reactivation of fetal gene expression during hypertrophy and heart failure. A: α-myosin heavy chain (MHC) and β-MHC gene expression decrease and increase respectively with heart failure in PAC RV compared with sham (*P < 0.05). There is also an increase in the expression of other fetal genes including atrial natriuretic peptide (ANP, B) and α-actin (C). D: SERCA expression was downregulated with hypertrophy and heart failure (ANOVA P < 0.05). E: expression of miR-208a and 208b parallel the changes in α-MHC and β-MHC gene expression. During early hypertrophy miR-208a predominates in the RV. As hypertrophy progresses, there is a shift from miR-208a to 208b. With the development of heart failure, there is a dramatic increase in miR-208b, while miR-208a remains downregulated (*P < 0.05). **P < 0.01, ***P < 0.001.
miRs common to hypertrophy and heart failure: enhanced apoptosis and fibrosis.
Of the five upregulated miRs common to the stages of hypertrophy and heart failure, miRs-199a, 210, 214, and 21 were associated with genes involved in myocyte survival and adaptation to stress (table 1). While all of these miRs increased during hypertrophy, their expression plateaued or fell with the development of heart failure. miR-199a targets ubiquitin conjugating enzymes, many of which were downregulated in our model. miR-21 was upregulated by >10-fold with concomitant downregulation of its target genes such as interleukin 12, Fas ligand belonging to the tumor necrosis factor superfamily, and Pten. Downregulation of Pten was associated with an increase in matrix metalloprotease-2 gene expression. This upregulation was not sustained as with the development of heart failure; miR-21 expression began to fall. Coinciding with this fall in miR-21 expression was the upregulation of proapoptotic miRs such as miR-34a, whose expression was further increased during the stage of heart failure. Eleven of 16 downregulated miRs showed a plateau in their expression from the stage of hypertrophy to heart failure. Among these, downregulation of the miR-30 family was associated with upregulation of sorting nexin, metadherin, and MAP kinase 8. In addition, downregulation of miR-133 family, miRs-149, and 26b was associated with upregulation of collagens. miRs-150, 29c, and 499 decreased by two - to fourfold during hypertrophy and further decreased with heart failure. These miRs were associated with upregulation in Tgf-β signaling, collagens, formin-binding protein and myosin 5. miRs-1 and 30d were downregulated at 4 days with their expression being less downregulated by 10 days. Muscle-specific miR-1 downregulation was associated with upregulation of DNA polymerase alpha 1, fibronectins, annexins, and ubiquitin conjugation. No direct targets of miR-378* were identified in our model.
Table 1.
miRs common to right and left ventricular hypertrophy and failure
| miRs | Target Genes | Function |
|---|---|---|
| Upregulated (n = 5) | Downregulated | |
| miR-199a-5p | Ube2 g1, Ube2i | ubiquitin proteosomal system |
| Gsk3b; CamK2a | prosurvival, antifibrotic, antiapoptotic, glycogen metabolism; calcium homeostasis | |
| miR-210 | Nmnat2; Scara3 | NAD synthesis; scavenges ROS |
| miR-214 | Mrpl17; BclII, Akt3 | mitochondrial protein synthesis; survival, antiapoptotic |
| miR-34a | Ccnd1 | cell cycle G1/S transition |
| miR-21 | IL12a, FasL Pten | ↑fibroblast proliferation/survival ECM breakdown |
| Downregulated (n = 16) | Upregulated | |
| miR-133a, 133a*, 133b | Col11a1, Col6a3; Mtap4 | fibrosis; microtubule stabilization |
| miR-149, 26b | Col6a3; Il17rd | fibrosis; FGF signaling |
| miR-30a, b, c, d, e, e* | Snx10; Lpp, Mtdh; Mapk8 | intracellular trafficking/cell adhesion; TNF-α-induced apoptosis |
| miR-378* | no targets | Metabolic shift lipid->glucose |
| miR-150 | Tgfbr2, Prkar1a | profibrotic, proapoptotic |
| miR-29c | collagens 1, III, IV, Fbn1 | profibrotic |
| miR-499 | Fnbp4, Cd2ap; Myo5a | actin and microtubule dynamics; cytoplasmic vesicle transport; TNF-α-induced apoptosis |
| Mapk8 | ||
| miR-1 | Pola1; Anxa4; Azin1; Fn1; Ube2 h, Trim2, Ankib1 | DNA replication; lsosomal metabolism; cell adhesion; ubiquitin conjugation |
Shown are microRNAs (miRs) altered in right ventricular hypertrophy-right ventricular failure (RVH/RVF) at 10 days that have been previously described in left ventricular hypertrophy-left ventricular failure (LVH/LVF). Dysregulated miRs common to both hypertrophy and heart failure are listed below with examples of their gene targets and related functions. (Fold change >2, Benjamini Hochberg corrected p valve <0.05 for miRs and target genes.)
RVH and RVF do not fully mirror LVH and LVF.
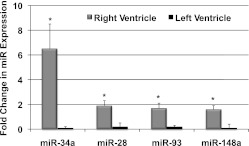
We next compared miRs dysregulated in RVH/RVF with LVH/LVF. Whereas many of the dysregulated miRs were similar to those differentially regulated during LVH/LVF (TAC), we identified four miRs that were uniquely upregulated in RVH/RVF but which have been reported as being downregulated in LVH/LVF. (12, 56, 57) These are miR-34a, 28, 93, and 148. We confirmed their upregulation in RVH using Taqman qRT-PCR. Although not previously reported to be upregulated with LVH, we confirmed this by qRT-PCR in a separate group of mice undergoing TAC (Fig. 7). miR expression was again correlated with gene microarray data at 10 days. In each case, key target genes listed were identified as being appropriately downregulated by gene microarray in the RV and upregulated in the LV, further evidence that these miRs played a differential role in the stressed RV vs. LV. Table 2 highlights examples of predicted, downregulated targets of these four unique miRs with their functions. The most differentially expressed, miR-34a was upregulated by sixfold in RVH. Cheng et al. (12) found miR-34a to be downregulated by 1.5-fold in LVH, and we confirmed that it was unchanged by RT-PCR in TAC using similar strain and age mice. We found coordinate downregulation of many miR-34a target genes in the RV, examples of which are shown in Table 2. Interestingly, one of these, Ccnd1, was found to be upregulated in LVH. miR-28 was upregulated by 1.9-fold in RVH but unchanged in LVH. There was associated downregulation of mitochondrial carrier genes and ubiquitin ligase. miR-148a was upregulated by 1.6-fold in RVH and was associated with downregulation of heat shock protein 40, protein phosphatase 1L, and citrate synthase. Upregulation of miR-93 by 1.7-fold was associated with downregulation of GTPase-activating protein, cyclin D1, Akt, and p38.
Fig. 7.
Unique RV-specific miRs. Comparison by RT-PCR for 4 miRs upregulated in RVH vs. sham but not in LVH vs. sham. For each of these miRs, there was excellent agreement between RT-PCR and array expression. *P < 0.01.
Table 2.
Unique right ventricle-specific miRs
| miRs Upregulated in RVH | Target Genes Downregulated in RVH | Function |
|---|---|---|
| miR-34a | Eno3, Acsl1, Aadacl1 | glucose/lipid oxidation |
| Ccnd1, Ppp1r11 | cell cycle G1/S transition, tumor suppressor | |
| miR-28 | Slc25a10 | mitochondrial carrier |
| Fbxl18 | ubiquitin ligase | |
| Zfp488 | transcriptional regulator | |
| Ppp1r9a, Iqsec2 | cytoskeletal and synaptic organization | |
| miR-148a | Dnajc13 (Hsp40) | ischemia-reperfusion protection |
| Ppm1l | antiapoptotic | |
| Map3k9 | cell proliferation, differentiation | |
| CamK2a | calcium homeostasis | |
| Cs | metabolic | |
| miR-93 | Arhgap12 | GTPase activation |
| Vldlr | metabolic | |
| Rnf6 | transcription activators | |
| Ccnd1, Pten | cell cycle G1/S transition, tumor suppressor | |
| Usp24 | deubiquitinating enzyme | |
| Akt3 | survival, antiapoptotic | |
| p38 (MAPK14) | transcription, cell cycle regulation |
miRs upregulated in RVH but downregulated in LVH and examples of their corresponding TargetScan and DIANA microT-predicted target genes. Target genes that are underlined are those that were not only downregulated in RVH but also upregulated in LVH. (miR fold change ≥1.5, corrected P valve <0.05; target gene fold change ≤1.5, corrected P value <0.05.)
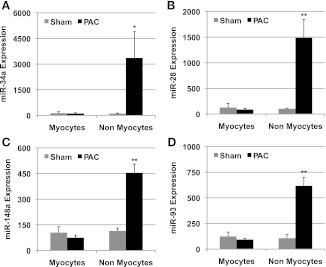
To further identify which cell type is responsible for the induction of these RV-specific miRs, we isolated RV myocytes and nonmyocytes from sham and PAC. qRT-PCR performed on these fractions demonstrated that the increase in all four RV-specific miRs was from the nonmyocyte fraction (Fig. 8). Protein expression of key target genes known to play a role in cardiac remodeling was also assessed and is shown in Fig. 9.
Fig. 8.
miR expression in myocytes vs. nonmyocytes. RV-specific miR expression by qRT-PCR normalized to U6 in the myocyte vs. the nonmyocyte fraction demonstrated a significantly higher expression in the nonmyocyte fraction in PAC vs. sham. There was no difference in expression in the myocyte fraction in PAC vs. sham. *P < 0.01, **P < 0.005.
Fig. 9.
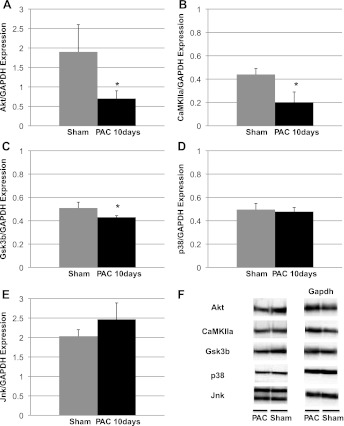
Protein expression of key miR targets, known regulators of cardiac remodeling, after 10 days' PAC. A: expression of the miR-93 and miR-214 target, Akt was decreased. B: expression of miR-148a and miR-199a-5p target, CaMKIIa was decreased. C: Gsk3β, an miR-199a-5p target, demonstrated decreased expression. These correspond with gene expression data. D, E: however, p38, an miR-93 target and Jnk, an miR-30 target, were not different between sham and PAC. *P < 0.05. F: representative blots for the above target proteins.
DISCUSSION
The molecular events that mark the transition from a stressed but compensated state, e.g., stable hypertrophy, to overt heart failure are still not completely understood. Both animal and human data have advanced our understanding of the molecular events that mark the progression from compensatory LVH to a maladaptive state of overt LV dysfunction and then to clinical heart failure (4, 15, 37, 54, 55). The role of miRs in LVH and LVF points to their potential involvement in disease progression with potential for clinical application as drug targets or as biomarkers. In contrast, there are no data on miR regulation of RV remodeling in response to hemodynamic stressors and the pathways leading to RVF (5, 29). Although the current study is mainly descriptive, it is an important and necessary first step toward evaluating the significance of differentially expressed miRs in the stressed RV.
The current study demonstrates, for the first time, alterations in miRs during the process of RV remodeling in response to afterload stress and the gene regulatory pathways leading to RVH and progressing to RVF. Many of these alterations are similar to those in the afterload-stressed LV. Key nodal points identified via pathway analysis in the afterload-stressed RV include cell growth, inflammation, hypertrophy pathways via Nfκb1; pathologic hypertrophy and heart failure via c-Jun-Akt and Raf1 pathways; injury and apoptosis (erbB2); extracellular matrix remodeling and fibrosis (Ep300); and angiogenesis (VegfA). Profiling the dynamic changes in miR expression associated with RV pressure overload demonstrates a balance between prosurvival and proapoptotic miRs during hypertrophy with the balance tilted toward proapoptosis and profibrosis during heart failure, as has been described with LVH/LVF. The expression of miRs maintaining the adaptation to hypertrophy such as miRs involved in cell growth, the ubiquitin proteosomal system, and cell adhesion are noted to plateau or decrease with the development of heart failure. Specifically, miRs dysregulated during the stage of hypertrophy target key mediators of hypertrophy to increase cardiomyocyte size (miR-199a-3p, Let-7) (20), enhance glucose uptake (miR-223) (36), enhance abnormal vascular tone (miR-143/145 cluster) (14), enhance resistance to apoptosis (miR-181a, Let 7) (11), and contribute to increased collagen synthesis (miR-30). miRs dysregulated during the phase of heart failure target genes involved in enhancing collagen synthesis (miR-133 and 30 family) (10, 16, 31, 39), enhancing apoptosis via cell cycle arrest (miR-34 family, miR-144/451 cluster) (59), and inhibiting endothelial cell proliferation and migration (miR-379, 503) (9). Reactivation of the fetal gene program is also noted with an increase in the expression of β-MHC and a decrease in α-MHC. miR-208b and 208a follow the expression of these genes, respectively (7).
The transition from hypertrophy to heart failure is marked by upregulation of miRs targeting genes critical to myocyte survival and adaptation to stress. Their expression subsequently decreases, signaling the development of heart failure (6, 22, 25, 32, 40). These include impairment in the ubiquitin proteosomal system (miR-199a-5p) (21, 48), decrease in antiapoptotic mechanisms, fibroblast proliferation, and survival and increased extracellular matrix breakdown (miR-21) (43, 45). Although miR-21 is also known to decrease PPARα protein levels thereby decreasing fatty acid oxidation, we found no change at the level of PPARα gene expression (30). Proapoptotic miRs begin to increase with heart failure (miR-34 family). There is upregulation of target genes involved in apoptosis and enhancing fibrosis (miRs-133 family, 30 family, 150, 149, 29c, 499), and enhancing intracellular trafficking, and cell adhesion (miR-30 family) (10, 31, 39). Muscle-specific miR-1 regulates an array of growth factor genes. This is associated with upregulation of genes involved in DNA replication, cell adhesion, and metabolism (22). An increase in miR-1 leads to a downregulation of these growth factors signaling a transition away from compensated hypertrophy (45). Excess of miR-378* has been implicated in the metabolic shift from fatty acid oxidation to glucose oxidation in cancer cells via PGC-1β. This is decreased in heart failure, but no direct targets have been identified in our model (17).
Although most of these alterations are similar to those in the afterload-stressed LV, there are important differences, highlighting differences in the adaptive mechanisms between the RV and LV response to pressure overload stress. We note unique RV-specific, upregulated miRs that are unchanged in LVH/LVF. miR-34a, the most upregulated miR in this group, downregulates target genes involved in glucose and lipid oxidation, enhancing cell cycle arrest and apoptosis in a p53-dependent and -independent fashion (34, 49). miR-34b and 34c are upregulated during heart failure in the RV and LV; however, miR-34a was upregulated in RVF only. Upregulation of miR-34a was found predominantly in the nonmyocyte fraction, which correlates with literature reports of miR-34a contributing to fibroblast and endothelial senescence (27, 38, 42). miR 28 downregulates target genes involved in mitochondrial transport, ubiquitin proteosomal system, and cytoskeletal organization. Upregulation of miR-148a downregulates genes important to ischemia-reperfusion injury protection, TCA cycle, cell survival and calcium homeostasis (41, 44, 50, 52). Upregulation of miR-93 is associated with downregulation of several of its target genes involved in GTPase and transcriptional activation, lipid metabolism, cell cycle regulation, and survival. Many of these processes have been described both in the progression of LVH and heart failure and in the RV subject to pressure-overload (1–3, 26, 28). In support of the validity of these upregulated miRs playing a role in RV remodeling, upregulation of RV-specific miRs in our model was associated with downregulation of genes involved in maintaining lipid oxidation, decreasing cell death, and senescence. Thus, although these pathways are not unique to the remodeling or failing RV, our data suggest that these processes may be differentially or more severely dysregulated in the RV than in the LV, thereby contributing to the increased susceptibility of the RV to heart failure compared with the afterload-stressed LV. Each of these processes will require further study to confirm a mechanistic role.
Our study has several limitations. Our model of PAC results in severe afterload stress to the RV, thereby causing a rapid progression from hypertrophy to heart failure. This is similar to most accepted models of LVF induced by TAC. Whether these alterations would be similar in a model of more moderate afterload stress with gradual development of RVH and RVF, as in patients with repaired or palliated congenital heart disease, remains to be determined in future studies. We recognize that upregulation of a miR alone doesn't prove its functional role and that overexpression and knock-down/antagomir studies will be needed to completely validate this data. Finally, TargetScan has limited ability to predict specific targets, thus a second target prediction program, DIANA microT, was used, and literature validation was performed where possible. However, in support of the significance of our findings, substantial numbers of the predicted target genes moved in the right direction with up- or downregulation in the RV and showed the exact opposite change in the LV.
In summary, this is first description of the dynamic changes in miR expression in the afterload-stressed RV as it transitions from hypertrophy to heart failure. Whereas many of these changes are similar to LVH/LVF, we identified four unique miRs that are upregulated in RVH/RVF that are not altered in LVH/LVF. These miRs regulate targets enhancing apoptosis, altering energy availability and impairing calcium handling, all of which may make the RV more vulnerable to failure when subject to afterload stress. RVH and RVF are important long-term sequel after repair of right-sided obstructive congenital heart lesions (tetralogy of Fallot, pulmonary atresia), in patients with systemic RV, and in those with pulmonary hypertension (35). Recent clinical trials suggest that medications used to treat LVF may not have a clear benefit when used to treat RVF (24, 46). In this study we have shown that miR regulation of RV remodeling represents a potential mechanism for differences between the RV and LV response to pressure overload stress.
GRANTS
The Pediatric Research Fund, Lucile Packard Children's Hospital; the Children's Heart Foundation; National Heart, Lung, and Blood Institute Grant HL-061535 to D. Bernstein. Grant support for Z. Ghosh, S. Hu, and J. C. Wu: DP2OD004437, R01HL-095571, and BWF CAMS (J. C. Wu).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: S.R., G.A.F., S.H., Z.G., J.C.W., and D.B. conception and design of research; S.R., M.Z., D.-Q.H., and V.R. performed experiments; S.R. analyzed data; S.R. interpreted results of experiments; S.R. and D.-Q.H. prepared figures; S.R. drafted manuscript; S.R., G.A.F., and D.B. edited and revised manuscript; S.R., M.Z., D.-Q.H., G.A.F., S.H., Z.G., V.R., J.C.W., and D.B. approved final version of manuscript.
REFERENCES
- 1. Bogaard HJ, Abe K, Vonk Noordegraaf A, Voelkel NF. The right ventricle under pressure: cellular and molecular mechanisms of right-heart failure in pulmonary hypertension. Chest 135: 794–804, 2009 [DOI] [PubMed] [Google Scholar]
- 2. Bogaard HJ, Natarajan R, Henderson SC, Long CS, Kraskauskas D, Smithson L, Ockaili R, McCord JM, Voelkel NF. Chronic pulmonary artery pressure elevation is insufficient to explain right heart failure. Circulation 120: 1951–1960, 2009 [DOI] [PubMed] [Google Scholar]
- 3. Braun MU, Szalai P, Strasser RH, Borst MM. Right ventricular hypertrophy and apoptosis after pulmonary artery banding: regulation of PKC isozymes. Cardiovasc Res 59: 658–667, 2003 [DOI] [PubMed] [Google Scholar]
- 4. Brener SJ, Duffy CI, Thomas JD, Stewart WJ. Progression of aortic stenosis in 394 patients: relation to changes in myocardial and mitral valve dysfunction. J Am Coll Cardiol 25: 305–310, 1995 [DOI] [PubMed] [Google Scholar]
- 5. Buermans HP, Redout EM, Schiel AE, Musters RJ, Zuidwijk M, Eijk PP, van Hardeveld C, Kasanmoentalib S, Visser FC, Ylstra B, Simonides WS. Microarray analysis reveals pivotal divergent mRNA expression profiles early in the development of either compensated ventricular hypertrophy or heart failure. Physiol Genomics 21: 314–323, 2005 [DOI] [PubMed] [Google Scholar]
- 6. Buscaglia LE, Li Y. Apoptosis and the target genes of microRNA-21. Chin J Cancer 30: 371–380, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, Willis MS, Selzman CH, Wang DZ. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest 119: 2772–2786, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Callis TE, Wang DZ. Taking microRNAs to heart. Trends Mol Med 14: 254–260, 2008 [DOI] [PubMed] [Google Scholar]
- 9. Caporali A, Meloni M, Vollenkle C, Bonci D, Sala-Newby GB, Addis R, Spinetti G, Losa S, Masson R, Baker AH, Agami R, le Sage C, Condorelli G, Madeddu P, Martelli F, Emanueli C. Deregulation of microRNA-503 contributes to diabetes mellitus-induced impairment of endothelial function and reparative angiogenesis after limb ischemia. Circulation 123: 282–291, 2011 [DOI] [PubMed] [Google Scholar]
- 10. Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MV, Hoydal M, Autore C, Russo MA, Dorn GW, 2nd, Ellingsen O, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G. MicroRNA-133 controls cardiac hypertrophy. Nat Med 13: 613–618, 2007 [DOI] [PubMed] [Google Scholar]
- 11. Chen G, Zhu W, Shi D, Lv L, Zhang C, Liu P, Hu W. MicroRNA-181a sensitizes human malignant glioma U87MG cells to radiation by targeting Bcl-2. Oncol Rep 23: 997–1003, 2010 [DOI] [PubMed] [Google Scholar]
- 12. Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol 170: 1831–1840, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cordes KR, Srivastava D. MicroRNA regulation of cardiovascular development. Circ Res 104: 724–732, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davis-Dusenbery BN, Chan MC, Reno KE, Weisman AS, Layne MD, Lagna G, Hata A. Down-regulation of Kruppel-like factor-4 (KLF4) by microRNA-143/145 is critical for modulation of vascular smooth muscle cell phenotype by transforming growth factor-beta and bone morphogenetic protein 4. J Biol 286: 28097–28110, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Douglas PS, Reichek N, Hackney K, Ioli A, Sutton MG. Contribution of afterload, hypertrophy and geometry to left ventricular ejection fraction in aortic valve stenosis, pure aortic regurgitation and idiopathic dilated cardiomyopathy. Am J Cardiol 59: 1398–1404, 1987 [DOI] [PubMed] [Google Scholar]
- 16. Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, van der Made I, Herias V, van Leeuwen RE, Schellings MW, Barenbrug P, Maessen JG, Heymans S, Pinto YM, Creemers EE. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res 104: 170–178, 2009 [DOI] [PubMed] [Google Scholar]
- 17. Eichner LJ, Perry MC, Dufour CR, Bertos N, Park M, -Pierre J, Giguere V. miR-378(*) mediates metabolic shift in breast cancer cells via the PGC-1beta/ERRgamma transcriptional pathway. Cell Metab 12: 352–361, 2010 [DOI] [PubMed] [Google Scholar]
- 18. El-Armouche A, Schwoerer AP, Neuber C, Emmons J, Biermann D, Christalla T, Grundhoff A, Eschenhagen T, Zimmermann WH, Ehmke H. Common microRNA signatures in cardiac hypertrophic and atrophic remodeling induced by changes in hemodynamic load. PLoS One 5: e14263, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ferreira JC, Koyanagi T, Palaniyandi SS, Fajardo G, Churchill EN, Budas G, Disatnik MH, Bernstein D, Brum PC, Mochly-Rosen D. Pharmacological inhibition of betaIIPKC is cardioprotective in late-stage hypertrophy. J Mol Cell Cardiol 51: 980–987, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fornari F, Milazzo M, Chieco P, Negrini M, Calin GA, Grazi GL, Pollutri D, Croce CM, Bolondi L, Gramantieri L. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res 70: 5184–5193, 2010 [DOI] [PubMed] [Google Scholar]
- 21. Haghikia A, Missol-Kolka E, Tsikas D, Venturini L, Brundiers S, Castoldi M, Muckenthaler MU, Eder M, Stapel B, Thum T, Petrasch-Parwez E, Drexler H, Hilfiker-Kleiner D, Scherr M. Signal transducer and activator of transcription 3-mediated regulation of miR-199a-5p links cardiomyocyte and endothelial cell function in the heart: a key role for ubiquitin-conjugating enzymes. Eur Heart J 32: 1287–1297, 2011 [DOI] [PubMed] [Google Scholar]
- 22. Han M, Toli J, Abdellatif M. MicroRNAs in the cardiovascular system. Curr Opin Cardiol 26: 181–189, 2011 [DOI] [PubMed] [Google Scholar]
- 23. Hines R. Right ventricular function and failure: a review. Yale J Biol Med 64: 295–307, 1991 [PMC free article] [PubMed] [Google Scholar]
- 24. Hsu DT, Zak V, Mahony L, Sleeper LA, Atz AM, Levine JC, Barker PC, Ravishankar C, McCrindle BW, Williams RV, Altmann K, Ghanayem NS, Margossian R, Chung WK, Border WL, Pearson GD, Stylianou MP, Mital S. Enalapril in infants with single ventricle: results of a multicenter randomized trial. Circulation 122: 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang X, Le QT, Giaccia AJ. MiR-210–micromanager of the hypoxia pathway. Trends Mol Med 16: 230–237, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ikeda S, Hamada M, Hiwada K. Cardiomyocyte apoptosis with enhanced expression of P53 and Bax in right ventricle after pulmonary arterial banding. Life Sci 65: 925–933, 1999 [DOI] [PubMed] [Google Scholar]
- 27. Ito T, Yagi S, Yamakuchi M. MicroRNA-34a regulation of endothelial senescence. Biochem Biophys Res Commun 398: 735–740, 2010 [DOI] [PubMed] [Google Scholar]
- 28. Jucker BM, Doe CP, Schnackenberg CG, Olzinski AR, Maniscalco K, Williams C, Hu TC, Lenhard SC, Costell M, Bernard R, Sarov-Blat L, Steplewski K, Willette RN. PPARdelta activation normalizes cardiac substrate metabolism and reduces right ventricular hypertrophy in congestive heart failure. J Cardiovasc Pharmacol 50: 25–34, 2007 [DOI] [PubMed] [Google Scholar]
- 29. Kaufman BD, Desai M, Reddy S, Osorio JC, Chen JM, Mosca RS, Ferrante AW, Mital S. Genomic profiling of left and right ventricular hypertrophy in congenital heart disease. J Card Fail 14: 760–767, 2008 [DOI] [PubMed] [Google Scholar]
- 30. Kida K, Nakajima M, Mohri T, Oda Y, Takagi S, Fukami T, Yokoi T. PPARalpha is regulated by miR-21 and miR-27b in human liver. Pharm Res 28: 2467–2476, 2011 [DOI] [PubMed] [Google Scholar]
- 31. Kriegel AJ, Liu Y, Fang Y, Ding X, Liang M. The miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol Genomics 44: 237–244, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li LM, Hou DX, Guo YL, Yang JW, Liu Y, Zhang CY, Zen K. Role of microRNA-214-targeting phosphatase and tensin homolog in advanced glycation end product-induced apoptosis delay in monocytes. J Immunol 186: 2552–2560, 2011 [DOI] [PubMed] [Google Scholar]
- 33. Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genom 8: 166, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, Wiggins JF, Bader AG, Fagin R, Brown D, Tang DG. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med 17: 211–215, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lopez L, Cohen MS, Anderson RH, Redington AN, Nykanen DG, Penny DJ, Deanfield JE, Eidem BW. Unnatural history of the right ventricle in patients with congenitally malformed hearts. Cardiol Young 20, Suppl 3: 107–112, 2010 [DOI] [PubMed] [Google Scholar]
- 36. Lu H, Buchan RJ, Cook SA. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc Res 86: 410–420, 2010 [DOI] [PubMed] [Google Scholar]
- 37. Lund O, Kristensen LH, Baandrup U, Hansen OK, Nielsen TT, Emmertsen K, Jensen FT, Flo C, Rasmussen BS, Pilegaard HK. Myocardial structure as a determinant of pre- and postoperative ventricular function and long-term prognosis after valve replacement for aortic stenosis. Eur Heart J 19: 1099–1108, 1998 [DOI] [PubMed] [Google Scholar]
- 38. Maes OC, Sarojini H, Wang E. Stepwise up-regulation of microRNA expression levels from replicating to reversible and irreversible growth arrest states in WI-38 human fibroblasts. J Cell Physiol 221: 109–119, 2009 [DOI] [PubMed] [Google Scholar]
- 39. Matkovich SJ, Wang W, Tu Y, Eschenbacher WH, Dorn LE, Condorelli G, Diwan A, Nerbonne JM, Dorn GW., 2nd MicroRNA-133a protects against myocardial fibrosis and modulates electrical repolarization without affecting hypertrophy in pressure-overloaded adult hearts. Circ Res 106: 166–175, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Narducci MG, Arcelli D, Picchio MC, Lazzeri C, Pagani E, Sampogna F, Scala E, Fadda P, Cristofoletti C, Facchiano A, Frontani M, Monopoli A, Ferracin M, Negrini M, Lombardo GA, Caprini E, Russo G. MicroRNA profiling reveals that miR-21, miR486 and miR-214 are upregulated and involved in cell survival in Sezary syndrome. Cell Death Dis 2: e151, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Okubo S, Wildner O, Shah MR, Chelliah JC, Hess ML, Kukreja RC. Gene transfer of heat-shock protein 70 reduces infarct size in vivo after ischemia/reperfusion in the rabbit heart. Circulation 103: 877–881, 2001 [DOI] [PubMed] [Google Scholar]
- 42. Raitoharju E, Lyytikainen LP, Levula M, Oksala N, Mennander A, Tarkka M, Klopp N, Illig T, Kahonen M, Karhunen PJ, Laaksonen R, Lehtimaki T. miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis 219: 211–217, 2011 [DOI] [PubMed] [Google Scholar]
- 43. Roy S, Khanna S, Hussain SR, Biswas S, Azad A, Rink C, Gnyawali S, Shilo S, Nuovo GJ, Sen CK. MicroRNA expression in response to murine myocardial infarction: miR-21 regulates fibroblast metalloprotease-2 via phosphatase and tensin homologue. Cardiovasc Res 82: 21–29, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Saito J, Toriumi S, Awano K, Ichijo H, Sasaki K, Kobayashi T, Tamura S. Regulation of apoptosis signal-regulating kinase 1 by protein phosphatase 2Cepsilon. Biochem J 405: 591–596, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res 100: 416–424, 2007 [DOI] [PubMed] [Google Scholar]
- 46. Shaddy RE, Boucek MM, Hsu DT, Boucek RJ, Canter CE, Mahony L, Ross RD, Pahl E, Blume ED, Dodd DA, Rosenthal DN, Burr J, LaSalle B, Holubkov R, Lukas MA, Tani LY. Carvedilol for children and adolescents with heart failure: a randomized controlled trial. JAMA 298: 1171–1179, 2007 [DOI] [PubMed] [Google Scholar]
- 47. Sheehy SP, Huang S, Parker KK. Time-warped comparison of gene expression in adaptive and maladaptive cardiac hypertrophy. Circ Cardiovasc Genet 2: 116–124, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Song XW, Li Q, Lin L, Wang XC, Li DF, Wang GK, Ren AJ, Wang YR, Qin YW, Yuan WJ, Jing Q. MicroRNAs are dynamically regulated in hypertrophic hearts, and miR-199a is essential for the maintenance of cell size in cardiomyocytes. J Cell Physiol 225: 437–443, 2010 [DOI] [PubMed] [Google Scholar]
- 49. Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett 582: 1564–1568, 2008 [DOI] [PubMed] [Google Scholar]
- 50. Takayama S, Reed JC, Homma S. Heat-shock proteins as regulators of apoptosis. Oncogene 22: 9041–9047, 2003 [DOI] [PubMed] [Google Scholar]
- 51. Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, Gross C, Engelhardt S, Ertl G, Bauersachs J. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation 116: 258–267, 2007 [DOI] [PubMed] [Google Scholar]
- 52. Tibiche C, Wang E. MicroRNA regulatory patterns on the human metabolic network. Open Syst Biol J 1: 1–8, 2008 [Google Scholar]
- 53. Urashima T, Zhao M, Wagner R, Fajardo G, Farahani S, Quertermous T, Bernstein D. Molecular and physiological characterization of RV remodeling in a murine model of pulmonary stenosis. Am J Physiol Heart Circ Physiol 295: H1351–H1368, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Uwabe K, Kitamura M, Hachida M, Endo M, Hashimoto A, Koyanagi H. Long-term outcome of left ventricular dysfunction after surgery for severe aortic stenosis. J Heart Valve Dis 4: 503–507, 1995 [PubMed] [Google Scholar]
- 55. Uwabe K, Kitamura M, Noji S, Aomi S, Hachida M, Endo M, Hashimoto A, Koyanagi H. [Surgical results of aortic stenosis with or without left ventricular dysfunction–postoperative change of left ventricular function]. Nippon Kyobu Geka Gakkai Zasshi 43: 1710–1715, 1995 [PubMed] [Google Scholar]
- 56. van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA 103: 18255–18260, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang N, Zhou Z, Liao X, Zhang T. Role of microRNAs in cardiac hypertrophy and heart failure. IUBMB Life 61: 566–571, 2009 [DOI] [PubMed] [Google Scholar]
- 58. Zhang X, Azhar G, Helms SA, Wei JY. Regulation of cardiac microRNAs by serum response factor. J Biomed Sci 18: 15, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang X, Wang X, Zhu H, Zhu C, Wang Y, Pu WT, Jegga AG, Fan GC. Synergistic effects of the GATA-4-mediated miR-144/451 cluster in protection against simulated ischemia/reperfusion-induced cardiomyocyte death. J Mol Cell Cardiol 49: 841–850, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhao M, Chow A, Powers J, Fajardo G, Bernstein D. Microarray analysis of gene expression after transverse aortic constriction in mice. Physiol Genomics 19: 93–105, 2004 [DOI] [PubMed] [Google Scholar]