Abstract
Common inbred strains of the laboratory rat can be divided into four different mitochondrial DNA haplotype groups represented by the SHR, BN, LEW, and F344 strains. In the current study, we investigated the metabolic and hemodynamic effects of the SHR vs. LEW mitochondrial genomes by comparing the SHR to a new SHR conplastic strain, SHR-mtLEW; these strains are genetically identical except for their mitochondrial genomes. Complete mitochondrial DNA (mtDNA) sequence analysis comparing the SHR and LEW strains revealed gene variants encoding amino acid substitutions limited to a single mitochondrial enzyme complex, NADH dehydrogenase (complex I), affecting subunits 2, 4, and 5. Two of the variants in the mt-Nd4 subunit gene are located close to variants known to be associated with exercise intolerance and diabetes mellitus in humans. No variants were found in tRNA or rRNA genes. These variants in mt-Nd2, mt-Nd4, and mt-Nd5 in the SHR-mtLEW conplastic strain were linked to reductions in oxidative and nonoxidative glucose metabolism in skeletal muscle. In addition, SHR-mtLEW conplastic rats showed increased serum nonesterified fatty acid levels and resistance to insulin stimulated incorporation of glucose into adipose tissue lipids. These results provide evidence that inherited variation in mitochondrial genes encoding respiratory chain complex I subunits, in the absence of variation in the nuclear genome and other confounding factors, can influence glucose and lipid metabolism when expressed on the nuclear genetic background of the SHR strain.
Keywords: mitochondria, insulin resistance, conplastic, spontaneously hypertensive rat
the inheritance of type 2 diabetes has been reported by some investigators to show bias toward the maternal lineage (1, 21). In addition, alterations in mitochondrial function have been observed in muscle of lean, insulin resistant offspring of subjects with Type 2 diabetes (15). These observations have provided indirect evidence suggesting that inherited variation in the mitochondrial genome may influence glucose metabolism and risk for Type 2 diabetes. Recently, in studies comparing two strains of spontaneously hypertensive rats (SHR) that differ only in their mitochondrial genomes, we found that naturally occurring variation in mitochondrial DNA (mtDNA) can be directly linked to inherited variation in known risk factors for Type 2 diabetes (16). In these studies, we compared the SHR progenitor strain to an SHR-mtBN conplastic strain harboring the mitochondrial genome of the BN strain. These two strains are genetically identical except for multiple sequence variants between their mitochondrial genomes. In addition, based on sequencing results from the mitochondrial genomes of many different strains of laboratory rats (12, 16, 20) we found that common laboratory rats can be divided into four different haplotype groups represented by the SHR, BN, LEW, and F344 strains.
In the current study, we report derivation and characterization of a new SHR-mtLEW conplastic strain in which we have transferred the LEW mitochondrial genome onto the genetic background of the SHR progenitor strain. Because the SHR and LEW mitochondrial genomes differ only with respect to three genes encoding subunits of NADH dehydrogenase, any metabolic differences between the SHR progenitor strain and the SHR-mtLEW conplastic strain can be directly traced to sequence variation in mtDNA for subunits of a single enzyme complex (NADH dehydrogenase). In these studies that control for variation in the nuclear genome and in other confounding factors, we have found that inherited variation in mitochondrial genes encoding subunits of NADH dehydrogenase can influence mitochondrial enzyme activity and insulin stimulated glucose metabolism in skeletal muscle and adipose tissue.
MATERIALS AND METHODS
Animals.
We selectively replaced the mitochondrial genome of a highly inbred strain of spontaneously hypertensive rats (SHR/OlaIpcv strain, hereafter referred to as SHR) with the mitochondrial genome of a highly inbred strain of Lewis (LEW/Ipcv) rats to create a novel SHR/OlaIpcv-mtLEW conplastic strain (hereafter referred to as SHR-mtLEW) harboring the LEW mitochondrial genome on the SHR nuclear genetic background. We derived the conplastic strain using the “supersonic” breeding strategy of Behringer (3) to accelerate the breeding process (13). In the current experiments, we studied male conplastic rats after 10–12 generations of backcrossing. According to Silver's formula, after 12 backcross generations the conplastic strain is homozygous at 99.95% of all loci. To test for identity of nuclear genomes between SHR and SHR-mtLEW conplastic strains, we genotyped them with polymorphic markers spaced at ∼10 cM intervals throughout the genomes as previously described (16). Of the markers tested, none was found to be heterozygous in the conplastic strain. Although no amount of marker genotyping can exclude the possibility of some residual heterozygosity with absolute certainty, there is a very high probability that the conplastic and progenitor strains used in this study are homozygous and identical at any given locus in the nuclear genome. All experiments were performed in agreement with the Animal Protection Law of the Czech Republic and were approved by the Ethics Committee of the Institute of Physiology, Academy of Sciences of the Czech Republic, Prague.
Metabolic phenotypes.
After weaning, we fed the conplastic and progenitor rats standard chow and at 10 wk of age, switched them to a semisynthetic diet (Hope Farms) containing 60% fructose for 2 wk prior to study. In previous studies in the SHR, we have found that administration of a high-fructose diet can increase susceptibility to metabolic disturbances in this strain. At the end of the study, rats in the ad libitum fed state were killed by decapitation and serum and tissues were collected for final biochemical analyses.
Oral glucose tolerance test.
Oral glucose tolerance tests (OGTT) were performed after 2 wk of fructose feeding using a glucose load of 300 mg/100 g body wt after overnight fasting. Blood was drawn from the tail without anesthesia before the glucose load (0 min time point) and at 30, 60, and 120 min thereafter.
Biochemical analyses.
Blood glucose levels were measured by the glucose oxidase assay (Pliva-Lachema, Brno, Czech Republic) using tail vein blood drawn into 5% trichloroacetic acid and promptly centrifuged. Serum triglyceride concentrations were measured by standard enzymatic methods (Pliva-Lachema). Nonesterified fatty acids (NEFA) levels were determined using an acyl-CoA oxidase-based colorimetric kit (Roche Diagnostics, Mannheim, Germany). Serum insulin concentrations were determined using the Mercodia Rat Insulin ELISA kit (Mercodia, Uppsala, Sweden).
Muscle triglyceride measurements.
For determination of triglycerides, soleus muscles were powdered under liquid N2 and extracted for 16 h in chloroform-methanol (2:1), after which 2% KH2PO4 was added, and the solution was centrifuged. The organic phase was removed and evaporated under N2. The resulting pellet was dissolved in isopropyl alcohol, and triglyceride content was determined by enzymatic assay (Pliva-Lachema).
Glucose utilization in skeletal muscles.
Glycogen synthesis was determined in isolated soleus muscle by measuring the incorporation of 14C-U glucose into glycogen as previously described (17). The muscles were attached to a stainless steel frame in situ at in vivo length by special clips and separated from other muscles and tendons and immediately incubated for 2 h in Krebs-Ringer bicarbonate buffer, pH 7.4, gaseous phase 95% O2 and 5% CO2 that contained 5.5 mM unlabeled glucose, 0.5 μCi/ml of 14C-U glucose (UVVR, Prague, Czech Republic), and 3 mg/ml bovine serum albumin (Armour, Fraction V) with or without 250 μU/ml insulin. After 2 h incubation, glycogen was extracted and glucose incorporation into glycogen determined.
Basal and insulin-stimulated glucose oxidation was determined in isolated gastrocnemius muscle by measuring the incorporation of 14C-U glucose into CO2. The gastrocnemius muscles were attached to a stainless steel frame in situ at in vivo length by special clips and separated from other muscles and tendons and immediately incubated for 2 h in Krebs-Ringer bicarbonate buffer, pH 7.4 that contained 5.5 mM unlabeled glucose, 0.5 μCi/ml of 14C-U glucose, and 3 mg/ml BSA (Armour, Fraction V) with or without 250 μU/ml insulin. After 2 h incubation, 0.3 ml of 1 M hyamine hydroxide was injected into central compartment of the incubation vessel and 0.5 ml of 1 M H2SO4 into the main compartment to liberate CO2. The vessels were incubated for another 30 min, and the hyamine hydroxide was then quantitatively transferred into the scintillation vial containing 10 ml of toluene-based scintillation fluid for counting of radioactivity.
Glucose utilization in isolated epididymal adipose tissue.
Distal parts of epididymal adipose tissue were rapidly dissected and incubated for 2 h in Krebs-Ringer bicarbonate buffer with 5 mmol/l glucose, 0.1 μCi 14C-U-glucose/ml and 2% bovine serum albumin, gaseous phase 95% O2, and 5% CO2 in the presence (250 μU/ml) or absence of insulin in incubation media. All incubations were performed at 37°C in sealed vials in a shaking water bath. Estimation of 14C-glucose incorporation into neutral lipids was performed as described previously (17). Briefly, adipose tissue was removed from incubation medium, rinsed in saline, and immediately put into chloroform. The pieces of tissue were dissolved using a Teflon pestle homogenizer, methanol was added (chloroform-methanol 2:1), and lipids were extracted at 4°C overnight. The remaining tissue was removed, KH2PO4 was added, and a clear extract was taken for further analysis. An aliquot was evaporated and reconstituted in scintillation liquid, and the radioactivity measured by scintillation counting. Incremental glucose utilization was calculated as the difference between the insulin stimulated and basal incorporation of glucose into neutral lipids.
Blood pressure measurements.
Arterial blood pressures were measured continuously by radiotelemetry in paired experiments between conscious, unrestrained male rats. All rats were allowed to recover for at least 7 days after surgical implantation of radiotelemetry transducers before the start of blood pressure recordings. Pulsatile pressures were recorded in 5-s bursts every 10 min throughout the day and night, and 24 h averages for systolic arterial blood pressure were calculated for each rat for 1 wk periods. After measuring blood pressure for 2 wk, all rats were given 1% NaCl for drinking instead of tap water for an additional week of blood pressure measurements to test for effects of the LEW vs. SHR mtDNA on blood pressure salt-sensitivity. The results from each rat in the same group were then averaged to obtain the group means.
Activities and protein content of mitochondrial oxidative phosphorylation enzymes, and adenine nucleotide levels.
Liver and heart ventricle homogenates (5%, wt/vol) were prepared in STE medium [0.25 M sucrose, 10 mM Tris·HCl, 1 mM EDTA, pH 7.4, 1 μg/ml PIC (protease inhibitor mixture Sigma P8340)], and gastrocnemius muscle homogenates were prepared in KCl medium (150 mM KCl, 2 mM EDTA, 50 mM Tris·HCl, pH 7.4, 1 μg/ml PIC) using a motor-driven glass-Teflon homogenizer, filtered through 200 μm nylon mesh and frozen at −80°C.
We used spectrophotometric methods (18, 25) to measure enzyme activities of citrate synthase (CS), cytochrome c oxidase (COX, complex IV), succinate DCPIP reductase (SDR, complex II), succinate cytochrome c reductase (SCCR, complexes II + III), rotenone-sensitive NADH coenzyme Q reductase (NQR, complex I), and rotenone-sensitive NADH cytochrome c reductase (NCCR, complexes I + III). Enzyme activity results are expressed per mg of homogenate protein.
We used SDS-PAGE and Western blotting of tissue homogenates to measure levels of a panel of mitochondrial respiratory chain proteins using specific antibodies and methods similar to those previously described (10). The levels of respiratory chain complexes I–V were determined using primary mouse monoclonal antibodies (MitoSciences, Eugene, OR) to complex I (NADH dehydrogenase) NDUFA9 subunit (MS111) and NDUFS3 subunit (MS112); complex II (succinate dehydrogenase) SDH70 subunit (MS204); complex III (bc1 complex) core 2 subunit (MS304); complex IV (cytochrome c oxidase) subunit Cox4 (MS407); and complex V (F1Fo ATP synthase) α-subunit (MS507), respectively. We quantified the protein levels by detection of infrared fluorescence using anti-mouse Alexa Fluor 680 or IRDye 800-labeled secondary antibodies and an Odyssey Infrared Imaging System (LI-COR Biotechnology, Lincoln, NE). Results are expressed as relative fluorescence units determined per mg homogenate protein.
Liquid nitrogen snap-frozen samples of skeletal muscle, liver, and heart were also processed for measurement of the content of adenine nucleotides by high-pressure liquid chromatography (7).
Tissue content of mitochondrial DNA.
We used real-time PCR to measure DNA copy number of mitochondrial genes mt-Co1, mt-Nd4, mt-Atp8, and mt-Cyb relative to the amount of the nuclear gene cytochrome c oxidase, subunit IV, isoform 1 (Cox4i1) as an internal control. For real time PCR, total DNA was isolated using standard methods and analyzed by real-time PCR testing using QuantiTect SYBR Green reagents (Qiagen, Valencia, CA) on an Opticon continuous fluorescence detector (MJ Research, Waltham, MA) with an annealing temperature of 58°C. Mitochondrial DNA levels were normalized relative to the internal genomic DNA control with relative amounts mtDNA to genomic DNA determined in triplicate using the preferred method of Muller et al. (14). The primers used to amplify mitochondrial genes were: mt-Co1 upstream 5′-gga gca gta ttc gcc atc at, mt-Co1 downstream 5′-cgg ccg taa gtg aga tga at; mt-Cyb upstream 5′-cat cag tca ccc aca tctg c, mt-Cyb downstream 5′-tgg atg gaa tgg gat ttt gt; mt-Nd4 upstream 5′-ctc aca aca cac ccc cta cc, mt-Nd4 downstream 5′-tcc cat aac ccc cta gct tt; mt-Atp8 upstream 5′-tgc cac aac tag aca cat cca, mt-Atp8 downstream 5′-tgt ggg ggt aat gaa aga gg. The primers used to amplify the nuclear gene Cox4i1 were upstream 5′-act acc cct tgc ctg atg tg and downstream 5′-aca gat ctg cgc tca cac ac.
LEW mtDNA sequence analysis.
We amplified the entire rat mitochondrial genome from DNA extracted from liver tissue of a 12 wk old LEW/Ipcv male using the PCR and 34 overlapping primer pairs. Sequencing was performed on an Applied Biosystems 3730xl DNA Analyzer using the BigDye Terminator v3.1 Cycle sequencing kit (Applied Biosystems, Foster City, CA). We then searched for variants by comparing the LEW sequence results to the published mitochondrial genome sequence (accession #AC000022) of the BN (BN/SsNHsdMCW) and SHR/Ola strains. We confirmed all variants causing amino acid changes by resequencing the specific regions of interest in at least three LEW/Ipcv rats.
Statistical analysis.
We used 10 SHR and 10 SHR-mtLEW rats for determination of metabolic traits. Blood pressure was determined in eight SHR and nine SHR-mtLEW rats, mitochondrial traits were measured in nine SHR and nine SHR-mtLEW rats. We used Student's t-test to analyze for differences between group means with statistical significance defined as P < 0.05. All results are expressed as means ± SE.
RESULTS
LEW mitochondrial DNA sequence analysis.
In contrast to previous comparisons showing multiple variants in the SHR vs. BN mitochondrial genomes (17), sequence analysis revealed a relatively small number of differences in the SHR vs. LEW mitochondrial genomes. Altogether, five amino acid substitutions were found in the mt-Nd2, mt-Nd4, and mt-Nd5 genes between the SHR and LEW strains. No variants were found in any other structural genes or in mitochondrial tRNA or rRNA genes between the SHR and LEW strains. The mtDNA sequence of the LEW/Ipcv strain was identical to previously published mtDNA sequence of the LEW/NCrlBR strain (12). Two mutations in the mt-Nd4 gene that resulted in SHR vs. LEW amino acid substitutions of Thr356Ala and Leu419Pro are close to known pathological mutations associated with exercise intolerance and diabetes mellitus in humans (Table 1). The alignment of these two amino acid substitutions in the SHR vs. LEW strains compared with other species is shown in Fig. 1. As can be seen, the Thr356 variant in the SHR appears unique since all other species tested, including human, bovine, dog and fugu fish have alanine in this position.
Table 1.
Sequence differences between LEW and SHR mitochondrial DNA
| AA Character Hydrophobic/-philic R-chain | ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene | AA No. | Rat LEW | Rat SHR | Human | Relevant Human mtDNA Site | Nearby Human Pathology Mutation, AA No. | LEW | SHR |
| mt-Nd2 | 18 | Val | Ala | Leu | hu 4522 | LHON | hydrophobic | tiny |
| 4640C>A, Ile57Met | nonpolar, aliphatic | nonpolar | ||||||
| 356 | Ala | Thr | Ala | hu 11825 | exercise intolerance/oncocytoma | tiny | hydrophilic | |
| 11832G>A, Trp358Ter | nonpolar | polar, OH | ||||||
| 401 | Val | Ile | Leu | hu 11960 | OAT | hydrophobic | hydrophobic | |
| 11994C>T, Thr412Ile | nonpolar, aliphatic | nonpolar, aliphatic | ||||||
| diabetes mellitus | hydrophobic | hydrophobic | ||||||
| mt-Nd4 | 419 | Pro | Leu | Leu | hu 12014 | 12026A>G, Ile423Val | nonpolar, cyclic | nonpolar, aliphatic |
| mitochondrial myopathy and renal failure | tiny | hydrophilic | ||||||
| mt-Nd5 | 37 | Ala | Thr | Lys | hu 12445 | 12425A-del, N30-frameshift | nonpolar | polar, OH |
AA, amino acid; LHON, Leber hereditary optic neuropathy; OAT, oligoasthenoteratozoospermia.
Fig. 1.
Alignment of mt-Nd4 variants across different species.
Mitochondrial oxidative phosphorylation enzyme activities.
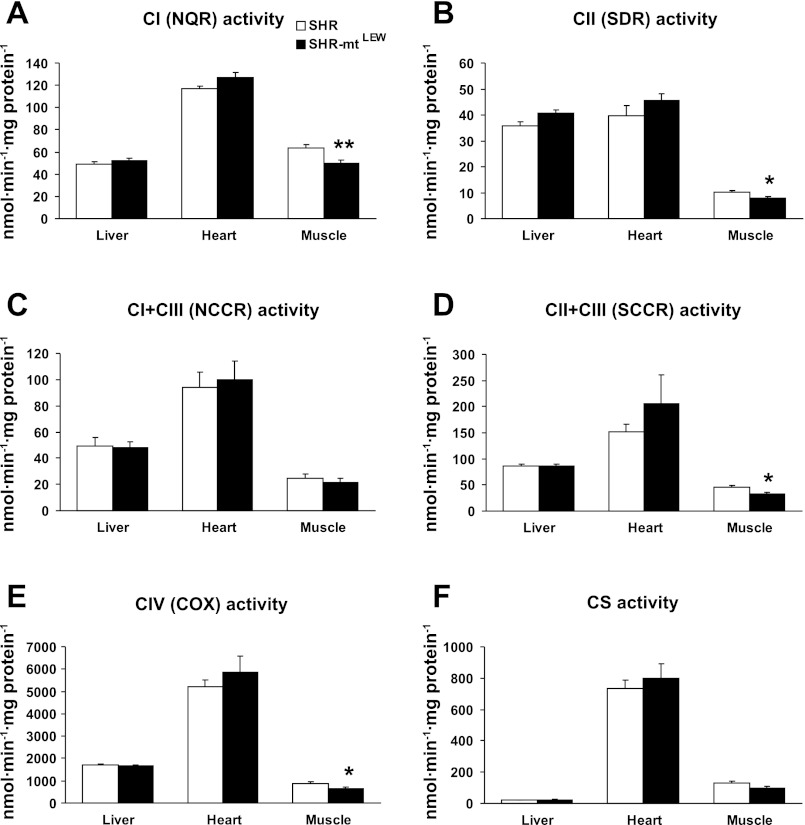
Analysis of enzyme activities of mitochondrial respiratory chain complexes (Fig. 2) was performed in tissue homogenates and included NQR, NCCR, SDR, SCCR, COX, and CS. In muscle homogenates, specific activities of all enzymes showed lower values in SHR-mtLEW conplastic strain (decreases of 13–27%) that resulted in 1.15- to 1.37-fold elevation of SHR/SHR-mtLEW ratios. The strain differences were statistically significant in the case of CI (50 ± 3 vs. 63 ± 3 nmol·min−1·mg protein−1, P < 0.01), CII (7.9 ± 0.6 vs. 10.2 ± 0.6 nmol·min−1·mg protein−1, P < 0.05), SCCR (33 ± 3 vs. 45 ± 3 nmol·min−1·mg protein−1, P < 0.05) and COX (643 ± 62 vs. 874 ± 76 nmol·min−1·mg protein−1, P < 0.05). No significant strain differences in enzyme activities were observed in liver and heart tissues.
Fig. 2.
Mitochondrial respiratory chain enzyme activities in tissue homogenates. Values are means ± SE from 9 rats in each group. *P < 0.05 and **P < 0.01. See text for abbreviations.
Mitochondrial oxidative phosphorylation enzyme content and mtDNA copy number.
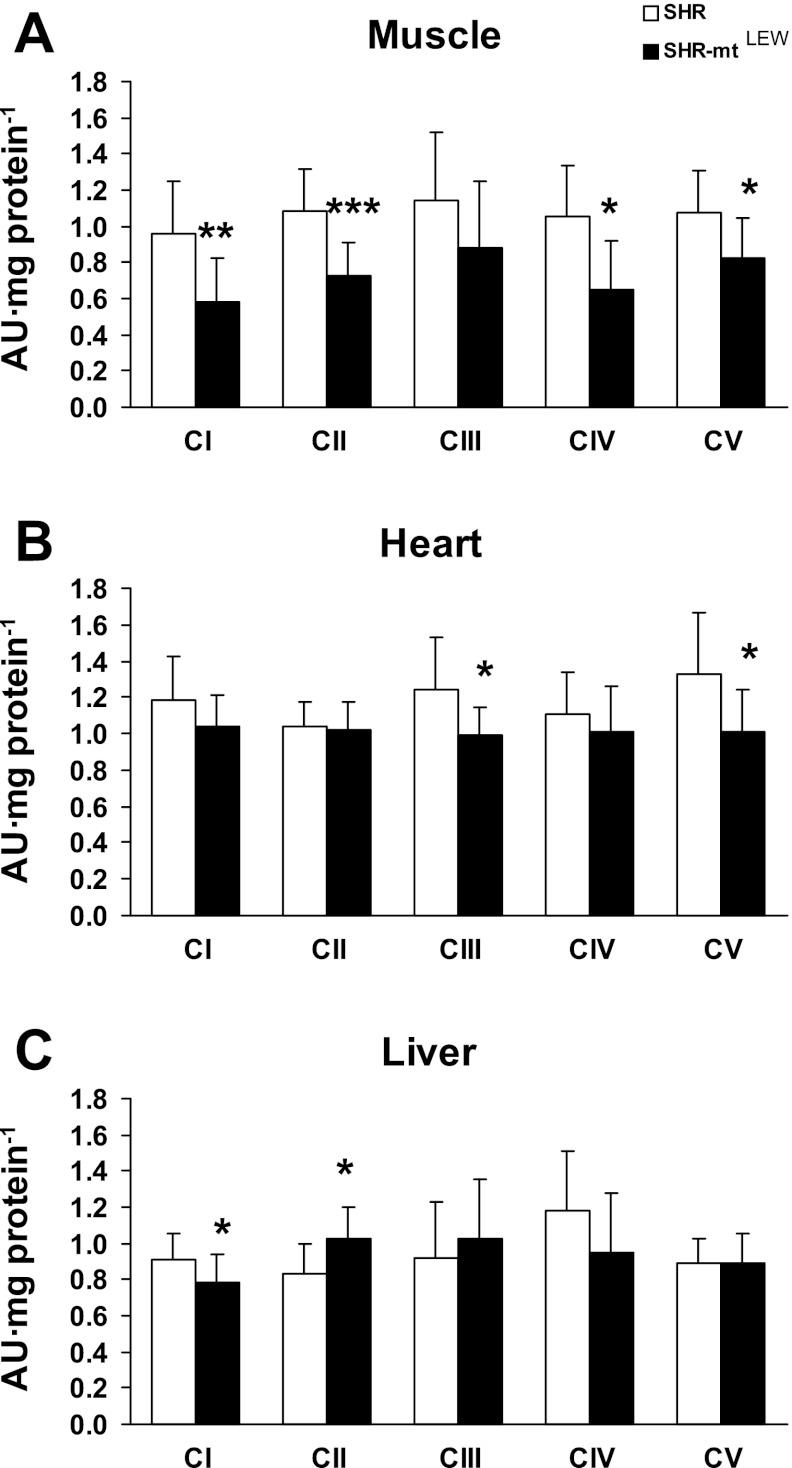
Tissue content of mitochondrial respiratory chain complexes CI (NADH dehydrogenase), CII (succinate dehydrogenase), CIII (bc1 complex), CIV (cytochrome c oxidase), and CV (ATP synthase) was analyzed by Western blot using specific monoclonal antibodies (Fig. 3). In muscle of the SHR-mtLEW conplastic strain, the specific content of all protein complexes was lower than in the SHR progenitor strain (decreases of 23–39%), and this decrease was significant in the case of CI (0.580 ± 0.246 vs. 0.954 ± 0.298 AU/mg protein, P < 0.01), CII (0.729 ± 0.183 vs. 1.081 ± 0.237 AU/mg protein, P < 0.001), CIV (0.645 ± 0.277 vs. 1.058 ± 0.273 AU/mg protein, P < 0.05), and CV (0.820 ± 0.229 vs. 1.073 ± 0.231 AU/mg protein, P < 0.05). Similar, but less pronounced decreases in SHR-mtLEW were found in the heart, with the decrease being significant in case of CIII (0.988 ± 0.160 vs. 1.237 ± 0.294 AU/mg protein, P < 0.05) and CV (1.011 ± 0.233 vs. 1.329 ± 0.341 AU/mg protein, P < 0.05). The liver of SHR-mtLEW conplastic rats showed a significant decrease in the specific content of CI (0.784 ± 0.153 vs. 0.910 ± 0.144 AU/mg protein, P < 0.05), while CII was increased (1.029 ± 0.174 vs. 0.835 ± 0.165 AU/mg protein, P < 0.05). Significant decrease of CI in muscle (0.729 ± 0.183 vs. 1.08 ± 0.237 AU/mg protein, P < 0.001) and liver (0.424 ± 0.171 vs. 1.08 ± 0.256 AU/mg protein, P < 0.001) of SHR-mtLEW conplastic rats was confirmed using antibody to another subunit NDUSF3. We found no reductions in mtDNA copy number relative to genomic DNA content between the SHR-mtLEW conplastic strain and the SHR progenitor strain (data not shown).
Fig. 3.
Mitochondrial respiratory chain enzyme levels in tissue homogenates. Values are means ± SE from 9 rats in each group. *P < 0.05; **P < 0.01; ***P < 0.001.
Adenine nucleotide levels.
Tissue content of ATP, ADP and AMP, determined by HPLC analysis in heart, liver, and muscle showed similar levels in the SHR vs. SHR-mtLEW conplastic strains as well as for the ATP/ADP ratio and the energy charge parameter (Table 2).
Table 2.
Adenine nucleotide levels
| Trait | SHR | SHR-mtLEW |
|---|---|---|
| Muscle | ||
| ATP | 1,100.9 ± 193.86 | 1,281.2 ± 258.53 |
| ADP | 369.2 ± 59.92 | 422.9 ± 90.04 |
| AMP | 22.03 ± 23.69 | 33.39 ± 36.01 |
| ATP/ADP | 3.01 ± 0.46 | 3.14 ± 0.84 |
| Energy charge | 0.86 ± 0.03 | 0.86 ± 0.05 |
| Heart | ||
| ATP | 615.5 ± 264.11 | 752.5 ± 138.59 |
| ADP | 641.5 ± 68.76 | 674.2 ± 104.99 |
| AMP | 563.6 ± 288.0 | 434.0 ± 153.5 |
| ATP/ADP | 0.96 ± 0.41 | 1.13 ± 0.23 |
| Energy charge | 0.52 ± 0.15 | 0.59 ± 0.07 |
| Liver | ||
| ATP | 501.9 ± 178.06 | 573.1 ± 121.26 |
| ADP | 488.6 ± 134.4 | 539.7 ± 107.69 |
| AMP | 359.5 ± 112.4 | 396.5 ± 141.0 |
| ATP/ADP | 1.02 ± 0.1783 | 1.08 ± 0.24 |
| Energy charge | 0.55 ± 0.06 | 0.56 ± 0.08 |
Data are means ± SD. Levels of ATP, ADP, and ATP are expressed in nmol/g per 100 mg wet wt.
Analysis of the mitochondrial oxidative phosphorylation (OXPHOS) system showed tissue-specific effects of the mtDNASHR replacement with mtDNALEW, suggesting a downregulation of OXPHOS enzyme content and activities in muscle and to a lesser extent also in livers of the conplastic rats. The observed changes in the OXPHOS system were not associated with detectable differences in the tissue content of adenine nucleotides AMP, ADP, and ATP or in the ATP/ADP ratio.
Metabolic phenotypes in SHR-mtLEW conplastic rats.
We found that glucose, insulin, and triglyceride levels were similar between the SHR-mtLEW conplastic and the SHR progenitor strain, and we did not detect any strain differences in glucose and insulin levels after oral glucose loading (Table 3). However, conplastic rats showed significantly increased nonfasting serum NEFA levels as well as higher NEFA levels during the OGTT (Table 3), suggesting reduced antilipolytic effects of insulin. The mean body weight of the conplastic strain was significantly greater than that of the SHR strain (369 ± 10 vs. 324 ± 5 g, P = 0.00004); however, there were no significant differences in relative weight of epididymal and subcutaneous fat or in relative liver weight (data not shown), which suggests that the greater body weight in conplastic rats was not due to increased adiposity.
Table 3.
Parameters of lipid and glucose metabolism
| Trait | SHR | SHR-mtLEW |
|---|---|---|
| Serum glucose, mmol/l | 3.8 ± 0.1 | 3.9 ± 0.1 |
| AUC for glucose during OGTT, mmol·l−1·2 h−1 | 550 ± 19 | 545 ± 9 |
| Serum NEFA at OGTT0 min, mmol/l | 1.93 ± 0.06 | 2.02 ± 0.08 |
| Serum NEFA at OGTT60 min, mmol/l | 0.86 ± 0.08 | 1.10 ± 0.07* |
| Serum NEFA, mmol/l | 0.48 ± 0.04 | 0.61 ± 0.03* |
| Serum insulin, mmol/l | 0.76 ± 0.11 | 0.71 ± 0.04 |
| Serum triglycerides, mmol/l | 2.1 ± 0.2 | 1.9 ± 0.2 |
| Muscle triglycerides, μmol/g | 5.0 ± 0.7 | 5.0 ± 0.6 |
Values are means ± SE, n = 10 per strain,
P < 0.05. AUC, area under the curve; NEFA, nonesterified fatty acids; OGTT, oral glucose tolerance test.
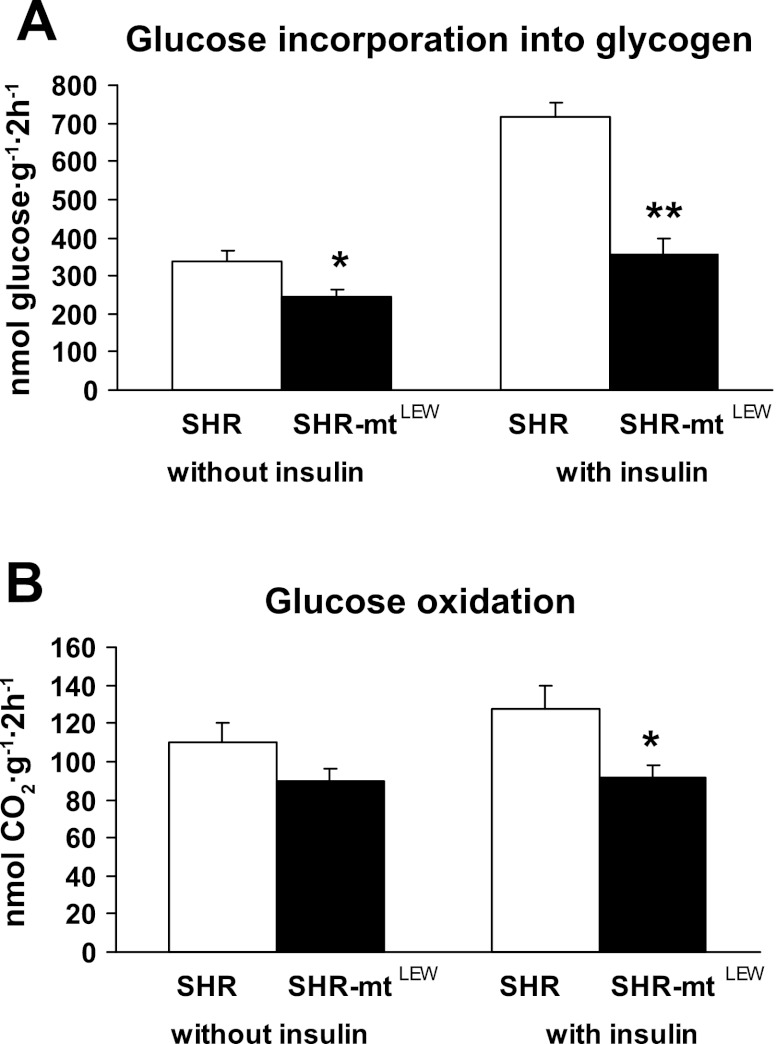
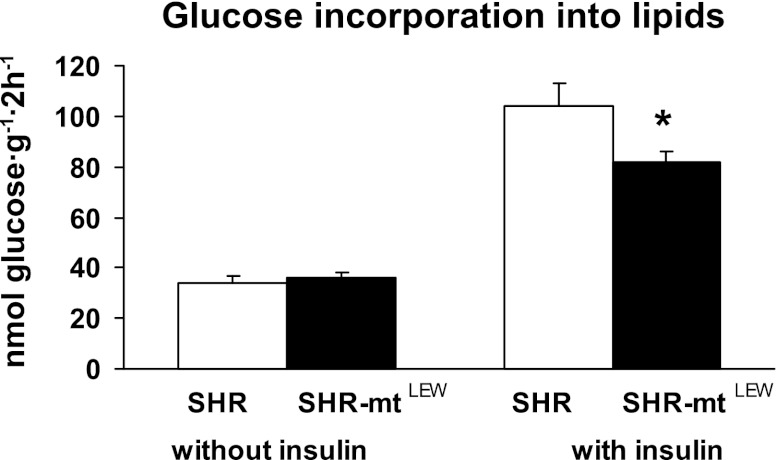
Figure 4A shows basal and insulin-stimulated incorporation of radioactively labeled glucose into skeletal muscle glycogen. In the SHR-mtLEW conplastic strain, glucose incorporation into skeletal muscle glycogen was significantly lower than in the SHR progenitor strain both before and after stimulation with insulin. The insulin-induced increase in nonoxidative glucose metabolism was significantly lower in the conplastic rats than in the SHR controls (110 ± 29 vs. 383 ± 49 nmol·g−1·2 h−1, P < 0.0005). The SHR-mtLEW conplastic rats also exhibited significantly lower glucose oxidation in skeletal muscle both in the absence and presence of insulin in the incubation media (Fig. 4B). In addition, the conplastic rats showed adipose tissue insulin resistance as judged by reduced insulin stimulated incorporation of glucose into adipose tissue lipids (Fig. 5). The insulin-induced increase in glucose incorporation into adipose tissue lipids was significantly lower in conplastic rats compared with SHR controls (46 ± 4 vs. 70 ± 6 nmol·g−1·2 h−1, P = 0.005).
Fig. 4.
Effect of insulin (250 μU/ml) on nonoxidative glucose metabolism (glucose incorporation into glycogen) (A) and glucose oxidation (B) in gastrocnemius muscles isolated from SHR and SHR-mtLEW conplastic rats. Values are means ± SE from 10 rats in each group. *P < 0.05; **P < 0.01.
Fig. 5.
Basal and insulin stimulated incorporation of glucose into lipids in epididymal adipose tissue. Values are means ± SE from 10 rats in each group. *P < 0.05.
Blood pressures and heart rates.
There were no significant differences in blood pressures and heart rates measured by radiotelemetry between the SHR and SHR-mtLEW conplastic rats. The average mean arterial pressure in conplastic rats compared with the SHR over a period of 8 days was 146 ± 1 vs. 146 ± 1 mmHg. The average heart rates were 318 ± 3 vs. 322 ± 2.
DISCUSSION
The results of the current study provide evidence that inherited variation in mitochondrial genes encoding respiratory chain complex I subunits of the LEW origin predispose the SHR conplastic strain to reduced OXPHOS protein content and enzyme activities and impaired sensitivity of muscle and adipose tissue to insulin action. Recently, we reported that SHR-mtBN conplastic rats exhibited insulin resistance similar to that observed in SHR-mtLEW conplastic rats compared with SHR controls (16). However, owing to the fact that multiple sequence differences exist between the mitochondrial genomes of the SHR and BN strains, it has been difficult to determine which particular strain variants in mtDNA might be contributing to the metabolic differences observed between the SHR progenitor and the SHR-mtBN conplastic strain. Sequence analysis of the SHR and LEW mtDNA revealed gene variants encoding amino acid substitutions only in subunits 2, 4, and 5 of NADH dehydrogenase (complex I).
Complex I (NADH:ubiquinone oxidoreductase; EC 1.6.5.3) is the largest of the five mitochondrial respiratory chain complexes (for review see Refs. 11, 24). It catalyzes oxidation of NADH by transfer of electrons to the lipid-soluble ubiquinone, and the transfer of two electrons is coupled to the translocation of four protons from the negative matrix side to the positive side of the intermembrane space. The complex I structure has an “L”-shape and comprises two arms that are perpendicular to each other (8): a hydrophobic arm embedded in the lipid membrane and a hydrophilic, peripheral arm protruding into the matrix. Intact complex I can be resolved into four subcomplexes: Iα, Iβ, Iγ, and Iλ. Mammalian complex I is ∼980 kDa in size and consists of at least 45 different subunits of which seven (ND1, ND2, ND3, ND4, ND4L, ND5, and ND6) are encoded by mtDNA (5). ND1, ND2, ND3, and ND4L reside in subcomplex Iγ; ND4 and ND5 in subcomplex Iβ. Subunits ND1 and ND2 are grouped together, as are subunits ND4 and ND5. ND1 has been implicated in the formation of the ubiquinone binding pocket and together with proton translocating subunit ND2 may be located near the thin stalk, connecting the peripheral and membrane arms (9). ND4 and ND5, the other suspected proton translocating subunits, are believed to be located at the distal part of the membrane domain (19).
ND4 and ND6 subunits have been shown to be essential for the assembly of other ND subunits into the complex (22). Loss of the ND4 subunit will prevent the formation of the complex I membrane arm (4). Subunit ND2 is also needed for the assembly and/or stability of complex I (2). In contrast, subunit ND5 is not essential for assembly of the other ND subunits into the complex, and loss of ND5 leads only to a lower efficiency of assembly or to instability of the membrane arm. ND5 is likely to be located at the periphery of the membrane arm, and therefore may be the last of the ND subunits to be assembled into the complex (4). Nevertheless, since synthesis of ND5 is rate limiting, ND5 is essential for complex I activity (6).
Isolated complex I deficiency is the most common cause of OXPHOS dysfunction (23), and presentation of the disease due to mtDNA mutations often occurs in late childhood or adolescence. mtDNA mutations have been identified in each of the seven mtDNA genes encoding complex I subunits, expressing various clinical symptoms, either restricted to a single organ or tissue, or presenting in a multisystemic manner.
Of the mtDNA genes of complex I exhibiting sequence differences between SHR and LEW haplotypes including ND2, ND4, and ND5, over a dozen different pathogenic mutations have been reported in each of the corresponding human forms of ND2, ND4, and ND5 that result in various mitochondrial disorders (http://www.mitomap.org). Two variants in the protein encoded by the mt-Nd4 subunit gene observed in these rat studies are located close to variants known to be associated with exercise intolerance and diabetes mellitus in humans, and both mutations are quite rare compared with other species (http://www.mitomap.org). For example, the Thr356Ala substitution in the SHR is located in a position close to the Trp358Ter substitution suggested to affect exercise tolerance in humans. The possible functional significance of the Thr356 variant in the SHR is supported by the observation that other species ranging from fugu fish to humans appear to carry only alanine at this position. Mitochondrial polymorphisms are generally regarded as deleterious in nature; however, our study suggests that some may provide a metabolic advantage as might be the case of this unusual Thr356Ala substitution in the SHR.
As indicated by the differences observed between SHR and SHR-mtLEW rats in the amount and activity of mitochondrial respiratory chain complexes, the complex I content and activity are lowered in muscle, while in liver and heart the changes are less pronounced or absent, indicating tissue-specific manifestation of the SHR/LEW mtDNA polymorphisms. While complex I changes appear to be a direct consequence of mtDNA polymorphisms, the changes in other OXPHOS complexes, again most pronounced in muscle, must be of secondary origin due to mitochondria-nuclear cross talk, possibly triggered by metabolic disturbances due to changes in complex I. We have not found any significant changes in tissue adenine nucleotides, their ratio, or energy charge parameter, but the approach used may be not sensitive enough to uncover subtle changes or the changes in different cellular compartments. The same holds for possible changes in mitochondrial reactive oxygen species (ROS) production, which is difficult to measure in situ, and the activities of antioxidative defense system represent only an indirect parameter of the intensity of ROS generation. Further studies will be required to determine the exact mechanism whereby the observed variants in NADH dehydrogenase affect systemic metabolic phenotypes. Results of the current study suggest that the amino acid substitutions we observed in complex I subunits might affect metabolic phenotypes by influencing overall respiratory chain function.
In summary, we have found that in the fructose-fed SHR model of the hypertension metabolic syndrome, inherited variation in mitochondrial genes encoding respiratory chain complex I subunits can influence mitochondrial function, oxidative and nonoxidative glucose metabolism in skeletal muscle, insulin-stimulated incorporation of glucose into adipose tissue lipids, and circulating levels of fatty acids. These results should serve to motivate future studies of the molecular and cellular mechanisms whereby the variants observed in NADH dehydrogenase subunits influence multiple complex metabolic traits relevant to diabetes and the metabolic syndrome.
GRANTS
This work was supported by National Heart, Lung, and Blood Institute Grants HL-35018, HL-56028, and HL-63709 to T. W. Kurtz; the European Community's Seventh Framework Program (FP7/2007-2013) under grant agreement HEALTH-F4-2010-241504 (EURATRANS); Grant ME10019 to P. Mlejnek, Grants OC08017 and 1M6837805002 to J. Houštěk and LH11049 to V. Zídek from the Ministry of Education of the Czech Republic; Grants MZO00023001, NR9359-3, and NR9387-3 from the Ministry of Health of the Czech Republic to L. Kazdová and M. Pravenec; Grants 301/08/0166 and P303/10/0505 to M. Pravenec and V. Zídek from the Grant Agency of the Czech Republic; and research project AV0Z 50110509.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: J.H. and M.P. conception and design of research; J.H., Z.D., L.K., and M.P. analyzed data; J.H. and M.P. interpreted results of experiments; J.H., L.K., T.W.K., and M.P. edited and revised manuscript; J.H., K.H., M.V., Z.D., V.L., V.Z., P.M., M.S., J.S., I.M., L.K., O.O., T.W.K., and M.P. approved final version of manuscript; K.H., M.V., Z.D., V.L., V.Z., P.M., M.S., J.S., I.M., L.K., and O.O. performed experiments; K.H. and M.P. prepared figures; M.P. drafted manuscript.
REFERENCES
- 1. Alcolado JC, Laji K, Gill-Randall R. Maternal transmission of diabetes. Diabet Med 19: 89–98, 2002 [DOI] [PubMed] [Google Scholar]
- 2. Antonicka H, Ogilvie I, Taivassalo T, Anitori RP, Haller RG, Vissing J, Kennaway NG, Shoubridge EA. Identification and characterization of a common set of complex I assembly intermediates in mitochondria from patients with complex I deficiency. J Biol Chem 278: 43081–43088, 2003 [DOI] [PubMed] [Google Scholar]
- 3. Behringer R. Supersonic congenics? Nat Genet 18: 108, 1998 [DOI] [PubMed] [Google Scholar]
- 4. Bourges I, Ramus C, Mousson de Camaret B, Beugnot R, Remacle C, Cardol P, Hofhaus G, Issartel JP. Structural organization of mitochondrial human complex I: role of the ND4 and ND5 mitochondria-encoded subunits and interaction with prohibitin. Biochem J 383: 491–499, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carroll J, Fearnley IM, Skehel JM, Shannon RJ, Hirst J, Walker JE. Bovine complex I is a complex of 45 different subunits. J Biol Chem 281: 32724–32727, 2006 [DOI] [PubMed] [Google Scholar]
- 6. Chomyn A. Mitochondrial genetic control of assembly and function of complex I in mammalian cells. J Bioenerg Biomembr 33: 251–257, 2001 [DOI] [PubMed] [Google Scholar]
- 7. Flachs P, Novotny J, Baumruk F, Bardova K, Bourova L, Miksik I, Sponarova J, Svoboda P, Kopecky J. Impaired noradrenaline-induced lipolysis in white fat of aP2-Ucp1 transgenic mice is associated with changes in G-protein levels. Biochem J 364: 369–376, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Friedrich T, Bottcher B. The gross structure of the respiratory complex I: a Lego System. Biochim Biophys Acta 1608: 1–9, 2004 [DOI] [PubMed] [Google Scholar]
- 9. Grigorieff N. Structure of the respiratory NADH:ubiquinone oxidoreductase (complex I). Curr Opin Struct Biol 9: 476–483, 1999 [DOI] [PubMed] [Google Scholar]
- 10. Jesina P, Tesarova M, Fornuskova D, Vojtiskova A, Pecina P, Kaplanova V, Hansikova H, Zeman J, Houstek J. Diminished synthesis of subunit a (ATP6) and altered function of ATP synthase and cytochrome c oxidase due to the mtDNA 2 bp microdeletion of TA at positions 9205 and 9206. Biochem J 383: 561–571, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koopman WJ, Nijtmans LG, Dieteren CE, Roestenberg P, Valsecchi F, Smeitink JA, Willems PH. Mammalian mitochondrial complex I: biogenesis, regulation, and reactive oxygen species generation. Antioxid Redox Signal 12: 1431–1470, 2010 [DOI] [PubMed] [Google Scholar]
- 12. Kumarasamy S, Gopalakrishnan K, Shafton A, Nixon J, Thangavel J, Farms P, Joe B. Mitochondrial polymorphisms in rat genetic models of hypertension. Mamm Genome 21: 299–306, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Landa V, Zidek V, Pravenec M. Generation of rat “supersonic” congenic/conplastic strains using superovulation and embryo transfer. Methods Mol Biol 597: 267–275, 2010 [DOI] [PubMed] [Google Scholar]
- 14. Muller PY, Janovjak H, Miserez AR, Dobbie Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 32: 1372–1374, 1376, 1378–1379, 2002 [PubMed] [Google Scholar]
- 15. Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med 350: 664–671, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pravenec M, Hyakukoku M, Houstek J, Zidek V, Landa V, Mlejnek P, Miksik I, Dudova-Mothejzikova K, Pecina P, Vrbacky M, Drahota Z, Vojtiskova A, Mracek T, Kazdova L, Oliyarnyk O, Wang J, Ho C, Qi N, Sugimoto K, Kurtz T. Direct linkage of mitochondrial genome variation to risk factors for type 2 diabetes in conplastic strains. Genome Res 17: 1319–1326, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qi N, Kazdova L, Zidek V, Landa V, Kren V, Pershadsingh HA, Lezin ES, Abumrad NA, Pravenec M, Kurtz TW. Pharmacogenetic evidence that cd36 is a key determinant of the metabolic effects of pioglitazone. J Biol Chem 277: 48501–48507, 2002 [DOI] [PubMed] [Google Scholar]
- 18. Rustin P, Chretien D, Bourgeron T, Gerard B, Rotig A, Saudubray JM, Munnich A. Biochemical and molecular investigations in respiratory chain deficiencies. Clin Chim Acta 228: 35–51, 1994 [DOI] [PubMed] [Google Scholar]
- 19. Sazanov LA, Peak-Chew SY, Fearnley IM, Walker JE. Resolution of the membrane domain of bovine complex I into subcomplexes: implications for the structural organization of the enzyme. Biochemistry 39: 7229–7235, 2000 [DOI] [PubMed] [Google Scholar]
- 20. Schlick NE, Jensen-Seaman MI, Orlebeke K, Kwitek AE, Jacob HJ, Lazar J. Sequence analysis of the complete mitochondrial DNA in 10 commonly used inbred rat strains. Am J Physiol Cell Physiol 291: C1183–C1192, 2006 [DOI] [PubMed] [Google Scholar]
- 21. Sun F, Cui J, Gavras H, Schwartz F. A novel class of tests for the detection of mitochondrial DNA-mutation involvement in diseases. Am J Hum Genet 72: 1515–1526, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ugalde C, Vogel R, Huijbens R, Van Den Heuvel B, Smeitink J, Nijtmans L. Human mitochondrial complex I assembles through the combination of evolutionary conserved modules: a framework to interpret complex I deficiencies. Hum Mol Genet 13: 2461–2472, 2004 [DOI] [PubMed] [Google Scholar]
- 23. Valsecchi F, Koopman WJ, Manjeri GR, Rodenburg RJ, Smeitink JA, Willems PH. Complex I disorders: causes, mechanisms, and development of treatment strategies at the cellular level. Dev Disabil Res Rev 16: 175–182, 2010 [DOI] [PubMed] [Google Scholar]
- 24. Vogel RO, Smeitink JA, Nijtmans LG. Human mitochondrial complex I assembly: a dynamic and versatile process. Biochim Biophys Acta 1767: 1215–1227, 2007 [DOI] [PubMed] [Google Scholar]
- 25. Wharton DC, Tzagoloff A. Cytochrome oxidase from beef heart mitochondria. Meth Enzymol 10: 245–253, 1967 [Google Scholar]