Abstract
The clinical significance of copy number variants (CNVs) in congenital heart disease (CHD) continues to be a challenge. Although CNVs including genes can confer disease risk, relationships between gene dosage and phenotype are still being defined. Our goal was to perform a quantitative analysis of CNVs involving 100 well-defined CHD risk genes identified through previously published human association studies in subjects with anatomically defined cardiac malformations. A novel analytical approach permitting CNV gene frequency “spectra” to be computed over prespecified regions to determine phenotype-gene dosage relationships was employed. CNVs in subjects with CHD (n = 945), subphenotyped into 40 groups and verified in accordance with the European Paediatric Cardiac Code, were compared with two control groups, a disease-free cohort (n = 2,026) and a population with coronary artery disease (n = 880). Gains (≥200 kb) and losses (≥100 kb) were determined over 100 CHD risk genes and compared using a Barnard exact test. Six subphenotypes showed significant enrichment (P ≤ 0.05), including aortic stenosis (valvar), atrioventricular canal (partial), atrioventricular septal defect with tetralogy of Fallot, subaortic stenosis, tetralogy of Fallot, and truncus arteriosus. Furthermore, CNV gene frequency spectra were enriched (P ≤ 0.05) for losses at: FKBP6, ELN, GTF2IRD1, GATA4, CRKL, TBX1, ATRX, GPC3, BCOR, ZIC3, FLNA and MID1; and gains at: PRKAB2, FMO5, CHD1L, BCL9, ACP6, GJA5, HRAS, GATA6 and RUNX1. Of CHD subjects, 14% had causal chromosomal abnormalities, and 4.3% had likely causal (significantly enriched), large, rare CNVs. CNV frequency spectra combined with precision phenotyping may lead to increased molecular understanding of etiologic pathways.
Keywords: congenital heart disease, copy number variation, genetics
structural congenital heart disease (CHD) is the most common form of congenital malformations, affecting 0.8% of live births (21). Other than infection, more children die from CHD in infancy than from all other forms of disease (25). In addition, it is estimated that at least 10% of early miscarriages are a consequence of severe cardiac malformations (10). The causes of congenital cardiac malformations are largely unknown. It is estimated that 18% are due to chromosomal causes or genetic structural abnormalities including trisomies (Trisomy 21, 13, and 18) as well as deletion syndromes; all of these are associated with significant disease risk for CHD (36). A small percentage of congenital cardiac malformations are disorders in which underlying single genes have been discovered such as TBX5 in Holt-Oram syndrome; JAG1 in Alagille syndrome; and PTPN11, SOS1, and KRAS in Noonan syndrome (36). Known environmental risk factors during pregnancy, such as maternal diabetes or prenatal exposure to drugs, viruses, and reduced folate intake account for a small percentage of CHD cases (16, 24). Although our understanding of molecular pathways in cardiac development has grown tremendously in the past few years, the etiology of human and clinically relevant CHD in the majority (∼75%) of cases cannot yet be identified or explained (14, 16).
The widespread use of microarray-based genomic technologies over the past 5–6 yr have implicated copy number variants (CNVs) in numerous disorders such as neuropsychiatric diseases (49), craniofacial phenotypes, cancer, and congenital anomalies including CHD (7, 18, 35, 36). Relative to sequence variations such as single base-pair mutations or single nucleotide polymorphisms (SNPs), rare and large CNVs are hypothesized to confer higher disease risk as entire genes are deleted or duplicated (12, 31). However, poor reproducibility between microarray platforms and the lack of standardized analytical tools highlight the importance of careful filtering in CNV detection studies (37). Nondisease-related copy number polymorphisms (CNPs and/or common CNVs ≥1%) are abundant, as evidenced by the growing Database of Genomic Variants (DGV) (22, 57). Similar to the challenges in the sequence analysis of unique genetic variants, the discovery of rare etiologic CNVs remains a challenge, both because it is more difficult to detect a rare event over another event seen many times and because of the intrinsic low prior probability of there being such a variant at any particular location in the genome in any individual (28).
Recently, an algorithm to clinically interpret CNVs in patients with CHD was described (6). This approach is primarily based on gene content and overlap with known causal CHD syndromes, rather than on CNV inheritance and size (6). We employed a parallel approach in this study and utilized a strict criteria to define “likely causal” duplications or deletions, in well-established human CHD risk genes. We chose 100 CHD risk genes or regions that were supported by published observations in human studies as a means to identify potentially disease-relevant CNVs. A majority of these known CHD risk genes were previously described or could be identified through the CHD WIKI portal (1, 36). In addition, genes associated with recognized causal chromosomal abnormalities in CHD were included, as well as recently identified candidate genes from association studies (see Table 1) (1, 42).
Table 1.
Known CHD risk genes
| Gene | Gene Name | Cytoband | Gene Start | Gene Size | ABI CN Assay # | CHD WIKI | OMIM ID | PubMed ID |
|---|---|---|---|---|---|---|---|---|
| ACP6 | ACID PHOSPHATASE 6, LYSOPHOSPHATIDE | 1q21.1 | 145585791 | 23467 | Hs00320736_cn | 611471 | 15117819, 19597493 | |
| ACTC1 | ACTIN, ALPHA, CARDIAC MUSCLE | 15q14 | 32867588 | 7631 | NS | 102540 | 17611253, 17947298 | |
| ACVR2B | ACTIVIN A RECEPTOR, TYPE IIB | 3p22.2 | 38470793 | 38844 | NS | 602730 | 20193066 | |
| ALDH1A2 | ALDEHYDE DEHYDROGENASE 1 FAMILY, MEMBER A2 | 15q22.1 | 56032918 | 112280 | NS | 603687 | 19886994 | |
| ANKRD1 | ANKYRIN REPEAT DOMAIN-CONTAINING PROTEIN 1 | 10q23.31 | 92661836 | 9176 | NS | 609599 | 18273862, 20193066 | |
| ASXL2 | ADDITIONAL SEX COMBS-LIKE 2 | 2p23.3 | 25815756 | 139060 | 612991 | 19597493 | ||
| ATRX | ATR-X GENE | Xq21.1 | 76647011 | 281364 | S | 300032 | 20193066 | |
| BCL9 | B-CELL CLL/LYMPHOMA 9 | 1q21.1 | 145479805 | 84834 | Hs01608359_cn | 602597 | 15117819, 19597493 | |
| BCOR | BCL6 COREPRESSOR | Xp11.4 | 39795442 | 46221 | Hs02764783_cn | S | 300485 | 15770227 |
| BRAF | V-RAF MURINE SARCOMA VIRAL ONCOGENE HOMOLOG B1 | 7q34 | 140080281 | 190752 | S | 164757 | 16474404, 19206169 | |
| CBL | CAS-BR-M MURINE ECOTROPIC RETROVIRAL TRANSFORMING SEQUENCE HOMOLOG | 11q23.3 | 118582199 | 101870 | 165360 | 15266616 | ||
| CFC1 | CRYPTIC PROTEIN | 2q21.1 | 131066804 | 6748 | NS, S | 605194 | 11062482 | |
| CHD1L | CHROMODOMAIN HELICASE DNA-BINDING PROTEIN 1-LIKE | 1q21.1 | 145180914 | 53153 | Hs00327255_cn | 613039 | 15117819, 19597493 | |
| CHD7 | CHROMODOMAIN HELICASE DNA-BINDING PROTEIN 7 | 8q12.2 | 61753892 | 188129 | Hs01604098_cn | S | 608892 | 15300250 |
| Hs01362863_cn | ||||||||
| CITED2 | CBP/p300-INTERACTING TRANSACTIVATOR, WITH GLU/ASP-RICH C-TERMINAL DOMAIN | 6q24.1 | 139735091 | 2387 | NS | 602937 | 16287139 | |
| COL2A1 | COLLAGEN, TYPE II, ALPHA-1 | 12q13.11 | 46653014 | 31538 | Hs00560273_cn | S | 120140 | 20193066 |
| CRELD1 | CYSTEINE-RICH PROTEIN WITH EGF-LIKE DOMAINS 1 | 3p25.3 | 9950505 | 11585 | NS | 607170 | 12632326 | |
| CRKL | V-CRK AVIAN SARCOMA VIRUS CT10 ONCOGENE HOMOLOG-LIKE | 22q11.21 | 19601713 | 36177 | Hs01301005_cn | 602007 | 20494672** | |
| CSDE1 | COLD-SHOCK DOMAIN-CONTAINING E1, RNA-BINDING | 1p13.2 | 115061059 | 41135 | S | 191510 | 20193066 | |
| EHMT1 | EUCHROMATIC HISTONE METHYLTRANSFERASE 1 | 9q34.3 | 139725237 | 125162 | Hs00150023_cn | S | 607001 | 16826528, 20193066 |
| ELN | ELASTIN | 7q11.23 | 73080362 | 41810 | Hs03073113_cn | NS, S | 130160 | 12952863 |
| EVC | ELLIS-VAN CREVELD SYNDROME | 4p16.1 | 5763824 | 103108 | S | 225500 | 12571802 | |
| EVC2 | EVC2 GENE | 4p16.1 | 5615052 | 146143 | S | 607261 | 12571802 | |
| FBN1 | FIBRILLIN 1 | 15q21.1 | 46487796 | 237414 | S | 134797 | 10441597, 18412115 | |
| FKBP6 | FK506-BINDING PROTEIN 6 | 7q11.23 | 72380235 | 30342 | Hs03635913_cn | 604839 | 12952863 | |
| Hs03630484_cn | ||||||||
| FLNA | FILAMIN A | Xq28 | 153230093 | 26107 | S | 300017 | 17190868 | |
| FMO5 | FLAVIN-CONTAINING MONOOXYGENASE 5 | 1q21.1 | 145124461 | 39085 | Hs02744463_cn | 603957 | 15117819, 19597493 | |
| FOXC1 | FORKHEAD BOX C1 | 6p25.3 | 1555679 | 3449 | Hs02241194_cn | S | 601090 | 15654696 |
| FOXH1 | FORKHEAD BOX H1 | 8q24.3 | 145670316 | 2210 | NS | 603621 | 18538293 | |
| FOXL2 | FORKHEAD TRANSCRIPTION FACTOR FOXL2 | 3q22.3 | 140145755 | 2736 | Hs01045878_cn | S | 605597 | 18642388, 20193066 |
| FOXL2 | FORKHEAD TRANSCRIPTION FACTOR FOXL2 | 3q22.3 | 140145755 | 2736 | Hs01045878_cn | S | 605597 | 18642388, 20193066 |
| GATA4 | GATA-BINDING PROTEIN 4 | 8p23.1 | 11599125 | 55793 | Hs01321405_cn | NS | 600576 | 16025100 |
| GATA6 | GATA-BINDING PROTEIN 6 | 18q11.2 | 18003413 | 32812 | Hs02615249_cn | NS | 601656 | 19666519 |
| GDF1 | GROWTH/DIFFERENTIATION FACTOR 1 | 19p13.11 | 18840360 | 27593 | Hs07489748_cn | NS | 602880 | 17924340 |
| GJA1 | GAP JUNCTION PROTEIN, ALPHA-1 | 6q22.31 | 121798443 | 14129 | S | 121014 | 11470490 | |
| GJA5 | GAP JUNCTION PROTEIN, ALPHA-5 | 1q21.1 | 145694955 | 17153 | Hs00597111_cn | NS | 121013 | 15117819 |
| GPC3 | GLYPICAN 3 | Xq26.2 | 132497441 | 449891 | Hs00702786_cn | S | 300037 | 10232747, 20193066 |
| GTF2IRD1 | GTF2I REPEAT DOMAIN-CONTAINING PROTEIN 1 | 7q11.23 | 73506055 | 148793 | 604318 | 12952863 | ||
| HAND1 | HEART- AND NEURAL CREST DERIVATIVES-EXPRESSED 1 | 5q33.2 | 153834724 | 3293 | 602406 | 10189962 | ||
| HEY2 | HAIRY/ENHANCER OF SPLIT-RELATED WITH YRPW MOTIF 2 | 6q22.31 | 126112424 | 11684 | NS | 604674 | 20193066 | |
| HOXA1 | HOMEOBOX A1 | 7p15.2 | 27099138 | 3012 | Hs00428080_cn | S | 142955 | 16155570 |
| HRAS | V-HA-RAS HARVEY RAT SARCOMA VIRAL ONCOGENE HOMOLOG | 11p15.5 | 522241 | 3309 | Hs00137975_cn | S | 190020 | 17054105 |
| ISL1 | ISL LIM HOMEOBOX 1 | 5q11.2 | 50714714 | 11606 | 600366 | 20520780 | ||
| JAG1 | JAGGED 1 | 20p12.2 | 10566331 | 36363 | NS, S | 601920 | 11152664 | |
| KIF3C | KINESIN FAMILY MEMBER 3C | 2p23.3 | 26002958 | 55989 | 602845 | 19597493 | ||
| KRAS | V-KI-RAS2 KIRSTEN RAT SARCOMA VIRAL ONCOGENE HOMOLOG | 12p12.1 | 25249446 | 45675 | S | 190070 | 16474405, 16474404 | |
| LBR | LAMIN B RECEPTOR | 1q42.12 | 223655826 | 27316 | S | 600024 | 20193066 | |
| LEFTY1 | LEFT-RIGHT DETERMINATION FACTOR 1 | 1q42.12 | 224140604 | 2855 | 603037 | 10053005 | ||
| LEFTY2 | LEFT-RIGHT DETERMINATION FACTOR 2 | 1q42.12 | 224190925 | 4618 | NS | 601877 | 10053005 | |
| MAP2K1 | MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 | 15q22.31 | 64466264 | 104672 | S | 176872 | 18042262 | |
| MAP2K2 | MITOGEN-ACTIVATED PROTEIN KINASE KINASE 2 | 19p13.3 | 4041319 | 33807 | S | 601263 | 18042262 | |
| MAP3K7IP2 | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 7 | 6q25.1 | 149680755 | 93687 | NS | 602614 | 20493459 | |
| MAPK1 | MITOGEN-ACTIVATED PROTEIN KINASE 1 | q11.21–22q11. | 20443946 | 108024 | Hs02937892_cn | 176948 | 21127295** | |
| MED13L | MEDIATOR COMPLEX SUBUNIT 13-LIKE | 12q24.21 | 114880763 | 318763 | NS | 608771 | 14638541 | |
| MGP | MATRIX GAMMA-CARBOXYGLUTAMIC ACID | 12p12.3 | 14926093 | 4002 | S | 154870 | 9916809, 20193066 | |
| MID1 | MIDLINE 1 | Xp22.2 | 10373595 | 388135 | Hs02158662_cn | S | 300552 | 12833403, 20193066 |
| Hs02784563_cn | ||||||||
| MLL2 | MYELOID/LYMPHOID OR MIXED LINEAGE LEUKEMIA 2 | 12q13.12 | 47699024 | 36350 | S | 602113 | 20711175 | |
| MYH11 | MYOSIN, HEAVY CHAIN 11, SMOOTH MUSCLE | 16p13.11 | 15704492 | 153896 | Hs00358138_cn | NS | 160745 | 16444274 |
| MYH6 | MYOSIN, HEAVY CHAIN 6, CARDIAC MUSCLE, ALPHA | 14q11.2 | 22921038 | 26284 | NS | 160710 | 15735645 | |
| MYH7 | MYOSIN, HEAVY CHAIN 7, CARDIAC MUSCLE, BETA | 14q11.2 | 22951786 | 22924 | NS | 160760 | 21604106, 18159245 | |
| NF1 | NEUROFIBROMATOSIS, TYPE I | 17q11.2 | 26446120 | 282701 | S | 162200 | 11078559, 20193066 | |
| NKX2-5 | NK2 HOMEOBOX 5 | 5q35.2 | 172591743 | 3125 | NS | 600584 | 9651244 | |
| NKX2-6 | NK2, DROSOPHILA, HOMOLOG OF, 6 | 8p21.1 | 23615909 | 3957 | NS | 611770 | 15649947 | |
| NODAL | NODAL, MOUSE, HOMOLOG OF | 10q22.1 | 71862076 | 9353 | NS | 601265 | 19064609 | |
| NOTCH1 | NOTCH, DROSOPHILA, HOMOLOG OF, 1 | 9q34.3 | 138508716 | 51343 | Hs00041764_cn | NS | 190198 | 16025100, 19597493 |
| NOTCH2 | NOTCH, DROSOPHILA, HOMOLOG OF, 2 | 1p12 | 120255698 | 158101 | S | 600275 | 16773578 | |
| NPHP3 | NEPHROCYSTIN 3 | 3q22.1 | 133882143 | 41823 | Hs02580407_cn | S | 608002 | 19177160 |
| NRAS | NEUROBLASTOMA RAS VIRAL ONCOGENE HOMOLOG | 1p13.2 | 115048600 | 12438 | S | 164790 | 20193066 | |
| NSD1 | NUCLEAR RECEPTOR-BINDING Su-var, ENHANCER OF ZESTE, AND TRITHORAX | 5q35.2–5q35.3 | 176492685 | 167135 | Hs00053100_cn | S | 606681 | 15742365, 20193066 |
| Hs00022652_cn | ||||||||
| PDGFRA | PLATELET-DERIVED GROWTH FACTOR RECEPTOR, ALPHA | 4q12 | 54790020 | 69149 | NS | 173490 | 20071345 | |
| PITX2 | PAIRED-LIKE HOMEODOMAIN TRANSCRIPTION FACTOR 2 | 4q25 | 111758028 | 19929 | 601542 | 16274491 | ||
| PPM1K | PROTEIN PHOSPHATASE, PP2C DOMAIN-CONTAINING, 1K | 4q22.1 | 89400555 | 24357 | 611065 | 19597493 | ||
| PRKAB2 | PROTEIN KINASE, AMP-ACTIVATED, NONCATALYTIC, BETA-2 | 1q21.1 | 145093308 | 17445 | Hs02605549_cn | 602741 | 15117819 | |
| PTPN11 | PROTEIN-TYROSINE PHOSPHATASE, NONRECEPTOR-TYPE, 11 | 12q24.13 | 111340918 | 91182 | S | 176876 | 17515436 | |
| RAB10 | RAS-ASSOCIATED PROTEIN RAB10 | 2p23.3 | 26110477 | 103305 | 612672 | 19597493 | ||
| RAF1 | V-RAF-1 MURINE LEUKEMIA VIRAL ONCOGENE HOMOLOG 1 | 3p25.1 | 12600099 | 80601 | Hs02645733_cn | 164760 | 17603483, 19597493 | |
| RAI1 | RETINOIC ACID-INDUCED GENE 1 | 17p11.2 | 17525511 | 129979 | S | 607642 | 16845274, 20193066 | |
| ROR2 | RECEPTOR TYROSINE KINASE-LIKE ORPHAN RECEPTOR 2 | 9q22.31 | 93524704 | 227561 | S | 602337 | 20193066 | |
| RUNX1 | RUNT-RELATED TRANSCRIPTION FACTOR 1 | 21q22.12 | 35081967 | 261498 | 151385 | 19863549, 19172993 | ||
| SALL4 | SAL-LIKE 4 | 20q13.2 | 49833989 | 18466 | Hs00139344_cn | S | 607343 | 12843316 |
| SEMA5A | SEMAPHORIN 5A | 5p15.2 | 9088137 | 511096 | Hs01709772_cn | 609297 | 9464278 | |
| SH3PXD2B | SH3 AND PX DOMAINS-CONTAINING PROTEIN 2B | 5q35.1 | 171693107 | 121025 | S | 613293 | 20137777 | |
| SHOC2 | SUPPRESSOR OF CLEAR, C. ELEGANS, HOMOLOG OF | 10q25.2 | 112713902 | 49511 | S | 602775 | 19684605, 20193066 | |
| SLC2A10 | SOLUTE CARRIER FAMILY 2 (FACILITATED GLUCOSE TRANSPORTER), MEMBER 10 | 20q13.12 | 44771685 | 26707 | S | 606145 | 16550171, 20193066 | |
| SOS1 | SON OF SEVENLESS, DROSOPHILA, HOMOLOG 1 | 2p22.1 | 39062193 | 138915 | S | 182530 | 17143285 | |
| SOX7 | SRY-BOX 7 | 8p23.1 | 10618687 | 6745 | Hs00923277_cn | 612202 | 19606479 | |
| STRA6 | STIMULATED BY RETINOIC ACID 6, MOUSE, HOMOLOG OF | 15q24.1 | 72258860 | 23385 | Hs01994903_cn | S | 610745 | 17273977 |
| TBX1 | T-BOX 1 | 22q11.21 | 18124225 | 26887 | Hs01313390_cn | NS, S | 602054 | 14585638 |
| TBX20 | T-BOX 20 | 7p14.3 | 35208566 | 51201 | Hs04957392_cn | NS | 606061 | 17668378, 19762328 |
| TBX3 | T-BOX 3 | 12q24.21 | 113592441 | 13911 | S | 601621 | 16892408 | |
| TBX5 | T-BOX 5 | 12q24.21 | 113276117 | 54513 | S | 601620 | 11376442 | |
| TDGF1 | TERATOCARCINOMA-DERIVED GROWTH FACTOR 1 | 3p21.31 | 46594183 | 4773 | NS | 187395 | 18538293, 20193066 | |
| TERT | TELOMERASE REVERSE TRANSCRIPTASE | 5p15.33 | 1306286 | 41876 | Hs03078158_cn | 187270 | ||
| TFAP2B | TRANSCRIPTION FACTOR AP2-BETA | 6p12.3 | 50894397 | 28888 | Hs01355864_cn | NS, S | 601601 | 10802654 |
| TGFBR2 | TRANSFORMING GROWTH FACTOR-BETA RECEPTOR, TYPE II | 3p24.1 | 30622997 | 87640 | 190182 | 15235604, 15731757 | ||
| TMEM40 | TRANSMEMBRANE PROTEIN 40 | 3p25.1 | 12750391 | 25417 | Hs01878707_cn | 19597493 | ||
| VEGFA | VASCULAR ENDOTHELIAL GROWTH FACTOR A | 6p21.1 | 43845930 | 16271 | NS | 192240 | 20420808 | |
| WHSC1 | WHS CANDIDATE 1 GENE | 4p16.3 | 1842920 | 110812 | Hs02237093_cn | 602952 | 9222965 | |
| ZEB2 | ZINC FINGER E BOX-BINDING HOMEOBOX 2 | 2q22.3 | 144862052 | 132334 | S | 605802 | 11595972 | |
| ZFPM2 | ZINC FINGER PROTEIN, MULTITYPE 2 | 8q23.1 | 106400322 | 485621 | NS | 603693 | 9927675, 10892744 | |
| ZIC3 | ZINC FINGER PROTEIN OF CEREBELLUM 3 | Xq26.3 | 136476011 | 5914 | Hs02692150_cn | NS | 300265 | 14681828, 10980576 |
NS, nonsyndromic; S, syndromic, NCBI Build 36.1/hg18.
Animal study.
CHD consists of heterogenous anatomy with distinct phenotypic subtypes. The European Paediatric Cardiac Coding (EPCC) System (17) has been cross mapped with the Society of Thoracic Surgeons/ European Association of Cardiothoracic Surgery (STS/EACTS) coding system through the International Society for Nomenclature of Paediatric and Congenital Heart Disease in the creation of the International Pediatric and Congenital Cardiac Code (IPCCC). We characterized cardiac malformations by subphenotyping according to both the EPCC and the STS/EACTS coding systems. We compared 945 CHD cases with a publicly available cohort of 2,026 disease-free primarily pediatric individuals (40). Cases and controls were genotyped on different platforms; therefore, a second cohort of 880 control subjects genotyped on the same platform and within the same facility as the CHD cohort was included in the analysis.
This study represents a quantitative analysis of CNVs in a large population of subjects with precisely phenotyped cardiac malformations involving 100 candidate CHD risk genes. We hypothesized that large rare CNVs that were statistically enriched against two control cohorts would be causal. A strict algorithm was employed to determine if subphenotypes were enriched in gains and losses within 100 recognized CHD risk genes selected based on gene content compared with two control cohorts. Finally, a novel analytical approach, permitting CNV gene frequency spectra to be computed as a proportion of each cohort containing a gain or a loss over the above prespecified regions, was employed to determine phenotype-gene dosage relationships.
METHODS
CHD Case Ascertainment and Confirmation
This study was reviewed and approved in accordance to institutionally approved research [Institutional Review Board (IRB)] protocols by the Children's Hospital of Wisconsin (CHW, Milwaukee, WI). Subjects were consented through the Congenital Heart Disease Tissue Bank (CHDTB) and the Wisconsin Pediatric Cardiac Registry (WPCR), IRB-approved research databases housed at CHW (20, 47). These two biobanks provide DNA samples from cases and family members, detailed maternal environmental exposure data, family history of CHD, and cardiac tissue discards.
Inclusion criteria.
Structural congenital cardiac abnormalities, as identified within the IPCCC, included abnormalities of the following: the atria and atrial septum; atrioventricular valves or atrioventricular septum; cardiac position and connections; chest wall; conduction system; coronary arteries, arterial duct, pericardium, or arteriovenous fistulae; great veins; ventricles or ventricular septum; and ventriculoarterial valves or great arteries.
Exclusion criteria.
All acquired forms of pediatric heart disease in the absence of CHD, and frequent nonpathologic structural variants when no other CHD is present, included: patent foramen ovale, patent ductus arteriosus (PDA) under 30 days of age, PDA in premature infants (<35 wk gestation) and mitral valve prolapse (in the absence of at least mild valve insufficiency).
Note: The presence of a known or suspected chromosomal abnormality or known sequence variant in a CHD risk gene did not preclude participation in the study. In addition, the presence or absence of known environmental exposures did not preclude participation in the study.
Anatomic cardiac malformations were carefully characterized by phenotyping and subphenotyping according to both the EPCC 2011 and the STS/EACTS 2011 coding systems. All phenotypes were initially reviewed by a coding specialist, a surgeon, and a cardiologist. All discrepancies were reconciled by review of source documents including operative notes, echocardiograms, and review of operative surgeon. Anatomic phenotypes and subphenotypes were reported using EPCC 2011 terms, and final confirmatory review of all cases was performed by a single pediatric cardiothoracic surgeon (17). In addition, information regarding additional diagnosis, accompanying conditions, demographics, and a limited number of genetic risk factors was obtained through the Herma Heart Center (HHC) cardiac database at CHW.
Children's Hospital of Philadelphia Control Cohort
DNA samples analyzed in this study were obtained from the whole blood of healthy subjects routinely seen at primary care and well-child clinic practices within the Children's Hospital of Philadelphia (CHOP) Health Care Network. Data using hg18/March 2006/build 36.1 genomic coordinates were downloaded from http://cnv.chop.edu/ (40). High-resolution mapping of copy number variations in 2,026 healthy individuals was performed using the Illumina HumanHap 550 BeadChip (Illumina, San Diego, CA) (40).
Milwaukee Family Heart Study Control Cohort
Control subjects were drawn from the Milwaukee Family Heart Study (MFHS) in accordance with Medical College of Wisconsin IRB protocols (MCW, Milwaukee, WI). Subjects were ascertained as a hospital-based cohort, referred to the catheterization laboratory for diagnostic coronary angiography. Inclusion criteria were the ability to consent and age >21 yr. The following were considered exclusion criteria: end-stage renal disease, current treatment for a malignancy, and a diagnosis of coronary artery disease or a myocardial infarction at age >69 yr. In addition, we excluded all participants with acute coronary syndrome and significant valvular disease. Individuals with a diagnosis of other cardiac structural abnormalities were excluded based on either the result of echocardiography prior to or as determined during the invasive cardiac procedure.
Genomic DNA Extraction
Genomic DNA for CHD and MFHS cohorts was obtained from peripheral blood using standard protocols for DNA isolation from Roche Diagnostics, Promega Biotech (Wizard), and Qiagen (Gentra Puregene). Purified genomic DNA was resuspended in 1.0 mM Tris HCl pH 8.0 and 0.1 mM EDTA. DNA quality was tested by optical density 260/280 ratios, quantified by UV spectrophotometry using a Nanodrop 2000 (Thermo Scientific, Wilmington, DE). DNA stocks were stored at −80°C, dilutions for microarray analysis were stored at 100 ng/μl at −20°C.
CHD Risk Gene Prioritization and Selection
Genes or regions with previously associated disease/syndrome variants as identified through the CHD WIKI website (searched 01/04/2011 and updated 07/28/2011) and/or supported by previously published observations in human studies were selected (1, 34, 36, 42, 48). These known CHD risk genes are outlined in Table 1.
Briefly, CHD WIKI offers an updated overview of genes implicated in human CHD, obtained by an OMIM search, and complemented with a study of the PubMed literature concerning mutation analysis of candidate genes for congenital heart defects (1). The level of support was defined by inheritance of the mutation (de novo or inherited and segregated with a phenotype) and the association of a variant in the investigated CHD population vs. a normal control population (1). A comprehensive list of 100 CHD risk genes was selected; the vast majority of these selected genes are known to be expressed in the human heart (3, 11, 43, 46, 50, 54). According to CHD WIKI, syndromic genes were defined as congenital heart defects that are associated with a second major malformation (i.e., renal defects, cleft palate, brain malformations), with developmental delay or mental handicap, and/or the presence of dysmorphism.
Genotyping
Genotyping for the CHD and MFHS control cohort was performed with the Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA) as previously described (30, 47). All samples were run in the Advanced Genomics (AGEN) laboratory core at the Children's Research Institute (CRI)/MCW (Milwaukee, WI). A reference genomic DNA control sample, ref 103, supplied by Affymetrix, was run with every batch of subjects (Santa Clara, CA).
CNV Analysis and Quality Control
The CHD subject cohort comprised 1,020 subjects consented through the CHDTB or WPCR. We evaluated the quality and suitability of the subject population for a genetic association study. The population was required to pass copy number analysis quality metrics as seen in Table 2.
Table 2.
Quality control of CHD case and MFHS control cohorts and genotyping data
| Subjects, n | Subjects, n | ||
|---|---|---|---|
| MFHS Control Cohort | CHD Case Cohort | ||
| Starting subjects | 950 | Starting subjects | 1,020 |
| Remaining subjects | 880 | Remaining subjects | 958 |
| QC Exclusions | % Total | % Total | |
| MAPD QC | 3.05 | MAPD QC | 2.35 |
| Segment QC | 4.32 | Segment QC | 2.15 |
| Consent QC | NA | Consent QC | 0.10 |
| Sex QC | NA | Sex QC | 0.59 |
Copy number analysis exclusions were as follows: median absolute pairwise difference (MAPD) quality control (QC) ≥0.35, number of copy number polymorphism (CNP) segments ≥250, 1 subject with a status change to his/her consent, and sex tracking QC. Congenital heart disease (CHD) cases were reduced to a final n = 945 after inclusion and exclusion criteria were met.
CNV identification of study subjects required the processing of Affymetrix intensity (CEL) files using Genotyping Console version 3.0.2 (GTC) software as previously described (20, 47). CEL files of subjects with a median absolute pairwise difference >0.35 and a CNV segmentation count ≥250, indicative of poor DNA quality, were excluded from the study.
A final number of 945 CHD subjects and 880 MFHS controls remained in the study after inclusion and exclusion criteria were met.
As summarized in Table 3, the cases and controls were stratified according to age, sex, and race/ethnicity.
Table 3.
CHD case, CHOP, and MFHS control cohort demographics
| CHD Case Cohort | CHOP Control Cohort | MFHS Control Cohort | |
|---|---|---|---|
| Race | |||
| Caucasian | 655 | 1,320 | 870 |
| African American | 92 | 694 | 5 |
| Native American | 14 | 5 | |
| Hispanic | 90 | ||
| Asian | 26 | 12 | |
| Other | 68 | ||
| Total | 945 | 2,026 | 880 |
| Sex, % | |||
| Female | 44.02 | 36.59 | |
| Male | 55.87 | 63.41 | |
| Age, yr | |||
| Median age | 0.62 | 67.00 | |
| Average age | 4.03 | 65.66 |
CHOP, Children's Hospital of Philadelphia ; MFHS, Milwaukee Family Heart Study.
Copy number state of those subjects who passed quality control thresholds were determined with reference to the GenomeWideSNP_6.hapmap270 file and copy number calls were determined using the Affymetrix GTC segmentation algorithm. To reduce the presence of false positive CNVs, the segmentation algorithm parameters were set to identify only those regions larger than 25 kb comprising at least 25 contiguous markers. It has been shown that CNVs smaller than this are frequently false positive detection (40). In addition, all segments were monitored for degree of overlap with previously identified common CNVs, annotated by the DGV (22, 57).
Using a BED file format (chromosome, gene starting position, gene ending position, gene name), copy number information was drawn from custom gene regions (Table 1) extracted from the processed segment data.
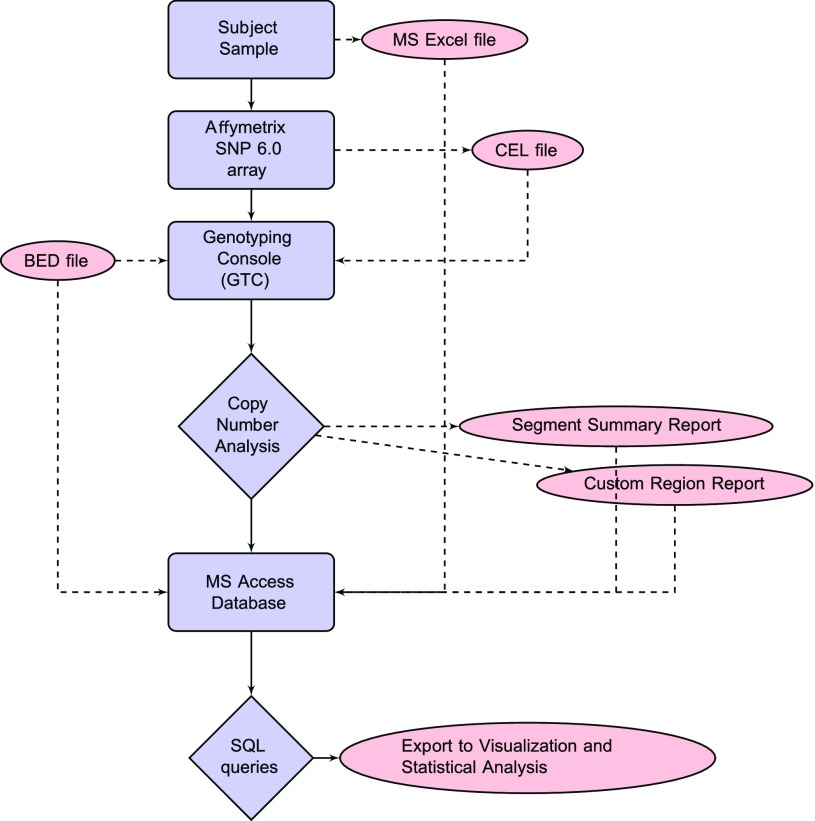
A flowchart for copy number analysis is presented in Fig. 1. A multipurpose Access database (Microsoft, Redmond, WA) served as a central repository for the cohort demographic data as well as the entire experimental set of copy number variant data. Database tables were populated with copy number data from the GTC analysis, detailed demographic data, and the annotated 100 CHD risk gene list (Table 1). Demographic data for CHD cases and MFHS controls were obtained via clinical and consent verification methods. SQL query results included aggregate CNV counts by phenotype or region for both CHD and MFHS controls. Graphical representation of the query results was accomplished using Excel (Microsoft) and R (45). Supplemental Table S1 includes a complete summary of all CNV profiles over the 100 CHD risk gene list for each subject as well as phenotypic and demographic information.1
Fig. 1.
CNV analysis flowchart from sample to statistics. Blue figures represent software used or a process/task performed. Red figures represent data files.
Overall CNV burden.
The total number of large CNVs throughout the genome was calculated by importing GTC segment files filtered by size (duplication ≥200 kb or deletion ≥100 kb) into an Access database. An external R program further filtered CNVs for all Build 36 annotated genes that did not occur as a CNP, defined as a normal variant (≥1%) in either the CHOP or MFHS control cohorts.
Algorithm for likely causal CNV determination.
A strict algorithm was employed to determine likely causal CNVs. Gains and losses were considered as potentially disease relevant if they fulfilled the following criteria: 1) size: duplication ≥200 kb or deletion ≥100 kb, 2) they did not occur as a CNP, defined as a normal variant (≥1%) in either CHOP or MHFS control cohort, and 3) CNV occurred over a gene region known to be associated with CHD (CHD 100 gene list).
A final step was taken because the MFHS cohort was aged and significantly different from CHD cases. Sex chromosome degradation in peripheral blood appears to be an age-related phenomenon (19). Studies have shown that a strong correlation exists between patient age and loss of the Y chromosome (52). Sex chromosome degradation is easily detected by the segment reports created by GTC because males have only one copy of Chr. X. To optimize the analysis of sex chromosomes, sex-matched references were employed; for X chromosome analysis, only females from all three cohorts were compared (55). Thus male MFHS controls were excluded from X chromosome results in all CNV analyses.
CNV frequency by phenotype.
CNVs fulfilling criteria 1–3 were analyzed for enrichment by subphenotypes.
CNV frequency by gene region.
CNV frequency “spectra” were computed as a proportion of each cohort containing a gain or a loss over the CHD associated gene list.
Complex CNV analysis.
To determine if subjects carried multiple CNVs, large rare CNVs outside of and in addition to the defined set of 100 disease-related CHD genes were screened using criteria 1 and 2 (see Ref. 56).
Confirmatory Studies
CNVs that were identified in the CHD cases were confirmed by either karyotype, FISH analysis, or TaqMan CN real-time quantitative PCR assays (Applied Biosystems). CNVs for one case asterisked in Table 5 was difficult to confirm and is currently pending, due to inconclusive TaqMAN copy number results. A representative set of identified CNVs within the CHOP cohort were previously validated (40), whereas CNVs identified in the MFHS cohort as part of this study were not confirmed. As a means of secondary CNV confirmation of CHD cases, microarray analysis was performed by an independent lab on a number of the CHD study subjects (n = 34). TaqMan copy number reactions (Table 1) were run in triplicate on an ABI HT7900 instrument (Applied Biosystems) under the following cycling conditions: 50°C for 2 min, 95°C for 10 min, then 40 cycles of 95°C for 15 s followed by 60°C for 1 min. Typically ∼20 ng of template genomic DNA was amplified in reaction volumes of 10 μl, as previously described (47). Copy number confirmations were assessed using a calibrator panel of six individuals with known copy number state over the gene of interest and analyzed using Copy Caller software version 1.0 (Applied Biosystems). If parents of subjects with confirmed CNVs were available, their DNA was analyzed to determine if CNVs were inherited or de novo, as noted in Table 5.
Table 5.
Case reports of likely causal CNVs
| Subject | Subphenotype | 100 CHD Gene Region | Exon(s) | LOSS_GAIN | Cytoband | CNV Start (Build 36, hg18) | CNV Size, kb | Markers, n | Inheritance | Gene Names on CNV Segment (100 CHD Genes in boldface) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | AS (valvar) | ACP6 BCL9 CHD1L FMO5 GJA5 PRKAB2 | all | Loss | 1q21.1 | 144643813 | 1654 | 684 | NBPF11 FAM108A3 PRKAB2 FMO5 CHD1L BCL9 ACP6 GJA5 GJA8 GPR89B NBPF11 | |
| 2 | AS (valvar) | CHD1L FMO5 PRKAB2 | all | Gain | 1q21.1 | 144943150 | 418 | 280 | PRKAB2 FMO5 CHD1L | |
| NSD1 | all | Gain | 5q35.2–5q35.3 | 175269980 | 1777 | 735 | THOC3 FAM153B C5orf25 KIAA1191 ARL10 HSPC111 HIGD2A CLTB FAF2 RNF44 PCDH24 GPRIN1 SNCB EIF4E1B TSPAN17 UNC5A HK3 UIMC1 ZNF346 FGFR4 NSD1 RAB24 PRELID1 MXD3 LMAN2 RGS14 SLC34A1 PFN3 F12 GRK6 PRR7 DBN1 PDLIM7 DOK3 DDX41 FLJ10404 TMED9 B4GALT7 | |||
| 3 | AS (valvar) | FOXC1 | all | Loss | 6p25.3–6p25.2 | 94649 | 2539 | 2130 | DUSP22 IRF4 EXOC2 HUS1B FOXQ1 FOXF2 FOXC1 GMDS C6orf195 MYLK4 | |
| 4 | ASD-SEC | MYH11 | all | Loss | 16p13.11–16p12.3 | 15186307 | 2903 | 1521 | MPV17L C16orf45 KIAA0430 NDE1 MYH11 C16orf63 ABCC1 ABCC6 NOMO3 LOC339047 XYLT1 | |
| 5 | ASD-SV | GATA4 | all | Loss | 8p23.1 | 11390744 | 304 | 213 | BLK GATA4 NEIL2 | |
| 6 | AVC (partial) | GATA4 SOX7 | all | Loss | 8p23.1 | 8055434 | 3844 | 3235 | PRAGMIN CLDN23 MFHAS1 THEX1 PPP1R3B TNKS MSRA UNQ9391 RP1L1 C8orf74 SOX7 PINX1 XKR6 MTMR9 AMAC1L2 FAM167A BLK GATA4 NEIL2 FDFT1 CTSB CTSB DEFB137 DEFB136 DEFB134 | |
| 7 | AVC (partial) | GDF1 | all | Gain | 19p13.11 | 18763592 | 378 | 163 | UPF1 GDF1 LASS1 COPE DDX49 HOMER3 SFRS14 ARMC6 SLC25A42 TMEM161A MEF2B | |
| 8 | AVC unbalanced + AVSD with ventricular imbalance | MID1 | 5′ UTR-i1 | Gain | Xp22.2 | 10714630 | 509 | 265 | MID1 HCCS ARHGAP6 AMELX | |
| 9 | AVSD with TOF | CRKL TBX1 | all | Gain | 22q11.21 | 17953160 | 1838 | 1106 | SEPT5 GP1BB TBX1 GNB1L C22orf29 TXNRD2 COMT ARVCF C22orf25 DGCR8 HTF9C RANBP1 ZDHHC8 RTN4R DGCR6L RIMBP3 ZNF74 SCARF2 KLHL22 MED15 PI4KA SERPIND1 SNAP29 CRKL AIFM3 LZTR1 THAP7 P2RX6 SLC7A4 | |
| 10 | CoA | ACP6 BCL9 CHD1L FMO5 GJA5 PRKAB2 | all | Gain | 1q21.1 | 144812585 | 1480 | 678 | PRKAB2 FMO5 CHD1L BCL9 ACP6 GJA5 GJA8 GPR89B GPR89C NBPF11 LOC728912 | |
| 11 | CoA | NOTCH1 | all | Loss | 9q34.3 | 138377108 | 229 | 105 | DNLZ CARD9 SNAPC4 SDCCAG3 PMPCA INPP5E SEC16A C9orf163 NO TCH1 | |
| 12 | DILV | HRAS | all | Gain | 11p15.5 | 354390 | 256 | 62 | B4GALNT4 PKP3 SIGIRR TMEM16J PTDSS2 RNH1 HRAS LRRC56 C11orf35 RASSF7 KIAA1542 IRF7 MUPCDH | |
| 13 | DORV | SEMA5A | i8-3′ UTR | Gain | 5p15.31–5p15.2 | 7119715 | 2152 | 1769 | ADCY2 C5orf49 FASTKD3 MTRR SEMA5A | |
| 14 | EBSTEIN'S | FKBP6 | 5′ UTR-i8 | Gain | 7q11.23 | 72073034 | 330 | 28 | TRIM74 STAG3L3 NSUN5 TRIM50 FKBP6 | |
| 15 | HLHS | EHMT1 | all | Gain | 9q34.3 | 139701521 | 264 | 142 | EHMT1 CACNA1B | |
| 16 | HLHS | FKBP6 | 5′ UTR-i8 | Gain | 7q11.23 | 72052197 | 348 | 34 | de novo | POM121 NSUN5C TRIM74 ST AG3L3 NSUN5 TRIM50 FKBP6 |
| 17 | HLHS | GATA4 | all | Gain | 8p23.1 | 11049252 | 1438 | 755 | unknown | XKR6 MTMR9 AMAC1L2 FAM167A BLK GATA4 NEIL2 FDFT1 CTSB DEFB137 DEFB136 DEFB134 DEFB130 ZNF705D DUB3 FAM86B1 DEFB130 |
| SOX7 | all | Gain | 8p23.1 | 8055434 | 2992 | 2584 | unknown | PRAGMIN CLDN23 MFHAS1 THEX1 PPP1R3B TNKS MSRA UNQ9391 RP1L1 C8orf74 SOX7 PINX1 XKR6 | ||
| 18 | HLHS | MYH11 | all | Gain | 16p13.11 | 14846829 | 1414 | 640 | inherited | NOMO1 NPIP PDXDC1 NTAN1 RRN3 MPV17L C16orf45 KIAA0430 NDE1 MYH11 C16orf63 ABCC1 ABCC6 NOMO3 |
| 19 | Other, Cardiac | CRKL TBX1 | all | Gain | 22q11.21 | 17161534 | 2634 | 1575 | DGCR6 PRODH DGCR2 DGCR14 TSSK2 GSC2 SLC25A1 CLTCL1 HIRA MRPL40 C22orf39 UFD1L CDC45L CLDN5 SEPT5 GP1BB TBX1 GNB1L C22orf29 TXNRD2 COMT ARVCF C22orf25 DGCR8 HTF9C RANBP1 ZDHHC8 RTN4R DGCR6L RIMBP3 ZNF74 SCARF2 KLHL22 MED15 PI4KA SERPIND1 SNAP29 CRKL AIFM3 LZTR1 THAP7 P2RX6 SLC7A4 | |
| 20 | PA, VSD | ACP6 BCL9 CHD1L FMO5 GJA5 PRKAB2 | all | Gain | 1q21.1 | 144812585 | 1480 | 678 | PRKAB2 FMO5 CHD1L BCL9 ACP6 GJA5 GJA8 GPR89B GPR89C NBPF11 LOC728912 | |
| 21 | Subaortic stenosis | CRKL | all | Gain | 22q11.21 | 19093207 | 699 | 626 | ZNF74 SCARF2 KLHL22 MED15 PI4KA SERPIND1 SNAP29 CRKL AIFM3 LZTR1 THAP7 P2RX6 SLC7A4 | |
| 22 | Subaortic stenosis | HRAS | all | Gain | 11p15.5 | 339238 | 271 | 63 | B4GALNT4 PKP3 SIGIRR TMEM16J PTDSS2 RNH1 HRAS LRRC56 C11orf35 RASSF7 KIAA1542 IRF7 MUPCDH | |
| 23 | DORV | FOXL2 NPHP3 | all | Gain | 3q22.1–3q26.1 | 131972967 | 32134 | 19750 | PIK3R4 ATP2C1 ATP2C1 ASTE1 NEK11 NUDT16 MRPL3 CPNE4 ACPP DNAJC13 ACAD11 CCRL1 UBA5 NPHP3 TMEM108 BFSP2 CDV3 TOPBP1 TF SRPRB RAB6B | |
| C3orf36 SLCO2A1 RYK AMOTL2 ANAPC13 CEP63 KY EPHB1 PPP2R3A MSL2L1 PCCB ST AG1 TMEM22 NCK1 IL20RB SOX14 CLDN18 DZIP1L A4GNT DBR1 ARMC8 TXNDC6 MRAS FAM62C CEP70 FAIM PIK3CB FOXL2 C3orf72 LOC389151 MRPS22 COPB2 RBP2 RBP1 NMNAT3 CLSTN2 TRIM42 SLC25A36 SPSB4 ACPL2 ZBTB38 RASA2 RNF7 GRK7 ATP1B3 TFDP2 GK5 XRN1 ATR PLS1 TRPC1 PCOLCE2 PAQR9 SR140 CHST2 SLC9A9 C3orf58 PLOD2 PLSCR4 PLSCR2 PLSCR1 PLSCR5 ZIC4 ZIC1 AGTR1 CPB1 CPA3 GYG1 HLTF HPS3 CP TM4SF18 TM4SF1 TM4SF4 WWTR1 COMMD2 RNF13 RNF13 PFN2 TSC22D2 SERP1 EIF2A SELT C3orf44 SIAH2 CLRN1 CLRN1 MED12L GPR171 P2RY14 GPR87 P2RY13 P2RY13 P2RY12 IGSF10 AADACL2 AADAC SUCNR1 MBNL1 TMEM14E P2RY1 RAP2B LOC152118 SGEF DHX36 GPR149 MME PLCH1 C3orf33 SLC33A1 GMPS KCNAB1 SSR3 TIPARP LEKR1 CCNL1 VEPH1 PTX3 C3orf55 SHOX2 RSRC1 MLF1 GFM1 LXN RARRES1 MFSD1 IQCJ SCHIP1 IL12A IFT80 SMC4 TRIM59 KPNA4 ARL14 PPM1L B3GALNT1 NMD3 C3orf57 OTOL1 SI SLITRK3 BCHE ZBBX SERPINI2 WDR49 PDCD10 SERPINI1 GOLIM4 EVI1 EVI1 MDS1 ARPM1 MYNN LRRC34 LRRIQ4 LRRC31 SAMD7 SEC62 GPR160 PHC3 PRKCI SKIL CLDN11 SLC7A14 RPL22L1 EIF5A2 SLC2A2 TNIK PLD1 FNDC3B GHSR TNFSF10 AADACL1 ECT2 SPATA16 NLGN1 NAALADL2 TBL1XR1 KCNMB2 ZMAT3 PIK3CA KCNMB3 ZNF639 MFN1 GNB4 ACTL6A MRPL47 NDUFB5 USP13 PEX5L TTC14 CCDC39 FXR1 DNAJC19 SOX2 ATP11B DCUN1D1 MCCC1 LAMP3 MCF2L2 B3GNT5 KLHL6 KLHL24 YEATS2 MAP6D1 PARL ABCC5 HTR3D HTR3C HTR3E | ||||||||||
| EIF2B5 DVL3 AP2M1 ABCF3 ALG3 ECE2 CAMK2N2 ECE2 PSMD2 EIF4G1 FAM131A CLCN2 POLR2H THPO CHRD EPHB3 MAGEF1 VPS8 C3orf70 EHHADH MAP3K13 TMEM41A LIPH SENP2 IGF2BP2 C3orf65 SFRS10 ETV5 DGKG CRYGS TBCCD1 DNAJB11 AHSG FETUB HRG KNG1 EIF4A2 RFC4 ADIPOQ ST6GAL1 RPL39L RTP1 MASP1 RTP4 SST RTP2 BCL6 LPP TPRG1 TP63 LEPREL1 SENP2 IGF2BP2 C3orf65 SFRS10 ETV5 DGKG CRYGS TBCCD1 DNAJB11 AHSG FETUB HRG KNG1 EIF4A2 RFC4 ADIPOQ ST6GAL1 RPL39L RTP1 MASP1 RTP4 SST RTP2 BCL6 LPP TPRG1 TP63 LEPREL1 | ||||||||||
| 24 | TOF | ACP6 BCL9 GJA5 | all | Gain | 1q21.1 | 145250193 | 1678 | 471 | BCL9 ACP6 GJA5 GJA8 GPR89B GPR89C NBPF11 LOC728912 PPIAL4 NBPF14 NBPF10 NBPF15 NBPF16 | |
| CHD1L FMO5 PRKAB2 | all | Gain | 1q21.1 | 144643813 | 600 | 223 | NBPF11 LOC728912 FAM108A3 PRKAB2 FMO5 CHD1L | |||
| 25 | TOF | CRKL | all | Loss | 22q11.21 | 18710744 | 1085 | 673 | RIMBP3 ZNF74 SCARF2 KLHL22 MED15 PI4KA SERPIND1 SNAP29 CRKL AIFM3 LZTR1 THAP7 P2RX6 SLC7A4 | |
| 26 | TOF | HOXA1 | all | Gain | 7p15.2–7p15.1 | 26113744 | 4718 | 3324 | NFE2L3 HNRNP A2B1 CBX3 SNX10 SKAP2 HOXA1 HOXA2 HOXA3 HOXA4 HOXA5 HOXA6 HOXA7 HOXA9 HOXA10 HOXA11 HOXA13 EVX1 HIBADH TAX1BP1 JAZF1 LOC402644 CREB5 KIAA0644 CPVL CHN2 PRR15 WIPF3 SCRN1 FKBP14 PLEKHA8 C7orf41 ZNRF2 NOD1 C7orf24 GARS CRHR2 INMT FLJ22374 | |
| TBX20 | all | Gain | 7p14.3–7p14.2 | 32897122 | 3321 | 2145 | KBTBD2 FKBP9 NT5C3 RP9 BBS9 BMPER NPSR1 DPY19L1 TBX20 HERPUD2 SEPT7 EEPD1 | |||
| 27 | TOF | MYH11 | all | Gain | 16p13.11 | 14805290 | 1455 | 642 | NOMO1 NPIP PDXDC1 NTAN1 RRN3 MPV17L C16orf45 KIAA0430 NDE1 MYH11 C16orf63 ABCC1 ABCC6 NOMO3 | |
| 28* | TOF | TERT | all | Loss | 5p15.33 | 80069 | 2948 | 1893 | PLEKHG4B LOC389257 CCDC127 SDHA PDCD6 LOC116349 EXOC3 SLC9A3 CEP72 TPPP ZDHHC11 BRD9 TRIP13 NKD2 SLC12A7 SLC6A19 SLC6A18 TERT CLPTM1L SLC6A3 LPCAT1 MRPL36 NDUFS6 IRX4 IRX2 C5orf38 | |
| 29 | TRI-AT | MAPK1 | all | Gain | 22q11.21–22q11.22 | 20264556 | 447 | 243 | UBE2L3 YDJC CCDC116 SDF2L1 PPIL2 YPEL1 MAPK1 PPM1F TOP3B | |
| 30 | TRI-AT | NSD1 | e24-3′ UTR | Gain | 5q35.3 | 176656286 | 330 | 133 | NSD1 RAB24 PRELID1 MXD3 LMAN2 RGS14 SLC34A1 PFN3 F12 GRK6 PRR7 DBN1 PDLIM7 DOK3 DDX41 FLJ10404 TMED9 B4GALT7 | |
| 31 | Truncus arteriosus | MAPK1 | all | Loss | 22q11.21–22q11.22 | 20055986 | 1237 | 863 | HIC2 RIMBP3B RIMBP3C UBE2L3 YDJC CCDC116 SDF2L1 PPIL2 YPEL1 MAPK1 PPM1F TOP3B VPREB1 ZNF280B ZNF280A PRAME | |
| GATA6 | all | Gain | 18q11.2 | 17749666 | 308 | 168 | GATA6 | |||
| 32 | Truncus arteriosus | SALL4 | all | Loss | 20q13.2 | 49428074 | 1839 | 1357 | NFATC2 ATP9A SALL4 ZFP64 TSHZ2 | |
| 33 | VSD (perimembranous) | CRKL | all | Gain | 22q11.21 | 19389671 | 406 | 451 | PI4KA SERPIND1 SNAP29 CRKL AIFM3 LZTR1 THAP7 P2RX6 SLC7A4 | |
| ACP6 BCL9 CHD1L FMO5 GJA5 PRKAB2 | all | Loss | 1q21.1 | 144723763 | 1574 | 683 | NBPF11 LOC728912 FAM108A3 PRKAB2 FMO5 CHD1L BCL9 ACP6 GJA5 GJA8 GPR89B GPR89C NBPF11 | |||
| 34 | VSD (perimembranous) | GATA4 SOX7 | all | Loss | 8p23.1 | 8027361 | 4456 | 3349 | PRAGMIN CLDN23 MFHAS1 THEX1 PPP1R3B TNKS MSRA UNQ9391 RP1L1 C8orf74 SOX7 PINX1 XKR6 MTMR9 AMAC1L2 FAM167A BLK GATA4 NEIL2 FDFT1 CTSB CTSB DEFB137 DEFB136 DEFB134 DEFB130 ZNF705D DUB3 FAM86B1 DEFB130 | |
| 35 | VSD (perimembranous) | GATA6 | all | Gain | 18q11.1–18q11.2 | 16795645 | 6118 | 3683 | ROCK1 ESCO1 SNRPD1 ABHD3 MIB1 GATA6 CTAGE1 RBBP8 CABLES1 C18orf45 RIOK3 C18orf8 NPC1 ANKRD29 LAMA3 TTC39C CABYR OSBPL1A IMPACT HRH4 ZNF521 SS18 PSMA8 TAF4B KCTD1 AQP4 AQP4 CHST9 |
Inconclusive TAQMAN results (see Subject 28). Boldface indicates confirmed genes. “Unknown” means one parental DNA was unavailable. “Other cardiac” phenotype (case 19) is double-chamber right ventricle (DCRV).
Statistical Analysis
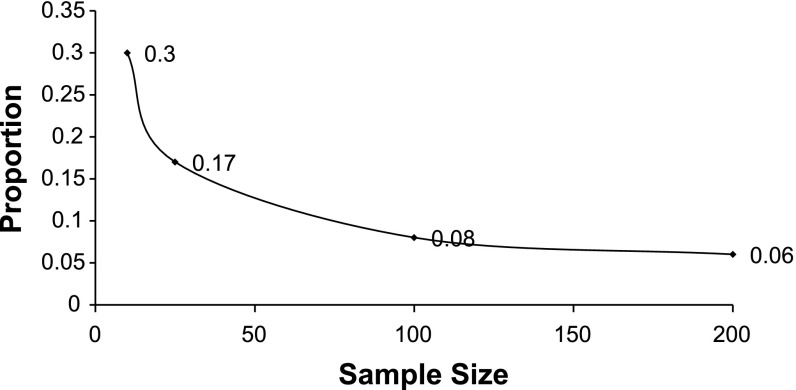
Since the expected incidence is very small (typically <5%) tests based on a normality assumption would be incorrect, therefore a one-tailed Barnard exact test was used for all comparisons of proportions of CNVs (8). A P ≤ 0.05 without adjustment is used for significance. A custom R program was used to calculate the P value and checked using Cytel StatXact (Cytel, Cambridge, MA) (15). StatXact was also used to calculate power. With a sample of 810, and a CNV incidence of 4.3%, we would have at least 90% power to detect a significant difference from 0.0196 (the CNV incidence of CHOP cohort's 39/2,026). We have given other power calculations for possible scenarios of subphenotypes (Fig. 2). We see that in an n = 100 sample group we would have ≥80% power if we had an 8% CNV incidence. For a cohort of 200 we would have ≥80% power to detect a difference of 6% CNV incidence.
Fig. 2.
Sample size (n) and copy number variant (CNV) proportion (fraction), required to detect difference from 0.0196 (CHOP control CNV fraction) at an alpha = 0.05, power at least 80%. CHOP, Children's Hospital of Philadelphia.
This figure demonstrates the sample size required (x-axis) with power of at least 80% under varying CNV proportions (y-axis) when the control cohort is 0.0196 (CHOP control CNV proportion) at an alpha = 0.05.
RESULTS
Phenotypes of CHD Study Subjects
Subjects diagnosed with congenital heart malformations (n = 945) and phenotyped in accordance with the EPCC terms were categorized into the 40 cardiac subphenotypes listed in Table 4 (17). The five largest phenotypes represented were as follows: hypoplastic left heart syndrome (HLHS) 14.8%, ventricular septal defect (VSD perimembranous) 7.7%, tetralogy of Fallot (TOF) 7.7%, coarctation of the aorta (CoA) 7.0%, and atrioventricular canal complete (AVC complete) 5.0%. The majority of subjects were represented by individual subphenotypes most of which contained <5.1% of the total CHD cohort.
Table 4.
CHD cohort by subphenotypes
| Diagnoses | Subjects | % of Total | Diagnoses | Subjects | % of Total |
|---|---|---|---|---|---|
| Aorto-pulmonary window + Patent Ductus Arteriosus (PDA)T21 | 5 | 0.53 | Mitral Valve Stenosis (MS, subvalvar, parachute)22q | 6 | 0.63 |
| AVSD + TOF (AVSD + TOF)T21 | 7 | 0.74 | Other, CardiacT21, 22q | 18 | 1.90 |
| Arrhythmias (Congenital Heart Block, Long QT, WPW) | 7 | 0.74 | Pulmonary Atresia (PA) | ||
| Aortic Stenosis (Valvar)T | 31 | 3.28 | - IVS-T21 | 18 | 1.90 |
| Atrial Septal Defect Secundum (ASD-SEC)T21 | 47 | 4.97 | - VSD-22q | 34 | 3.60 |
| Atrial Septal Defect Sinus Venosus (ASD-SV) | 13 | 1.38 | PAPVR | 12 | 1.27 |
| A-V Canal Complete (AVC Complete)T21 | 48 | 5.08 | Pulmonary Stenosis (Valvar) | 9 | 0.95 |
| A-V Canal Intermediate (AVC Intermediate)T21 | 7 | 0.74 | Shone's | 8 | 0.85 |
| A-V Canal Partial (AVC Partial)T21 | 17 | 1.80 | Subaortic stenosisT21 | 12 | 1.27 |
| A-V Canal Unbalanced + AVSD with ventricular imbalanceT21 | 14 | 1.48 | Supravalvar aortic stenosis (supravalvar AS) | 4 | 0.42 |
| Cardiomyopathy (DILATED) | 13 | 1.38 | Total Anomalous Venous Connection (TAPVC) | 15 | 1.59 |
| Cardiomyopathy (HYPERTROPHIC) | 4 | 0.42 | Tetralogy of Fallot (TOF)T21, 22q | 73 | 7.72 |
| Chest Wall | 4 | 0.42 | Transposition of Great Arteries (TGA) | ||
| Coarctation of the Aorta (CoA)T | 66 | 6.98 | - IVS - | 21 | 2.22 |
| Coronary Arteries (COR ART) | 10 | 1.06 | - VSD - | 21 | 2.22 |
| Double Inlet Left Ventricle (DILV) | 19 | 2.01 | Tricuspid Atresia (TRI-AT) | 29 | 3.07 |
| Double Outlet Right Ventricle (DORV)22q | 41 | 4.34 | Truncus Arteriosus (TA)22q | 29 | 3.07 |
| Ebstein's Anomaly (EBSTEINS) | 9 | 0.95 | Vascular ring and PA slingT21, 22q | 14 | 1.48 |
| Hypoplastic Left Heart Syndrome (HLHS)T | 140 | 14.81 | VSD inletT21 | 4 | 0.42 |
| Interrupted Aortic Arch (IAA)22q | 11 | 1.16 | VSD multiple + muscular | 10 | 1.06 |
| L-TGA | 7 | 0.74 | VSD perimembranousT21, 22q | 73 | 7.72 |
| Dilated Ascending Aorta (MARFAN) | 8 | 0.85 | VSD subarterialT21 | 7 | 0.74 |
The following individual phenotypes were included in the “other cardiac” subphenotype category: single ventricle other, absent left pulmonary artery (LPA), absent pulmonary valve, aorto-left ventricular tunnel, bicuspid aortic valve (BAV), cor triatriatum, double-chamber right ventricle (DCRV), left ventricular aneurysm, tricuspid regurgitation, true cleft of mitral leaflet (without AVSD). Superscripts were used to denote phenotypes where causal chromosomal as shown abnormalities were observed (see results, where T21 = Trisomy 21, 22q = 22qDS, and T = Turner's Syndrome). PAPVR, partial anomalous pulmonary venous return; VSD, ventricular septal defect.
Subjects With Recognized Causal Chromosomal Abnormalities
We ascribed 135 subjects to known CHD-related chromosomal abnormalities [T21 (n = 80), T18 (n = 1), 22qDS (n = 42), Turner (n = 8), William's (n = 3), and XXX (n = 1)] (36, 44). The syndromes and their associated phenotypes were as follows: T21: aorto-pulmonary window with PDA n = 2; AVSD + TOF n = 5; ASD-SEC n = 4; AVC complete n = 35; AVC intermediate n = 5; AVC partial n = 2; AVC unbalanced + AVSD with ventricular imbalance n = 1; other cardiac n = 1; pulmonary atresia (PA), IVS n = 1; subaortic stenosis n = 1; TOF n = 6; vascular ring + PA sling n = 1; VSD (inlet) n = 2; VSD (perimembranous) n = 13 and VSD (subarterial) n = 1, T18: TOF n = 1, 22qDS: DORV n = 1; IAA n = 4; mitral stenosis, subvalvar, parachute + mitral stenosis n = 1; other cardiac n = 1; PA, VSD n = 10; TOF n = 9; truncus arteriosus n = 12; vascular ring + PA sling n = 1 and VSD (perimembranous) n = 3, Turner: aortic stenosis (valvar) n = 1; CoA n = 4 and HLHS n = 2, mosaic Turner: CoA n = 1, William's: supravalvar aortic stenosis and XXX: PA, IVS.
Overall CNV Burden
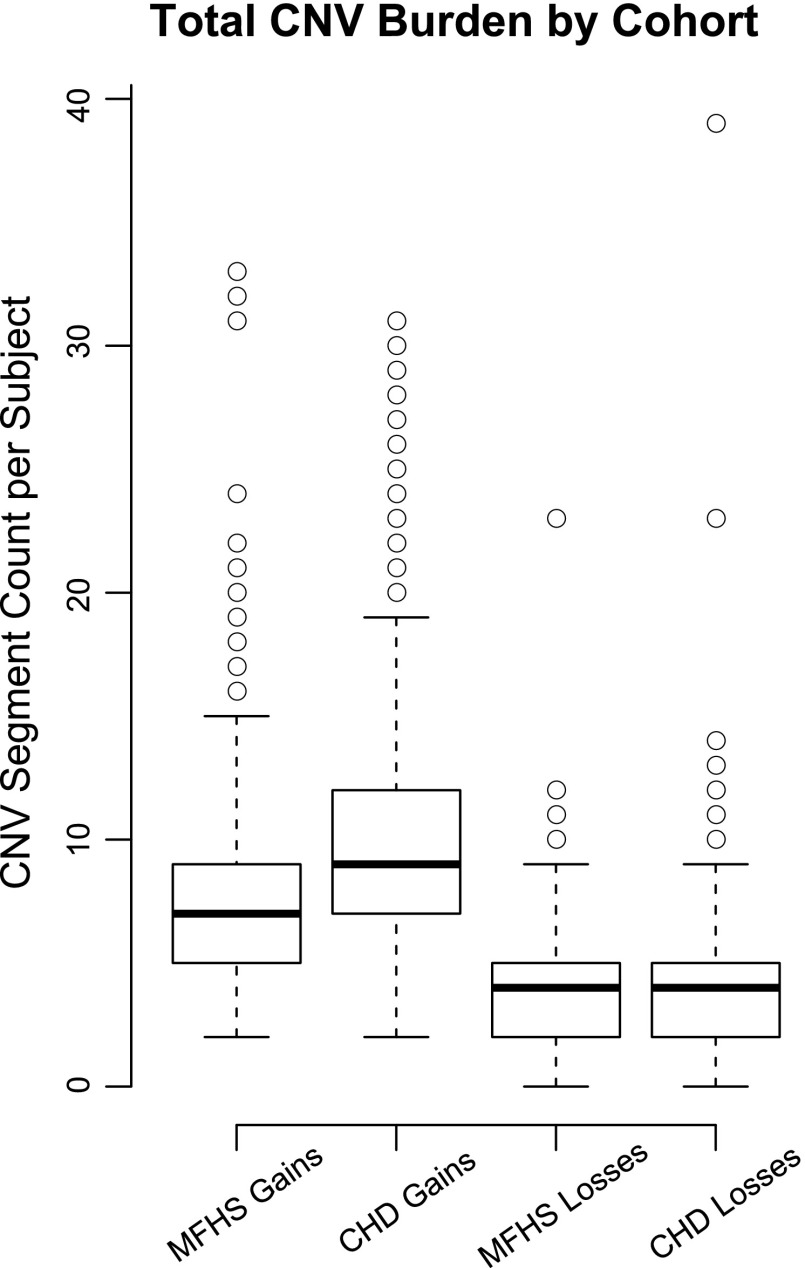
The total number of large CNVs (≥100 kb loss, ≥200 kb gain) throughout the genome were similar in both CHD and MFHS cohorts. When subjects with chromosomal abnormalities such as Trisomy 21 and 18, Turner, 22qDS, William's, and XXX were excluded, a significant number of the CHD cohort, 567 out of 810, carried a large rare CNV over a gene somewhere in their genome, while in the MFHS control cohort, this number was 391 of 880. Gains were twofold more common than losses in both cohorts despite the requirement to be twice as long (Fig. 3).
Fig. 3.
Total CNV burden by cohort. Standard box-and-whiskers plot for the distribution of large rare CNV segment count per subject in each of 4 cases: congenital heart disease (CHD) vs. Milwaukee Family Heart Study (MFHS) and gains vs. losses. Boxes represent the 1st and 3rd quartiles of each distribution, thick horizontal lines represent the median value, circles represent outliers, or the CHD cohort, major syndromes would significantly skew the distribution, so those subjects were excluded, leaving 810 syndrome-free subjects. Trisomy 21 and 18, Turner, 22qDS, William's and XXX chromosomal abnormalities were therefore excluded.
CHD Case Reports
Likely etiologic large, rare CNVs were identified in 35 CHD subjects. Table 5 summarizes the complete list of CHD subjects with CNVs over the known CHD risk gene regions (excluding the 135 subjects with known CHD-related chromosomal abnormalities). Three HLHS subjects (cases 16, 17, and 18) were studied for inheritance, a gain over FKBP6 was found to be a de novo event, a gain involving GATA4 and SOX7 was not present in one parent and the status of the other parent was unknown, and the MYH11 gain was inherited. Table 5 reports all of the known genes within each CNV segment, including our selected 100 CHD-associated genes.
Statistical Analysis of CNVs
Subphenotype analysis.
The CHD cohort, even after excluding genes involved in the known CHD-related chromosomal abnormalities, was enriched in large, rare CNVs involving CHD risk genes, where 35 of 810 subjects carried such a CNV (P ≤ 0.05 vs. both CHOP with 39 of 2,026 and MFHS with 14 of 880). Breaking this cohort into subgroups by specific phenotype often resulted in groups too small for statistical significance. Different subdivision schemes may achieve nominal significance. The entries in Table 6 where the frequency of CNV was significantly (P ≤ 0.05) different from the CHOP and MFHS cohorts are marked with a double asterisk. The CHD cohort, after excluding known causal chromosomal abnormalities, showed a frequency of CNV at 4.3%, and a power calculation is performed in Fig. 2 showing the difficulty in detecting a difference from the control's 1.9%. For subgroups of 10–25 individuals, the power to detect a difference from 1.9% (CHOP) required a proportion of 30 and 17%, respectively. Phenotypes showing significant (P ≤ 0.05) enrichment of large CNV events were aortic stenosis (valvar), AV canal (partial), AVSD with TOF, subaortic stenosis, TOF, and truncus arteriosus. Although HLHS was the most common phenotype in the CHD case cohort, this phenotype did not demonstrate significant large rare CNV enrichment.
Table 6.
CNV frequency by subphenotype
| Totals Including Causal Chromosomal Abnormalities |
Totals Excluding Causal Chromosomal Abnormalities |
|||||||
|---|---|---|---|---|---|---|---|---|
| Phenotype/Subphenotype | Subjects | Subjects with CNV Loss | Subjects with CNV Gain | Subjects with CNV Loss or Gain | Subjects | Subjects with CNV Loss | Subjects with CNV Gain | Subjects with CNV Loss or Gain |
| CHOP Cohort | 2,026 | 19 (0.94) | 20 (0.99) | 39 (1.92) | 2,026 | 19 (0.94) | 20 (0.99) | 39 (1.92) |
| MFHS Cohort | 880 | 3 (0.34) | 11 (1.25) | 14 (1.59) | 880 | 3 (0.34) | 11 (1.25) | 14 (1.59) |
| CHD Cohort | 945 | 66 (6.98) | 110 (11.64) | 172 (18.20)** | 810 | 12 (1.48) | 23 (2.84) | 35 (4.32)** |
| Turner | 8 | 8 (0.84) | 1 (0.10) | 8 | ||||
| Trisomy18 (T18) | 1 | 0 (0.00) | 1 (0.10) | 1 | ||||
| Trisomy21 (T21) | 80 | 0 (0.00) | 80 (8.35) | 80 | ||||
| Williams | 3 | 3 (0.31) | 0 (0.00) | 3 | ||||
| XXX | 1 | 0 (0.00) | 1 (0.10) | 1 | ||||
| 22qDS | 42 | 42 (4.38) | 1 (0.10) | 42 | ||||
| Aorto-pulmonary window + PDA | 5 | 0 (0.00) | 2 (40.00) | 2 (40.00)** | 3 | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Trisomy21 | 2 | 2 | 2 | |||||
| 22qDS | 0 | 0 | ||||||
| AVSD + TOF (AVSD + TOF) | 7 | 0 (0.00) | 6 (85.71) | 6 (85.71)** | 2 | 0 (0.00) | 1 (50.00) | 1 (50.00)** |
| Trisomy21 | 5 | 5 | 5 | |||||
| 22qDS | 0 | 0 | ||||||
| Aortic Stenosis (Valvar) | 31 | 3 (9.68) | 1 (3.23) | 4 (12.90)** | 30 | 2 (6.67) | 1 (3.33) | 3 (10.00)** |
| Turner | 1 | 1 | 1 | |||||
| Trisomy21 | 0 | 0 | ||||||
| 22qDS | 0 | 0 | ||||||
| Atrial Septal Defect Secundum (ASD-SEC) | 47 | 1 (2.13) | 4 (8.51) | 5 (10.64)** | 43 | 1 (2.33) | 0 (0.00) | 1 (2.33) |
| Trisomy21 | 4 | 4 | 4 | |||||
| 22qDS | 0 | 0 | ||||||
| Atrial Septal Defect Sinus Venosus (ASD-SV) | 13 | 1 (7.69) | 0 (0.00) | 1 (7.69) | 13 | 1 (7.69) | 0 (0.00) | 1 (7.69) |
| Trisomy21 | 0 | 0 | ||||||
| 22qDS | 0 | 0 | ||||||
| A-V Canal Complete (AVC Complete) | 48 | 0 (0.00) | 35 (72.92) | 35 (72.92)** | 13 | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Trisomy21 | 35 | 35 | 35 | |||||
| 22qDS | 0 | 0 | ||||||
| A-V Canal Intermediate (AVC Intermediate) | 7 | 0 (0.00) | 5 (71.43) | 5 (71.43)** | 2 | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Trisomy21 | 5 | 5 | 5 | |||||
| 22qDS | 0 | 0 | ||||||
| A-V Canal Partial (AVC Partial) | 17 | 1 (5.88) | 3 (17.65) | 4 (23.53)** | 15 | 1 (6.67) | 1 (6.67) | 2 (13.33)** |
| Trisomy21 | 2 | 2 | 2 | |||||
| 22qDS | 0 | 0 | ||||||
| A-V Canal Unbalanced + AVSD with ventricular imbalance | 14 | 0 (0.00) | 2 (14.29) | 2 (14.29)** | 13 | 0 (0.00) | 1 (7.69) | 1 (7.69) |
| Trisomy21 | 1 | 1 | 1 | |||||
| 22qDS | 0 | 0 | ||||||
| Coarctation of the Aorta (CoA) | 66 | 6 (9.09) | 2 (3.03) | 8 (12.12)** | 61 | 1 (1.64) | 1 (1.64) | 2 (3.28) |
| Turner | 5 | 5 | 1 | 5 | ||||
| Trisomy21 | 0 | 0 | ||||||
| 22qDS | 0 | 0 | ||||||
| Double Inlet Left Ventricle (DILV) | 19 | 0 (0.00) | 1 (5.26) | 1 (5.26) | 19 | 0 (0.00) | 1 (5.26) | 1 (5.26) |
| Trisomy21 | 0 | 0 | ||||||
| 22qDS | 0 | 0 | ||||||
| Double Outlet Right Ventricle (DORV) | 42 | 1 (2.38) | 3 (7.14) | 4 (9.52)** | 41 | 0 (0.00) | 2 (4.88) | 2 (4.88) |
| Trisomy21 | 0 | 0 | ||||||
| 22qDS | 1 | 1 | 1 | 1 | ||||
| Ebstein's Anomaly (EBSTEINS) | 9 | 0 (0.00) | 1 (11.11) | 1 (11.11) | 9 | 0 (0.00) | 1 (11.11) | 1 (11.11) |
| Trisomy21 | 0 | 0 | ||||||
| 22qDS | 0 | 0 | ||||||
| Hypoplastic Left Heart Syndrome (HLHS) | 140 | 2 (1.43) | 5 (3.57) | 7 (5.00)** | 138 | 0 (0.00) | 4 (2.90) | 4 (2.90) |
| Turner | 2 | 2 | 2 | |||||
| Trisomy21 | 0 | 0 | ||||||
| 22qDS | 0 | 0 | ||||||
| Interrupted Aortic Arch (IAA) | 11 | 4 (36.36) | 0 (0.00) | 4 (36.36)** | 7 | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Trisomy21 | 0 | 0 | ||||||
| 22qDS | 4 | 4 | 4 | |||||
| Mitral Valve Stenosis (MS, subvalvar, parachute) | 6 | 1 (16.67) | 0 (0.00) | 1 (16.67) | 5 | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Trisomy21 | 0 | 0 | ||||||
| 22qDS | 1 | 1 | 1 | |||||
| Other, Cardiac | 18 | 1 (5.56) | 2 (11.11) | 3 (16.67)** | 16 | 0 (0.00) | 1 (6.25) | 1 (6.25) |
| Trisomy21 | 1 | 1 | 1 | |||||
| 22qDS | 1 | 1 | 1 | |||||
| Pulmonary Atresia (PA) | 16 | 0 (0.00) | 0 (0.00) | 0 (0.00) | ||||
| -IVS- | 18 | 0 (0.00) | 2 (11.11) | 2 (11.11)** | ||||
| Trisomy21 | 1 | 1 | 1 | |||||
| XXX | 1 | 1 | 1 | |||||
| 22qDS | 0 | 0 | ||||||
| -VSD- | 34 | 10 (29.41) | 1 (2.94) | 11 (32.35)** | 24 | 0 (0.00) | 1 (4.17) | 1 (4.17) |
| Trisomy21 | 0 | 0 | ||||||
| 22qDS | 10 | 10 | 10 | |||||
| Subaortic stenosis | 12 | 0 (0.00) | 3 (25.00) | 3 (25.00)** | 11 | 0 (0.00) | 2 (18.18) | 2 (18.18)** |
| Trisomy21 | 1 | 1 | 1 | |||||
| 22qDS | 0 | 0 | ||||||
| Supravalvar AS | 4 | 3 (75.00) | 0 (0.00) | 3 (75.00)** | 1 | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Trisomy21 | 0 | 0 | ||||||
| Williams | 3 | 3 | 3 | |||||
| 22qDS | 0 | 0 | ||||||
| Tetralogy of Fallot (TOF) | 73 | 11 (15.07) | 10 (13.70) | 21 (28.77)** | 57 | 2 (3.51) | 3 (5.26) | 5 (8.77)** |
| Trisomy18 | 1 | 1 | 1 | |||||
| Trisomy21 | 6 | 6 | 6 | |||||
| 22qDS | 9 | 9 | 9 | |||||
| Tricuspid Atresia (TRI-AT) | 29 | 0 (0.00) | 2 (6.90) | 2 (6.90) | 29 | 0 (0.00) | 2 (6.90) | 2 (6.90) |
| Trisomy21 | 0 | 0 | ||||||
| 22qDS | 0 | 0 | ||||||
| Truncus Arteriosus (TA) | 29 | 14 (48.28) | 1 (3.45) | 14 (48.28)** | 17 | 2 (11.76) | 1 (5.88) | 2 (11.76)** |
| Trisomy21 | 0 | 0 | ||||||
| 22qDS | 12 | 12 | 12 | |||||
| Vascular ring and PA sling | 14 | 1 (7.14) | 1 (7.14) | 2 (14.29)** | 12 | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Trisomy21 | 1 | 1 | 1 | |||||
| 22qDS | 1 | 1 | 1 | |||||
| VSD inlet | 4 | 0 (0.00) | 2 (50.00) | 2 (50.00)** | 2 | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Trisomy21 | 2 | 2 | 2 | |||||
| 22qDS | 0 | 0 | ||||||
| Ventricular Septal Defect (VSD perimembranous) | 73 | 5 (6.85) | 15 (20.55) | 19 (26.03)** | 57 | 2 (3.51) | 2 | 3 (5.26) |
| Trisomy21 | 13 | 13 | 13 | |||||
| 22qDS | 3 | 3 | 3 | |||||
| Ventricular Septal Defect (VSD subarterial) | 7 | 0 (0.00) | 1 (14.29) | 1 (14.29) | 6 | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Trisomy21 | 1 | 1 | 1 | |||||
| 22qDS | 0 | 0 | ||||||
Significance over both CHOP and MFHS controls (P ≤ 0.05). Four patients had both gains and losses but are only counted once in the column “Subjects with CNV Loss or Gain”. The following subphenotypes contained 0 subjects with a CNV and were therefore removed from the table: Arrhythmias (Congenital Heart Block, Long QT, WPW), 7; Cardiomyopathy (DILATED), 13; Cardiomyopathy (HYPERTROPHIC), 4; Chest Wall, 4; Coronary Arteries (COR ART), 10; L-TGA, 7; Dilated Ascending Aorta (MARFAN), 8; Partial Anomalous Pulmonary Venous Return (PAPVR), 12; Pulmonary Stenosis (Valvar), 9; Shone's, 8; Total Anamolous Pulmonary Venous Connection (TAPVC; infracardiac, intracardiac, mixed, supracardiac), 15; Transposition of Great Arteries (IVS), 21; (VSD), 20; and Ventricular Septal Defect (VSD multiple + muscular), 10 (n = 161 total).
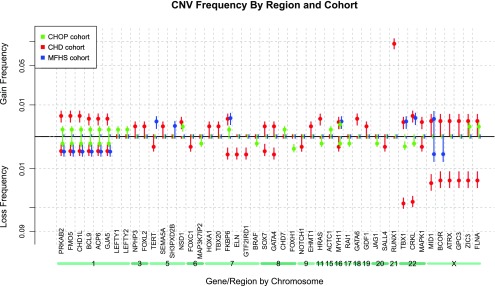
CNV gene frequency analysis and gene enrichment.
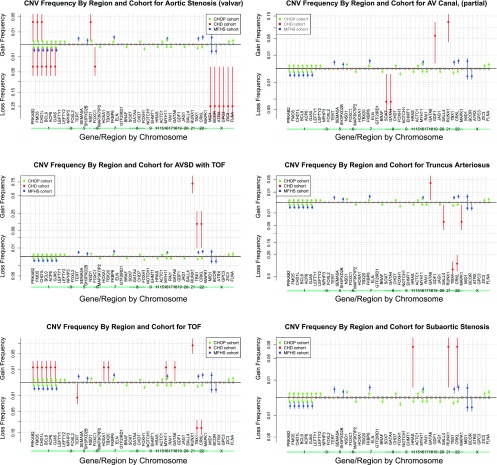
In addition, CNV frequency “spectra” were computed as a proportion of each cohort containing a gain or a loss over 100 CHD genes of interest (Fig. 4). (Spectra for individual CHD subphenotypes with statistically higher CNV frequencies are represented in Fig. 5.) The frequency of genes with gain or loss was compared with both control cohorts and significantly enriched genes are listed in Table 7. In addition, Supplemental Table S1 includes a complete summary of all CNV profiles over the 100 CHD risk gene list for each CHD subject, and a heatmap (Supplemental Fig. S1) illustrates the clustering of various groups of multiple subjects who share contiguous blocks of deleted or duplicated genes.
Fig. 4.
CNV frequency spectrum. Note that any gene showing 0% CNV frequency in all 3 cohorts was omitted from this figure due to space considerations. Vertical error bars drawn represent 1 SD from the mean in the estimated sampling distribution. From this visualization it is clear that gains over gene FKBP6 on chromosome 7 occur in all 3 cohorts, while losses of the same gene are only seen in the CHD cohort, implying a loss could cause CHD.
Fig. 5.
CNV frequency spectra of significantly enriched phenotypes. Note that any gene showing 0% CNV frequency in all 3 cohorts was omitted from this figure due to space considerations. Vertical error bars drawn represent 1 SD in the estimated sampling distribution. Significantly enriched phenotypes included: aortic stenosis (valvar), atrioventricular canal (partial), atrioventricular septal defect (AVSD) with tetralogy of Fallot (TOF), subaortic stenosis, TOF, and truncus arteriosus.
Table 7.
CHD-associated gene regions significantly enriched with large, rare CNVs
| Gains, % |
Losses, % |
Enriched For |
||||||
|---|---|---|---|---|---|---|---|---|
| Gene | CHD | CHOP | MFHS | CHD | CHOP | MFHS | Gains | Losses |
| PRKAB2 | 0.4 | 0.1 | 0.0 | 0.2 | 0.1 | 0.2 | √ | |
| FMO5 | 0.4 | 0.1 | 0.0 | 0.2 | 0.1 | 0.2 | √ | |
| CHD1L | 0.4 | 0.1 | 0.0 | 0.2 | 0.1 | 0.2 | √ | |
| BCL9 | 0.3 | 0.1 | 0.0 | 0.2 | 0.1 | 0.2 | √ | |
| ACP6 | 0.3 | 0.1 | 0.0 | 0.2 | 0.1 | 0.2 | √ | |
| GJA5 | 0.3 | 0.1 | 0.0 | 0.2 | 0.1 | 0.2 | √ | |
| FKBP6 | 0.3 | 0.1 | 0.3 | 0.3 | 0.0 | 0.0 | √ | |
| ELN | 0.0 | 0.0 | 0.0 | 0.3 | 0.0 | 0.0 | √ | |
| GTF2IRD1 | 0.0 | 0.0 | 0.0 | 0.3 | 0.0 | 0.0 | √ | |
| GATA4 | 0.1 | 0.0 | 0.0 | 0.3 | 0.0 | 0.0 | √ | |
| HRAS | 0.3 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | √ | |
| GATA6 | 0.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | √ | |
| RUNX1 | 8.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | √ | |
| CRKL | 0.4 | 0.0 | 0.3 | 4.2 | 0.1 | 0.0 | √ | |
| TBX1 | 0.2 | 0.0 | 0.2 | 4.4 | 0.1 | 0.0 | √ | |
| ATRX | 0.2 | 0.0 | 0.0 | 1.9 | 0.0 | 0.0 | √ | |
| GPC3 | 0.2 | 0.0 | 0.0 | 1.9 | 0.0 | 0.0 | √ | |
| BCOR | 0.2 | 0.0 | 0.0 | 1.9 | 0.0 | 0.3 | √ | |
| ZIC3 | 0.2 | 0.1 | 0.0 | 1.9 | 0.0 | 0.0 | √ | |
| FLNA | 0.2 | 0.1 | 0.0 | 1.9 | 0.0 | 0.0 | √ | |
| MID1 | 0.2 | 0.0 | 0.3 | 2.1 | 0.0 | 0.3 | √ | |
The statistical test applied was the Barnard's exact test. Of our 100 candidate genes, 21 were found to be significantly enriched for CNVs (null hypothesis rejected P ≤ 0.05 in both cohorts: CHD vs. CHOP and CHD vs. MFHS, see boldface). We used the full cohorts for genes in autosomal chromosomes, and only the female portion for any genes on chromosomes (Chr.) X or Y. This leaves 322/880 for MFHS, 416/945 for CHD, and an estimated 1,013/2,026 for CHOP, usable for testing on the Chr. X genes.
Numerous genes were identified as significantly enriched (P ≤ 0.05 against both control cohorts), including losses, FKBP6, ELN, GTF2IRD1, GATA4, CRKL, TBX1, ATRX, GPC3, BCOR, ZIC3, FLNA and MID1, and gains, PRKAB2, FMO5, CHD1L, BCL9, ACP6, GJA5, HRAS, GATA6, and RUNX1. These genes are identified in Table 7.
The authors recognize that syndromic forms of congenital heart disease are relatively well understood; therefore, genes in chromosomal abnormalities known to be causally related to CHD were intentionally kept on the 100 candidate CHD risk gene list to contrast with CNVs found elsewhere. For instance, haploinsufficiency of the genes associated with William's Syndrome, FKBP6, ELN, and GTF2IRD1, identified the three William's Syndrome patients in the study (1). Losses of the TBX1 and CRKL genes are associated with 22qDS and were observed in deleted subjects (32, 53). Turner syndrome subjects carrying losses on the chromosome X genes involving MID1, BCOR, ATRX, GPC3, ZIC3, and FLNA were identified, as well as a female subject (XXX) who was identified with gains over these chromosome X gene regions. In addition, duplications involving RUNX1 were primarily Trisomy 21 subjects.
Gains at 1q21.1 including PRKAB2, FMO5, CHD1L, BCL9, ACP6, and GJA5 were significantly enriched in this study; however, losses that were observed in both control cohorts as well as the CHD cohort were not. Interestingly, gains at 1q21.1 were previously reported in isolated sporadic TOF (18). In our case cohort we observed one subject (case 24) with TOF (2 contiguous CNVs, 0.6 and 1.6 Mb), one subject (case 20) with PA-VSD (1.5 Mb), and another (case 10) with CoA (1.5 Mb). One complex subject (case 2) with AS valvar and Shone's had a shorter gain (418 kb) involving only PRKAB2, FMO5, and CHD1L in conjunction with a 1.8 Mb gain at 5q35.2, which included the NSD1 gene.
Chromosome 8p23.1 deletions involving GATA4 were enriched and have been reported as a cause of complex congenital heart defects and diaphragmatic hernia (51). These included subjects with AVC partial (case 6, 3.8 Mb loss), VSD perimembranous (case 34, 4.5 Mb loss), and ASD-SV (case 5, 304 kb loss).
Three subjects had gains involving the HRAS gene. The first was found in a complex subject with coarctation of the aorta: in addition to a 284 kb duplication involving the HRAS gene the subject had Turner syndrome. The remaining two gains (case 12, 256 kb; case 22, 271 kb) were found in subjects with DILV and subaortic stenosis, respectively (Table 5). Cardiovascular malformations are known to be related to Ras/MAPK pathway syndromes, and previous literature findings have reported associations of HRAS mutations in Costello Syndrome and with the subaortic stenosis phenotype (29). These gains involving HRAS appear to expand phenotypes related to the Ras/MAPK pathway.
Enriched CNVs identified in Table 7 are previously reported or can be found in the Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources (DECIPHER) with the exception of the gains involving GATA6. One of the three gains involving GATA6 was in a subject with Trisomy 18 with TOF. The remaining two subjects with CNV gains involving GATA6 were 1) a subject (case 31) with truncus arteriosus with a complex CNV over two CHD genes of interest, a 308 kb gain including GATA6, and a 1.2 Mb 22q11.2 distal deletion involving MAPK1 (losses in the distal region of 22q11.2 have previously been reported in subjects with truncus arteriosus) (2), and 2) a subject (case 35) with VSD perimembranous with two neighboring 6.1 and 6.9 Mb gains involving a gain on GATA6. Although sequence variants in GATA6 have been previously found to be associated with cardiac outflow tract defects (27), these gains have not been reported and suggest possible GATA6 triple sensitivity to conotruncal defects.
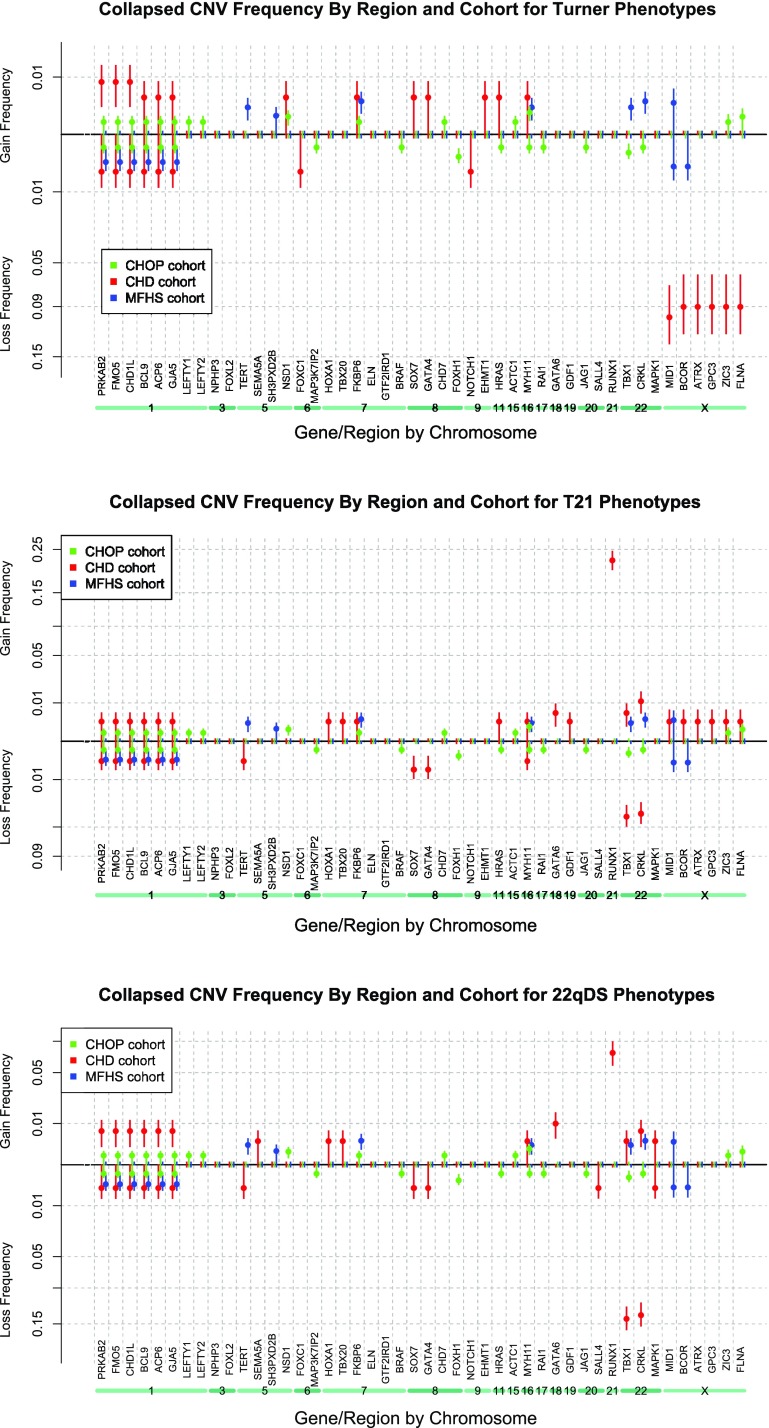
Collapsing groups of phenotypes by recognized causal chromosomal abnormalities.
To increase statistical power, a strategy for summing cohorts was employed; subphenotypes associated with T21, 22qDS, and Turner Syndrome (see Tables 4 and 6) were collapsed into three groups, respectively (33). We hypothesized collapsing subphenotypes into genetically related groups would increase power to detect additional related CNVs by phenotype. The three collapsed groups each demonstrated significant enrichment (P ≤ 0.05) of additional CNVs compared with both control cohorts (see Table 8 - Enriched Syndrome Genes and Fig. 6 - Spectra). Large, rare CNVs were significantly more frequent (P ≤ 0.05) in the groups of T21 subphenotypes and included gains involving GATA6 and RUNX1 and losses involving GATA4, SOX7, TBX1, and CRKL. Likewise, collapsing the HLHS, CoA, and AS (valvar) subphenotypes, which made up the Turner syndrome group, indicated significant gains involving the 1q21.1 gene regions, enriched losses involving the Chr. X genes, as well as gains involving GATA4, SOX7, EHMT1 (case 15), and HRAS and losses involving FOXC1 (case 3) and NOTCH1 (case 11). Although the T21 and 22qDS subclasses share some overlap of phenotypes (other cardiac, TOF, vascular ring/PA sling, and VSD perimembranous), it is interesting to note that the 22qDS grouping also included gains involving the 1q21.1 genes as well as GATA6 and RUNX1. Significant CNV losses within the 22qDS subclasses involved TBX1 and CRKL. All CNVs identified through the collapsed phenotypes are listed in Table 8 and are reported in DECIPHER.
Table 8.
Enriched syndrome genes
| 22q Like |
T21 Like |
Turner Like |
CHOP |
MFHS |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr. | Gene | Gain, % | Loss, % | Gain, % | Loss, % | Gain, % | Loss, % | Gain, % | Loss, % | Gain, % | Loss, % | ||||||
| 1 | ACP6 | √ | 0.67 | 0.33 | 0.27 | 0.27 | √ | 0.42 | 0.42 | 0.05 | 0.05 | 0.00 | 0.23 | ||||
| 1 | BCL9 | √ | 0.67 | 0.33 | 0.27 | 0.27 | √ | 0.42 | 0.42 | 0.05 | 0.05 | 0.00 | 0.23 | ||||
| 1 | CHD1L | √ | 0.67 | 0.33 | 0.27 | 0.27 | √ | 0.84 | 0.42 | 0.05 | 0.05 | 0.00 | 0.23 | ||||
| 1 | FMO5 | √ | 0.67 | 0.33 | 0.27 | 0.27 | √ | 0.84 | 0.42 | 0.05 | 0.05 | 0.00 | 0.23 | ||||
| 1 | GJA5 | √ | 0.67 | 0.33 | 0.27 | 0.27 | √ | 0.42 | 0.42 | 0.05 | 0.05 | 0.00 | 0.23 | ||||
| 1 | PRKAB2 | √ | 0.67 | 0.33 | 0.27 | 0.27 | √ | 0.84 | 0.42 | 0.05 | 0.05 | 0.00 | 0.23 | ||||
| 6 | FOXC1 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | √ | 0.42 | 0.00 | 0.00 | 0.00 | 0.00 | |||||
| 8 | GATA4 | 0.00 | 0.33 | 0.00 | √ | 0.55 | √ | 0.42 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||
| 8 | SOX7 | 0.00 | 0.33 | 0.00 | √ | 0.55 | √ | 0.42 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||
| 9 | EHMT1 | 0.00 | 0.00 | 0.00 | 0.00 | √ | 0.42 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |||||
| 9 | NOTCH1 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | √ | 0.42 | 0.00 | 0.00 | 0.00 | 0.00 | |||||
| 11 | HRAS | 0.00 | 0.00 | 0.27 | 0.00 | √ | 0.42 | 0.00 | 0.00 | 0.05 | 0.00 | 0.00 | |||||
| 18 | GATA6 | √ | 1.00 | 0.00 | √ | 0.55 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||
| 21 | RUNX1 | √ | 7.36 | 0.00 | √ | 22.25 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||
| 22 | CRKL | 0.67 | √ | 13.38 | 1.10 | √ | 3.57 | 0.00 | 0.00 | 0.00 | 0.05 | 0.34 | 0.00 | ||||
| 22 | TBX1 | 0.33 | √ | 14.05 | 0.55 | √ | 3.85 | 0.00 | 0.00 | 0.00 | 0.10 | 0.23 | 0.00 | ||||
| X | ATRX | 0.00 | 0.00 | 0.27 | 0.00 | 0.00 | √ | 8.99 | 0.00 | 0.00 | 0.00 | 0.00 | |||||
| X | BCOR | 0.00 | 0.00 | 0.27 | 0.00 | 0.00 | √ | 8.99 | 0.00 | 0.00 | 0.00 | 0.31 | |||||
| X | FLNA | 0.00 | 0.00 | 0.27 | 0.00 | 0.00 | √ | 8.99 | 0.10 | 0.00 | 0.00 | 0.00 | |||||
| X | GPC3 | 0.00 | 0.00 | 0.27 | 0.00 | 0.00 | √ | 8.99 | 0.00 | 0.00 | 0.00 | 0.00 | |||||
| X | MID1 | 0.00 | 0.00 | 0.27 | 0.00 | 0.00 | √ | 10.11 | 0.00 | 0.00 | 0.31 | 0.31 | |||||
| X | ZIC3 | 0.00 | 0.00 | 0.27 | 0.00 | 0.00 | √ | 8.99 | 0.10 | 0.00 | 0.00 | 0.00 | |||||
Boldface indicates significant values.
Fig. 6.
CNV frequency spectra of collapsed phenotypes by syndrome. Note that any gene showing 0% CNV frequency in all 3 cohorts was omitted from this figure due to space considerations. Vertical error bars drawn represent 1 SD in the estimated sampling distribution. Turner phenotypes, T21 phenotypes, and 22qDS phenotypes.
Additional findings of note include a gain involving TBX20 and loss involving SALL4. Three losses including TBX20 have been previously reported in subjects with CHD (ASD and VSDs) (26, 38). We identified a subject (case 26) with TOF with a 3.3 Mb gain involving TBX20 and an adjacent 4.7 Mb gain involving HOXA1, which has been reported in DECIPHER. Finally, we report a subject (case 32) with truncus arteriosus with a 1.8 Mb loss over the SALL4 gene, which has not been previously reported. This segment included a loss over NFATC2, a regulator of cardiac transcription factors but was not included in our 100 gene list because likely causal variants have not previously been reported in humans in this gene(9).
Distribution of CNVs by subject.
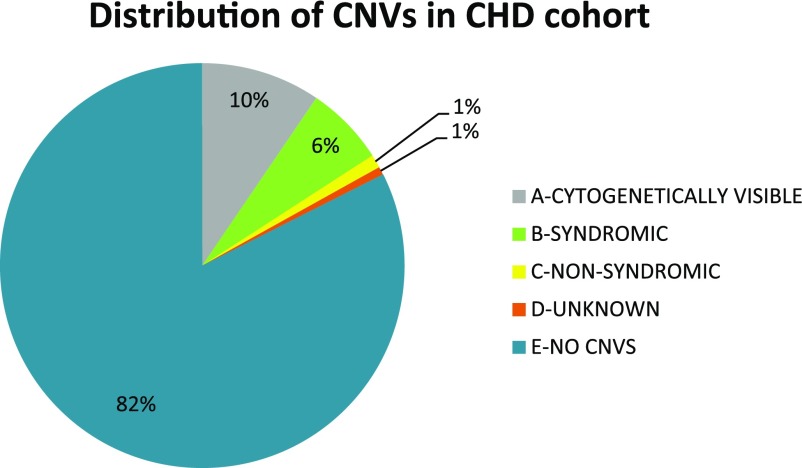
To characterize CHD study subjects with an approach more typically used in clinical genetics, CNVs were separated by size (whether or not they would be cytogenetically visible) and then the CHD WIKI site was employed to determine if remaining CNVs should be classified as involving a “syndromic” (two or more clinical features) or a “nonsyndromic” gene (1). Cytogenetically visible CNVs (category A) included chromosomal abnormalities ≥3 Mbps. This category contained subjects with Trisomy 21, 18; Turner; and XXX syndrome and represented ∼9% of the CHD cohort. Category B, contributing 6% to the overall CNV distribution, were those subjects with a CNV over a “syndromic-associated” CHD gene as reported by CHD WIKI (1). This subset contained 22qDS subjects (n = 42) with losses over the TBX1 gene, William's Syndrome subjects, all with a phenotype of supravalvar aortic stenosis (n = 3) with losses over the ELN, GTF2IRD1, and FKBP6 genes. The “nonsyndromic” segment (category C) representing 1% of the CHD cohort was also defined by the CHD WIKI portal. Six CHD case subjects, contributing 1% to the total, had a CNV over one of the 100 CHD-associated genes; however, their category was considered unknown. Category E represented individuals with no CNV over our predefined 100 CHD risk gene list. An individual could only fit into one category where D>A>B or C (see Fig. 7).
Fig. 7.
Distribution of CNVs in CHD cohort. Type A represents cytogenetically visible chromosomal abnormalities (≥3 Mbp), type B are those subjects with a CNV over a syndromic-associated CHD gene as reported by the CHD WIKI portal, type C are those recognized through CHD WIKI as nonsyndromic, type D are CNVs with an unknown category, and type E represents subjects with no CNV over our predefined 100 CHD-associated genes. An individual can only fit into 1 category where D>A>B or C. Numbers are rounded to the nearest percentage.
Complex CNVs.
Four basic mechanisms are involved in the generation of a majority of CNVs: deletion, duplication, inversion, and related combinations (56). We were interested if CHD subjects were at increased risk for carrying multiple CNVs. In the current study, 125 CHD subjects were defined as complex (methods). We identified 100 of those with known CHD-associated syndromes (T21, 59; T18, 1; 22qDS, 31; Turner, 6; William's, 2; XXX Syndrome, 1). Of the remaining 25, 24 contained likely causal CNVs for CHD as outlined in Table 5, whereas one subject contained a nonconfirmed CNV over a CHD-associated gene. Three complex subjects had CNVs on different chromosomes over two of our CHD associated genes of interest: subjects 2, 31, and 33 (Table 5). In addition, two subjects from the CHD cohort were both syndromic with their additional CNV over a second gene of interest: a Turner syndrome subject had a gain involving the HRAS gene and a 22qDS subject had an additional CNV involving a gain over the MAPK1 gene.
It is interesting to note that applying the “complex” criteria to the MFHS control cohort also identified 10 subjects from the controls that met the complex analysis requirements. These subjects had gains over the genes FKBP6, MYH11, TERT, TBX1, CRKL, SH3PXD2B, and losses over the 1q21.1 gene region and MID1.
DISCUSSION
CHD is a complex disease with demonstrated genetic etiology in a subset of patients. CNVs, viewed as an evolutionary driving force for new gene function resulting in improved survival and/or adaption to new environments and disease, contribute the largest component of natural human variation between any two individuals; indeed, CNVs contribute significantly more to inter-individual variation than SNPs (35, 39, 41). There is a broad range of CNV lengths. In this study we focused on large CNVs that can be detected with high accuracy and are relatively straightforward to confirm. It has previously been estimated that ∼65–80% of individuals have a large CNV (≥100 kb) and approximately three to seven CNV segments per individual (56). The average number of CNVs per subject in our CHD cohort supports these previous observations (Fig. 3). It is apparent that as CNV data continue to grow, the development of higher-resolution approaches will permit smaller CNV detection with better accuracy. This will potentially lead to additional disease association discoveries (23). However, data suggest that common CNVs (CNPs) are likely to be lower penetrance risk factors, whereas rare CNV variants are more likely to carry highly penetrant disease risk factors (13).
Significant challenges remain in CNV disease-association studies at both the platform and analysis levels (37). The relationship between phenotype and gene dosage is complex. Our study represents a comprehensive data curation and filtering of CNVs involving 100 recognized CHD risk genes detected in a large, anatomically phenotyped CHD population. We employed a strict algorithm to determine frequencies of CNVs involving regions that encompassed these CHD risk genes. The algorithm employed was very similar to a recent recommendation by Breckpot et al. (6) for determining if CNVs detected in CHD patients are clinically relevant; herein we performed a comparison against two different control populations and an analysis primarily based on known chromosomal abnormalities and gene content rather than more commonly used CNV detection approaches that prioritize by size. CNVs over these predefined gene regions were then used to search for relationships between cardiac phenotype and gene dosage.
The novel analytical approach described herein identified known causal chromosomal abnormalities (including T21, T18, 22qDS, Turner, William's, and XXX Syndromes), which represent 14% of CHD subjects in this study, similar to previous observations (36). Overall, this descriptive study suggests that (after excluding well-established causal chromosomal abnormalities) large, rare CNVs in 100 well-defined CHD risk genes confers significant risk of CHD and is likely etiologic in 4.3% of CHD cases, similar to previous observations (6). Cardiac subphenotypes showing the most significant (P ≤ 0.05) enrichment of large CNV events were aortic stenosis (valvar), AV canal (partial), AVSD with TOF, subaortic stenosis, TOF, and truncus arteriosus. CNV frequency spectra analysis identified enriched genes (P ≤ 0.05): losses: FKBP6, ELN, GTF2IRD1, GATA4, CRKL, TBX1, ATRX, GPC3, BCOR, ZIC3, FLNA, and MID1; and gains: PRKAB2, FMO5, CHD1L, BCL9, ACP6, GJA5, HRAS, GATA6, and RUNX1. 1q21.1 gains were enriched in subjects with conotruncal defects and coarctation of the aorta. 8p23.1 losses were enriched in subjects with septal defects and gains involving HRAS were observed in subaortic stenosis and DILV. Cardiovascular malformations are known to be related to Ras/MAPK pathway syndromes and previous literature findings have reported associations of HRAS mutations resulting in increased hRAS signaling with the subaortic stenosis phenotype. Other common phenotypes occurring in patients with hRAS mutations (also known as Costello syndrome) are cardiac hypertrophy (usually typical hypertrophic cardiomyopathy) and arrhythmia (usually supraventricular tachycardia, especially chaotic atrial rhythm/multifocal atrial tachycardia or ectopic atrial tachycardia) (43). Although DILV sometimes are associated with pulmonary stenosis we have not found any previous reports of hRAS mutations linked to this phenotype. Thus, our data appear to expand phenotypes related to the Ras/MAPK pathway.
We hypothesized that CNV frequency spectra combined with detailed anatomic classes would define the impact of gene dosage in etiologic molecular pathways. One set of clues when searching for genetic causes of CHD is given by the enrichment of CHD cases in various recognized causal chromosomal abnormalities such as T21, T18, 22qDS, Turner syndrome, and William's Syndrome (36). For instance, three of four (75%) subjects in our study with supravalvar aortic stenosis had deletions involving FKBP6, ELN, and GTF2IRD1 genes; all of these subjects had William's syndrome. In Turner syndrome, the incidence of CHD can be as high as 50% and include phenotypes such as BAV, CoA, ASD-VSD partially anomalus pulmonary vena cava, and HLHS, but these data vary (4). The specific cause for CHD in patients with Turner syndrome is currently unknown; several genes have been implicated but for the most part do not quite match Turner syndrome phenotypes or have only been associated with the syndrome by animal models (4, 5). In the present study, eight Turner syndrome subjects were easily identified from the total CHD cohort by CNV frequency spectra analysis. In these subjects, copy number losses were present on all six of the Chr. X genes (MID1, BCOR, ATRX, GPC3, ZIC3, and FLNA) that were selected as CHD-associated from our list of 100 genes. Two out of eight Turner cases in the study had HLHS, five had coarctation of the aorta, and one had aortic stenosis (valvar). Turner syndrome-associated phenotype percentages for the CHD cohort were in good agreement with published reports (4).
To test for additional CNV gene enrichment with increased power, subphenotypes associated with T21, 22qDS, and Turner Syndrome were collapsed in these groups, respectively. The three collapsed groups of subphenotypes each demonstrated enrichment (P ≤ 0.05) in additional CNVs compared with both control cohorts. Large, rare CNVs significantly increased (P ≤ 0.05) in the groups of T21 subphenotypes included gains over GATA6 and RUNX1 and losses over GATA4, SOX7, TBX1 and CRKL. Likewise, collapsing the HLHS, CoA, and AS (valvar) subphenotypes that made up the Turner syndrome group indicated significant gains over the 1q21.1 gene regions, enriched losses over the Chromosome X genes, as well as gains over likely etiologic genes such as GATA4, SOX7, EHMT1, and HRAS and losses over FOXC1 and NOTCH1. Although the T21 and 22qDS collapsed groups share some overlap of phenotypes, it is interesting to note that the 22qDS grouping also included gains over the 1q21.1 genes, as well as GATA6 and RUNX1. Significant CNV losses within the 22qDS subclasses were over TBX1 and CRKL. Incorporating gene dosage with detailed phenotyping into current molecular cardiogenesis models may allow future models of development to fine tune and increase our understanding of etiologic pathways.
By narrowing our focus on a select set of 100 well-known CHD risk genes, we limited the study by design. We focused on large rare CNVs in the current study; therefore, smaller CNVs have not yet been examined. Furthermore, the number of subjects per phenotype was small because of detailed anatomic groupings; collapsing into fewer groups (with larger n) according to developmental models would increase power and may permit identification of additional enriched genes. An additional foreseeable limitation was that CNVs may have been enriched in genes, but because the analysis required statistical significance with two control cohorts (where CNVs were not confirmed and may have been inflated and manifested as false positives), the study may not have been sufficiently powered to detect smaller but true differences.
To our knowledge, this is the first paper to curate a large and diverse CHD population with regard to subphenotype and CNV frequency by gene region. This appears to be a useful approach to visualize and eventually, given sufficient numbers, to quantify relative risk of CNVs for specific subphenotypes. Broadening to encompass the entire genome and performing the copy number spectra analysis at higher resolution should identify additional candidate genes in CHD. The ability to quantify risk of particular cardiac malformations by gene dosage should offer insight into critical molecular pathways impacted during human cardiogenesis. Furthermore, overlaying CNV data and details of resulting cardiac phenotype with known functional pathways of cardiogenesis should lead to increased understanding of the molecular etiology of heart malformations.
GRANTS
This study was supported in part by National Institutes of Health (NIH) Grant NIH1R21HD-060309-01 (A. Tomita-Mitchell), the CRI (A. Tomita-Mitchell and M. E. Mitchell), the Department of Surgery (A. Tomita-Mitchell, J. S. Tweddell, M. E. Mitchell), and in part by NIH Grant NIH5R01HL-089655 (U. Broeckel).
DISCLOSURES
A. Tomita-Mitchell, M. E. Mitchell, and C. A. Struble have conflicts of interest (significant financial interest in Aria Diagnostics, a molecular diagnostics company). M. Hidestrand and M. A. Goetsch have conflicts of interest (financial interest in Aria Diagnostics, a molecular diagnostics company).
AUTHOR CONTRIBUTIONS
Author contributions: A.T.-M., D.K.M., S.E.H., D.P.B., U.B., A.N.P., J.S.T., and M.E.M. conception and design of research; A.T.-M., D.K.M., C.A.S., M.E.T., K.D.S., M.H., S.E.H., M.A.G., P.M.S., D.P.B., U.B., A.N.P., and M.E.M. analyzed data; A.T.-M., D.K.M., C.A.S., M.E.T., K.D.S., M.H., P.M.S., D.P.B., A.N.P., J.S.T., and M.E.M. interpreted results of experiments; A.T.-M., D.K.M., M.E.T., K.D.S., M.A.G., and P.M.S. prepared figures; A.T.-M., D.K.M., M.E.T., M.H., P.M.S., and M.E.M. drafted manuscript; A.T.-M., D.K.M., C.A.S., M.E.T., K.D.S., M.H., S.E.H., M.A.G., P.M.S., D.P.B., U.B., A.N.P., and M.E.M. edited and revised manuscript; A.T.-M., D.K.M., C.A.S., M.E.T., K.D.S., M.H., S.E.H., M.A.G., P.M.S., D.P.B., U.B., A.N.P., J.S.T., and M.E.M. approved final version of manuscript; D.K.M., D.P.B., and U.B. performed experiments.
Supplementary Material
ACKNOWLEDGMENTS
First and foremost, we thank the families of the HHC from the CHW. We also acknowledge the significant contributions from the WPCR. We thank M. Krolikowski for excellent assistance with IRB contributions; the cardiothoracic surgical team including D. A. Jones, M. E. Barnes, J. L. Dunham-Ingle, T. A. Fehrenbacher, M. R. Madrzak, L. E. May, and R. G. Smith for research coordination; and the OR nursing staff for making this study possible. We gratefully acknowledge A. Klemm for assistance with the HHC database at CHW; the AGEN laboratory, especially R. B. Lorier, A. Mahter, A. Turner, B. Chirempes, and J. Wittke for technical assistance in the genetic laboratory; E. Semina for assistance in validations; S. Ghanta, J. Schroeder, A. Stoddard for helpful bioinformatic discussions; H. Liang for Tissue Bank support; and J. M. Routes, J. Lough, D. Beard, and B. Aronow for very helpful discussions and editing of this manuscript.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1. Barriot R, Breckpot J, Thienpont B, Brohee S, Van Vooren S, Coessens B, Tranchevent LC, Van Loo P, Gewillig M, Devriendt K, Moreau Y. Collaboratively charting the gene-to-phenotype network of human congenital heart defects. Genome Med 2: 16, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ben-Shachar S, Ou Z, Shaw CA, Belmont JW, Patel MS, Hummel M, Amato S, Tartaglia N, Berg J, Sutton VR, Lalani SR, Chinault AC, Cheung SW, Lupski JR, Patel A. 22q11.2 distal deletion: a recurrent genomic disorder distinct from DiGeorge syndrome and velocardiofacial syndrome. Am J Hum Genet 82: 214–221, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bittel DC, Butler MG, Kibiryeva N, Marshall JA, Chen J, Lofland GK, O'Brien JE., Jr Gene expression in cardiac tissues from infants with idiopathic conotruncal defects. BMC Med Genom 4: 1, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bondy CA. Congenital cardiovascular disease in Turner syndrome. Congenit Heart Dis 3: 2–15, 2008. [DOI] [PubMed] [Google Scholar]
- 5. Bondy CA, Cheng C. Monosomy for the X chromosome. Chromosome Res 17: 649–658, 2009. [DOI] [PubMed] [Google Scholar]
- 6. Breckpot J, Thienpont B, Arens Y, Tranchevent LC, Vermeesch JR, Moreau Y, Gewillig M, Devriendt K. Challenges of interpreting copy number variation in syndromic and non-syndromic congenital heart defects. Cytogenet Genome Res 135: 251–259, 2011. [DOI] [PubMed] [Google Scholar]
- 7. Breckpot J, Thienpont B, Peeters H, de Ravel T, Singer A, Rayyan M, Allegaert K, Vanhole C, Eyskens B, Vermeesch JR, Gewillig M, Devriendt K. Array comparative genomic hybridization as a diagnostic tool for syndromic heart defects. J Pediatr 156: 810–817, 2011. [DOI] [PubMed] [Google Scholar]
- 8. Chan ISF. Exact tests of equivalence and efficacy with a non-zero lower bound for comparative studies. Statist Med 1403–1413, 1998. [DOI] [PubMed] [Google Scholar]
- 9. Chang CP, Neilson JR, Bayle JH, Gestwicki JE, Kuo A, Stankunas K, Graef IA, Crabtree GR. A field of myocardial-endocardial NFAT signaling underlies heart valve morphogenesis. Cell 118: 649–663, 2004. [DOI] [PubMed] [Google Scholar]
- 10. Chinn A, Fitzsimmons J, Shepard TH, Fantel AG. Congenital heart disease among spontaneous abortuses and stillborn fetuses: prevalence and associations. Teratology 40: 475–482, 1989. [DOI] [PubMed] [Google Scholar]
- 11. Conti A, Fabbrini F, D'Agostino P, Negri R, Greco D, Genesio R, D'Armiento M, Olla C, Paladini D, Zannini M, Nitsch L. Altered expression of mitochondrial and extracellular matrix genes in the heart of human fetuses with chromosome 21 trisomy. BMC Genomics 8: 268, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat Genet 40: 1199–1203, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, Robson S, Vukcevic D, Barnes C, Conrad DF, Giannoulatou E, Holmes C, Marchini JL, Stirrups K, Tobin MD, Wain LV, Yau C, Aerts J, Ahmad T, Andrews TD, Arbury H, Attwood A, Auton A, Ball SG, Balmforth AJ, Barrett JC, Barroso I, Barton A, Bennett AJ, Bhaskar S, Blaszczyk K, Bowes J, Brand OJ, Braund PS, Bredin F, Breen G, Brown MJ, Bruce IN, Bull J, Burren OS, Burton J, Byrnes J, Caesar S, Clee CM, Coffey AJ, Connell JM, Cooper JD, Dominiczak AF, Downes K, Drummond HE, Dudakia D, Dunham A, Ebbs B, Eccles D, Edkins S, Edwards C, Elliot A, Emery P, Evans DM, Evans G, Eyre S, Farmer A, Ferrier IN, Feuk L, Fitzgerald T, Flynn E, Forbes A, Forty L, Franklyn JA, Freathy RM, Gibbs P, Gilbert P, Gokumen O, Gordon-Smith K, Gray E, Green E, Groves CJ, Grozeva D, Gwilliam R, Hall A, Hammond N, Hardy M, Harrison P, Hassanali N, Hebaishi H, Hines S, Hinks A, Hitman GA, Hocking L, Howard E, Howard P, Howson JM, Hughes D, Hunt S, Isaacs JD, Jain M, Jewell DP, Johnson T, Jolley JD, Jones IR, Jones LA, Kirov G, Langford CF, Lango-Allen H, Lathrop GM, Lee J, Lee KL, Lees C, Lewis K, Lindgren CM, Maisuria-Armer M, Maller J, Mansfield J, Martin P, Massey DC, McArdle WL, McGuffin P, McLay KE, Mentzer A, Mimmack ML, Morgan AE, Morris AP, Mowat C, Myers S, Newman W, Nimmo ER, O'Donovan MC, Onipinla A, Onyiah I, Ovington NR, Owen MJ, Palin K, Parnell K, Pernet D, Perry JR, Phillips A, Pinto D, Prescott NJ, Prokopenko I, Quail MA, Rafelt S, Rayner NW, Redon R, Reid DM, Renwick Ring SM, Robertson N, Russell E, Clair D, Sambrook JG, Sanderson JD, Schuilenburg H, Scott CE, Scott R, Seal S, Shaw-Hawkins S, Shields BM, Simmonds MJ, Smyth DJ, Somaskantharajah E, Spanova K, Steer S, Stephens J, Stevens HE, Stone MA, Su Z, Symmons DP, Thompson JR, Thomson W, Travers ME, Turnbull C, Valsesia A, Walker M, Walker NM, Wallace C, Warren-Perry M, Watkins NA, Webster J, Weedon MN, Wilson AG, Woodburn M, Wordsworth BP, Young AH, Zeggini E, Carter NP, Frayling TM, Lee C, McVean G, Munroe PB, Palotie A, Sawcer SJ, Scherer SW, Strachan DP, Tyler-Smith C, Brown MA, Burton PR, Caulfield MJ, Compston A, Farrall M, Gough SC, Hall AS, Hattersley AT, Hill AV, Mathew CG, Pembrey M, Satsangi J, Stratton MR, Worthington J, Deloukas P, Duncanson A, Kwiatkowski DP, McCarthy MI, Ouwehand W, Parkes M, Rahman N, Todd JA, Samani NJ, Donnelly P. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 464: 713–720, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cronk CE, Gangnon R, Cossette S, McElroy JA, Pelech AN. Modeling geographic risk of complex congenital heart defects in Eastern Wisconsin. Birth Defects Res A Clin Mol Teratol 91: 631–641, 2011. [DOI] [PubMed] [Google Scholar]
- 15. Erguler K. Barnard: Barnard's Unconditional Test. R package version 1.2. http://CRAN.R-project.org/package=Barnard.
- 16. Ferencz C, Correa-Villasenor A, Loffredo CA, Wilson PD. Malformations of the cardiac outflow tract. In: Genetic and Environmental Risk Factors of Major Cardiovascular Malformations: The Baltimore-Washington Infant Study: 1981–1989. Armonk: Futura Publishing, 1997, p. 59–102. [Google Scholar]
- 17. Franklin RC, Jacobs JP, Tchervenkov CI, Beland MJ. Bidirectional crossmap of the Short Lists of the European Paediatric Cardiac Code and the International Congenital Heart Surgery Nomenclature and Database Project. Cardiol Young 12: 431–435, 2002. [DOI] [PubMed] [Google Scholar]
- 18. Greenway SC, Pereira AC, Lin JC, DePalma SR, Israel SJ, Mesquita SM, Ergul E, Conta JH, Korn JM, McCarroll SA, Gorham JM, Gabriel S, Altshuler DM, Quintanilla-Dieck Mde L, Artunduaga MA, Eavey RD, Plenge RM, Shadick NA, Weinblatt ME, De Jager PL, Hafler DA, Breitbart RE, Seidman JG, Seidman CE. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet 41: 931–935, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guttenbach M, Koschorz B, Bernthaler U, Grimm T, Schmid M. Sex chromosome loss and aging: in situ hybridization studies on human interphase nuclei. Am J Hum Genet 57: 1143–1150, 1995. [PMC free article] [PubMed] [Google Scholar]
- 20. Harris S, Cronk C, Cassidy L, Simpson PM, Tomita-Mitchell A, Pelech AN. Exploring the environmental and genetic etiologies of congenital heart defects: the Wisconsin Pediatric Cardiac Registry. J Registry Manag 38: 24–29, 2011. [PubMed] [Google Scholar]
- 21. Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol 39: 1890–1900, 2002. [DOI] [PubMed] [Google Scholar]
- 22. Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet 36: 949–951, 2004. [DOI] [PubMed] [Google Scholar]
- 23. Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, Krauss RM, Myers RM, Ridker PM, Chasman DI, Mefford H, Ying P, Nickerson DA, Eichler EE. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet 84: 148–161, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jenkins KJ, Correa A, Feinstein JA, Botto L, Britt AE, Daniels SR, Elixson M, Warnes CA, Webb CL. Noninherited risk factors and congenital cardiovascular defects: current knowledge: a scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation 115: 2995–3014, 2007. [DOI] [PubMed] [Google Scholar]
- 25. Kathiriya IS, Srivastava D. Left-right asymmetry and cardiac looping: implications for cardiac development and congenital heart disease. Am J Med Genet 97: 271–279, 2000. [DOI] [PubMed] [Google Scholar]
- 26. Kirk EP, Sunde M, Costa MW, Rankin SA, Wolstein O, Castro ML, Butler TL, Hyun C, Guo G, Otway R, Mackay JP, Waddell LB, Cole AD, Hayward C, Keogh A, Macdonald P, Griffiths L, Fatkin D, Sholler GF, Zorn AM, Feneley MP, Winlaw DS, Harvey RP. Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am J Hum Genet 81: 280–291, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kodo K, Nishizawa T, Furutani M, Arai S, Yamamura E, Joo K, Takahashi T, Matsuoka R, Yamagishi H. GATA6 mutations cause human cardiac outflow tract defects by disrupting semaphorin-plexin signaling. Proc Natl Acad Sci USA 106: 13933–13938, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, Hubbell E, Veitch J, Collins PJ, Darvishi K, Lee C, Nizzari MM, Gabriel SB, Purcell S, Daly MJ, Altshuler D. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet 40: 1253–1260, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin AE, Alexander ME, Colan SD, Kerr B, Rauen KA, Noonan J, Baffa J, Hopkins E, Sol-Church K, Limongelli G, Digilio MC, Marino B, Innes AM, Aoki Y, Silberbach M, Delrue MA, White SM, Hamilton RM, O'Connor W, Grossfeld PD, Smoot LB, Padera RF, Gripp KW. Clinical, pathological, and molecular analyses of cardiovascular abnormalities in Costello syndrome: a Ras/MAPK pathway syndrome. Am J Med Genet A 155A: 486–507, 2011. [DOI] [PubMed] [Google Scholar]
- 30. Magri C, Sacchetti E, Traversa M, Valsecchi P, Gardella R, Bonvicini C, Minelli A, Gennarelli M, Barlati S. New copy number variations in schizophrenia. PLoS One 5: e13422, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature 461: 747–753, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Momma K. Cardiovascular anomalies associated with chromosome 22q11.2 deletion syndrome. Am J Cardiol 105: 1617–1624, 2010. [DOI] [PubMed] [Google Scholar]
- 33. Morgenthaler S, Thilly WG. A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: a cohort allelic sums test (CAST). Mutat Res 615: 28–56, 2007. [DOI] [PubMed] [Google Scholar]
- 34. Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, Lee C, Turner EH, Smith JD, Rieder MJ, Yoshiura K, Matsumoto N, Ohta T, Niikawa N, Nickerson DA, Bamshad MJ, Shendure J. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet 42: 790–793, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pang AW, MacDonald JR, Pinto D, Wei J, Rafiq MA, Conrad DF, Park H, Hurles ME, Lee C, Venter JC, Kirkness EF, Levy S, Feuk L, Scherer SW. Towards a comprehensive structural variation map of an individual human genome. Genome Biol 11: R52, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pierpont ME, Basson CT, Benson DW, Jr, Gelb BD, Giglia TM, Goldmuntz E, McGee G, Sable CA, Srivastava D, Webb CL. Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation 115: 3015–3038, 2007. [DOI] [PubMed] [Google Scholar]
- 37. Pinto D, Darvishi K, Shi X, Rajan D, Rigler D, Fitzgerald T, Lionel AC, Thiruvahindrapuram B, Macdonald JR, Mills R, Prasad A, Noonan K, Gribble S, Prigmore E, Donahoe PK, Smith RS, Park JH, Hurles ME, Carter NP, Lee C, Scherer SW, Feuk L. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotechnol 29: 512–520, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Posch MG, Gramlich M, Sunde M, Schmitt KR, Lee SH, Richter S, Kersten A, Perrot A, Panek AN, Al Khatib IH, Nemer G, Megarbane A, Dietz R, Stiller B, Berger F, Harvey RP, Ozcelik C. A gain-of-function TBX20 mutation causes congenital atrial septal defects, patent foramen ovale and cardiac valve defects. J Med Genet 47: 230–235, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M. Large-scale copy number polymorphism in the human genome. Science 305: 525–528, 2004. [DOI] [PubMed] [Google Scholar]
- 40. Shaikh TH, Gai X, Perin JC, Glessner JT, Xie H, Murphy K, O'Hara R, Casalunovo T, Conlin LK, D'Arcy M, Frackelton EC, Geiger EA, Haldeman-Englert C, Imielinski M, Kim CE, Medne L, Annaiah K, Bradfield JP, Dabaghyan E, Eckert A, Onyiah CC, Ostapenko S, Otieno FG, Santa E, Shaner JL, Skraban R, Smith RM, Elia J, Goldmuntz E, Spinner NB, Zackai EH, Chiavacci RM, Grundmeier R, Rappaport EF, Grant SF, White PS, Hakonarson H. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res 19: 1682–1690, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stankiewicz P, Sen P, Bhatt SS, Storer M, Xia Z, Bejjani BA, Ou Z, Wiszniewska J, Driscoll DJ, Maisenbacher MK, Bolivar J, Bauer M, Zackai EH, McDonald-McGinn D, Nowaczyk MM, Murray M, Hustead V, Mascotti K, Schultz R, Hallam L, McRae D, Nicholson AG, Newbury R, Durham-O'Donnell J, Knight G, Kini U, Shaikh TH, Martin V, Tyreman M, Simonic I, Willatt L, Paterson J, Mehta S, Rajan D, Fitzgerald T, Gribble S, Prigmore E, Patel A, Shaffer LG, Carter NP, Cheung SW, Langston C, Shaw-Smith C. Genomic and genic deletions of the FOX gene cluster on 16q241 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. Am J Hum Genet 84: 780–791, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stevens KN, Hakonarson H, Kim CE, Doevendans PA, Koeleman BP, Mital S, Raue J, Glessner JT, Coles JG, Moreno V, Granger A, Gruber SB, Gruber PJ. Common variation in ISL1 confers genetic susceptibility for human congenital heart disease. PLoS One 5: e10855, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA 101: 6062–6067, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tartaglia NR, Howell S, Sutherland A, Wilson R, Wilson L. A review of trisomy X (47,XXX). Orphanet J Rare Dis 5: 8, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Team RDC. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing, http://www.R-project.org, 2011. [Google Scholar]
- 46. Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, Gross C, Engelhardt S, Ertl G, Bauersachs J. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation 116: 258–267, 2007. [DOI] [PubMed] [Google Scholar]
- 47. Tomita-Mitchell A, Mahnke DK, Larson JM, Ghanta S, Feng Y, Simpson PM, Broeckel U, Duffy K, Tweddell JS, Grossman WJ, Routes JM, Mitchell ME. Multiplexed quantitative real-time PCR to detect 22q11.2 deletion in patients with congenital heart disease. Physiol Genomics 42A: 52–60, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. van Engelen K, Postma AV, van de Meerakker JB, Roos-Hesselink JW, Helderman-van den Enden AT, Vliegen HW, Rahman T, Baars MJ, Sels JW, Bauer U, Pickardt T, Sperling SR, Moorman AF, Keavney B, Goodship J, Klaassen S, Mulder BJ. Ebstein's anomaly may be caused by mutations in the sarcomere protein gene MYH7. Neth Heart J [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 320: 539–543, 2008. [DOI] [PubMed] [Google Scholar]
- 50. Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wat MJ, Shchelochkov OA, Holder AM, Breman AM, Dagli A, Bacino C, Scaglia F, Zori RT, Cheung SW, Scott DA, Kang SH. Chromosome 8p23.1 deletions as a cause of complex congenital heart defects and diaphragmatic hernia. Am J Med Genet A 149A: 1661–1677, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wong AK, Fang B, Zhang L, Guo X, Lee S, Schreck R. Loss of the Y chromosome: an age-related or clonal phenomenon in acute myelogenous leukemia/myelodysplastic syndrome? Arch Pathol Lab Med 132: 1329–1332, 2008. [DOI] [PubMed] [Google Scholar]
- 53. Yagi H, Furutani Y, Hamada H, Sasaki T, Asakawa S, Minoshima S, Ichida F, Joo K, Kimura M, Imamura S, Kamatani N, Momma K, Takao A, Nakazawa M, Shimizu N, Matsuoka R. Role of TBX1 in human del22q11.2 syndrome. Lancet 362: 1366–1373, 2003. [DOI] [PubMed] [Google Scholar]
- 54. Yanai I, Benjamin H, Shmoish M, Chalifa-Caspi V, Shklar M, Ophir R, Bar-Even A, Horn-Saban S, Safran M, Domany E, Lancet D, Shmueli O. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21: 650–659, 2005. [DOI] [PubMed] [Google Scholar]
- 55. Yatsenko SA, Shaw CA, Ou Z, Pursley AN, Patel A, Bi W, Cheung SW, Lupski JR, Chinault AC, Beaudet AL. Microarray-based comparative genomic hybridization using sex-matched reference DNA provides greater sensitivity for detection of sex chromosome imbalances than array-comparative genomic hybridization with sex-mismatched reference DNA. J Mol Diagnost 11: 226–237, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Ann Rev Genom Hum Genet 10: 451–481, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang J, Feuk L, Duggan GE, Khaja R, Scherer SW. Development of bioinformatics resources for display and analysis of copy number and other structural variants in the human genome. Cytogenet Genome Res 115: 205–214, 2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.