Abstract
Elevated heart rate (HR) is a risk factor for cardiovascular diseases. The goal of the study was to map HR trait in mice using quantitative trait locus (QTL) analysis followed by genome-wide association (GWA) analysis. The first approach provides mapping power and the second increases genome resolution. QTL analyses were performed in a C3HeB×SJL backcross. HR and systolic blood pressure (SBP) were measured by the tail-cuff plethysmography. HR was ∼80 beats/min higher in SJL compared with C3HeB. There was a wide distribution of the HR (536–763 beats/min) in N2 mice. We discovered a highly significant QTL (logarithm of odds = 6.7, P < 0.001) on chromosome 7 (41 cM) for HR in the C3HeB×SJL backcross. In the Hybrid Mouse Diversity Panel (58 strains, n = 5–6/strain) we found that HR (beats/min) ranged from 546 ± 12 in C58/J to 717 ± 7 in MA/MyJ mice. SBP (mmHg) ranged from 99 ± 6 in strain I/LnJ to 151 ± 4 in strain BXA4/PgnJ. GWA analyses were done using the HMDP, which revealed a locus (64.2–65.1 Mb) on chromosome 7 that colocalized with the QTL for elevated HR found in the C3HeB×SJL backcross. The peak association was observed for 17 SNPs that are localized within three GABAA receptor genes. In summary, we used a combined genetic approach to fine map a novel elevated HR locus on mouse chromosome 7.
Keywords: quantitative trait locus, genome-wide association, chromosome 7, GABAA
cardiovascular disease (CVD) is the leading cause of death in the United States (22). Hypertension affects >30% of Americans. Increases in systolic blood pressure (SBP) strongly correlate with a greater risk of cardiovascular events including myocardial infarction, congestive heart failure, and renal disease. Increases in SBP are often accompanied by elevations in heart rate (HR) (26). However, elevated HR can be a CVD risk factor independent of increased SBP (9, 17, 24–26, 30a). Genetic approaches have been very helpful in understanding gene contributions to hemodynamic parameters both in rodents and humans. Specifically, quantitative trait locus (QTL) analyses and genome-wide association (GWA) studies have become powerful tools for discovering genomic regions that house causal genes for SBP and HR.
Genetic studies in mice have led to the discovery of many important loci for SBP that have homology to human chromosomes. QTL analyses on progeny from inbred mouse crosses have identified several genomic regions that contribute to changes in SBP. Woo and Kurtz (35) identified four QTLs in a backcross of A/J (A) and C57BL/6J (C57BL) mice. One QTL, Abbp4, on mouse chromosome (chr) 11, was homologous to a confirmed locus found on human chr17 (15). In another cross between normotensive C3H/HeJ (C3H) and hypertensive SWR/J (SWR) mice two QTLs for SBP were found on chrs1 and 16 (7). The QTL on chr1, Bpq8, was found to be in concordance with a human QTL (23). Similar approaches have also been used for identifying QTLs for HR. Utilization of a small panel of inbred and recombinant inbred mouse strains for linkage analyses found genomic loci for HR and HR variability on chrs6 and 5, respectively (14). A QTL was also found on chr4 in an electrocardiographic study of a mouse model of sodium channelopathy with cardiac conduction dysfunction (28), which coincides with another locus found on chr4 in a cross of DBA/2J (DBA) and C57BL mouse strains (5). Such studies in mice have the potential to reveal pathways contributing to common forms of hypertension in humans. Unfortunately, because of the poor mapping resolution of QTL analyses, such studies have not led to the identification of causal genes and pathways (11).
Recently, Bennett et al. (4) showed that GWA analyses of a panel of inbred strains of mice, the Hybrid Mouse Diversity Panel (HMDP), exhibited mapping resolution that was at least an order of magnitude better than linkage analysis. The improved resolution is due to the fact that association takes advantage of historical recombination whereas linkage resolution relies on recombination in the genetic cross. Thus, GWA studies have been important in identifying genes for complex traits in humans. For example, a locus on human chr9 was discovered using a GWA study in a population of American Indians, leading to the identification of a new gene, KIAA1797, that regulates HR (25).
In this study we combined both genetic linkage and association approaches to identify and fine map loci that control hemodynamic traits. For linkage analyses we used a recently described cross between C3HeB/FeJ (C3HeB) and SJL/J (SJL) strains for vascular remodeling traits (19). We then performed GWA for HR in the HMDP (58 inbred strains) for greater genomic resolution of the genetic loci. We successfully identified a novel locus on chr7 that regulates elevated HR in mice using two independent genetic approaches.
MATERIALS AND METHODS
Animals.
Details of C3HeB×SJL backcross were recently described (19). Briefly, we used males and females in our experiments between 9 and 12 wk of age. We were able to measure hemodynamic phenotypes in all experimental mice: C3HeB = 17, SJL = 17, F1 = 13, N2 = 134. Male mice (9–10 wk old) of 58 inbred mouse strains (n = 322) were purchased from the Jackson Laboratory (Table 1). The University of Rochester Animal Care Committee approved all procedures on animals, which were conducted according to guidelines of the National Institutes of Health for the Use of Laboratory Animals.
Table 1.
List of inbred mice
| Strain # | Strain Name (abbreviation; n) | Strain # | Strain Name (abbreviation; n) |
|---|---|---|---|
| 1 | 129X1/SvJ (129X1; 5) | 31 | AXB-1/PgnJ (AXB1; 6) |
| 2 | A/J (A; 6) | 32 | AXB-2/PgnJ (AXB2; 6) |
| 3 | AKR/J (AKR; 5) | 33 | AXB-4/PgnJ (AXB4; 6) |
| 4 | BALB/cJ (BALB; 5) | 34 | AXB-5/PgnJ (AXB5; 6) |
| 5 | BTBRT+tf/J (BTBRT; 6) | 35 | AXB-6/PgnJ (AXB6; 6) |
| 6 | BUB/BnJ (BUB; 6) | 36 | AXB-8/PgnJ (AXB8; 6) |
| 7 | C3H/HeJ (C3H; 5) | 37 | AXB-10/PgnJ (AXB10; 6) |
| 8 | C3HeB/FeJ (C3HeB; 5) | 38 | AXB-12/PgnJ (AXB12; 5) |
| 9 | C57BL/6J (C57BL; 5) | 39 | AXB-13/PgnJ (AXB13; 6) |
| 10 | C57L/J (C57L; 5) | 40 | AXB-15/PgnJ (AXB15; 6) |
| 11 | C58/J (C58; 6) | 41 | AXB-19/PgnJ (AXB19; 5) |
| 12 | CBA/J (CBA; 6) | 42 | AXB-19a/PgnJ (AXB19a; 6) |
| 13 | CE/J (CE; 5) | 43 | AXB-19b/PgnJ (AXB19b; 6) |
| 14 | DBA/2J (DBA; 5) | 44 | AXB-23/PgnJ (AXB23; 4) |
| 15 | FVB/NJ (FVB; 6) | 45 | AXB-24/PgnJ (AXB24; 4) |
| 16 | I/LnJ (I; 5) | 46 | BXA-1/PgnJ (BXA1; 6) |
| 17 | KK/HIJ (KK; 6) | 47 | BXA-2/PgnJ (BXA2; 6) |
| 18 | LG/J (LG; 5) | 48 | BXA-4/PgnJ (BXA4; 5) |
| 19 | LP/J (LP; 5) | 49 | BXA-7/PgnJ (BXA7; 6) |
| 20 | MA/MyJ (MA; 6) | 50 | BXA-8/PgnJ (BXA8; 5) |
| 21 | NOD/LtJ (NOD; 6) | 51 | BXA-11/PgnJ (BXA11; 5) |
| 22 | NON/LtJ (NON; 5) | 52 | BXA-12/PgnJ (BXA12; 6) |
| 23 | NZB/BINJ (NZB; 5) | 53 | BXA-13/PgnJ (BXA13; 6) |
| 24 | NZW/LacJ (NZW; 6) | 54 | BXA-14/PgnJ (BXA14; 6) |
| 25 | PL/J (PL; 5) | 55 | BXA-16/PgnJ (BXA16; 6) |
| 26 | RIIIS/J (RIIIS; 6) | 56 | BXA-24/PgnJ (BXA24; 6) |
| 27 | SEA/GnJ (SEA; 5) | 57 | BXA-25/PgnJ (BXA25; 6) |
| 28 | SJL/J (SJL; 6) | 58 | BXA-26/PgnJ (BXA26; 6) |
| 29 | SM/J (SM; 5) | ||
| 30 | SWR (SWR; 6) |
n, Number of mice in each strain.
Physiological measurements.
Mice from the backcross and the HMDP (Table 1) were trained for tail-cuff plethysmography (Visitech System), and SBP and HR were measured as described (18). In brief, mice were trained for 5 days, and recordings from the last 2 days were used.
Linkage analysis in the C3HeB×SJL backcross.
Genotyping of the backcrossed mice was recently published by our group (19). Briefly, we used 80 microsatellite markers (ABI). We used MapManager QTX for linkage analyses as described (19).
GWA.
GWA mapping was done using efficient mixed-model association (EMMA) method, which corrects for population structure and genetic relatedness in model organism association mapping (16). Genome-wide significance P value thresholds (10−6) were used based on power simulations in a similar number of strains (16). The GWA analysis and EMMA algorithm were performed on the UCLA web-based server (http://mouse.cs.ucla.edu/emmaserver). The current HMDP genotype variation includes about 4 million single nucleotide polymorphisms (SNPs). We used a publically available Genome Browser from the University of California Santa Cruz web-server (http://uswest.ensembl.org) to visualize the significant peaks on the mouse genome.
Linkage disequilibrium.
Genotypes for linkage disequilibrium (LD) and identical by descent (IBD) analyses were obtained from Perlegen database (http://mouse.cs.ucla.edu/mousehapmap/full.html). Imputed genotypes covering 5.2 Mb with high-confidence (error rate for 8 million SNPs at 0.37%) were used for chr7 for 30 inbred strains (out of 94 included in total panel) and 28 RI strains (out of 40 in total panel). Statistical analyses and data visualizations were carried out using the R (version 2.13.1) statistical software (available at http://cran.r-project.org/). For LD, pair-wise correlation coefficients between pairs of SNPs were calculated using the “corFast” function in the WGCNA R package (21), which provides computationally more efficient alternative to the “cor” function to compute Pearson's correlation coefficients. The calculated correlation coefficients were then raised to the power of 2 to calculate the r-squared values. The resulting r-squared matrix was used to generate the LD plot using the “LDheatmap” R package (29). To calculate IBD between C3HeB and SJL strains, we counted the number of polymorphic markers within 10 Kb blocks using 2 Kb sliding windows beginning from the start of the 0.9 Mb region on chr7. For candidate gene analyses, functional variations in candidate genes were obtained from Ensembl genome browser (http://www.ensembl.org/index.html), and each variation was compared against the list of Perlegen's polymorphic SNPs for strains C3HeB and SJL. The liver expression [expression QTL (eQTL)] data from the cross between C57BL and DBA was obtained from http://geneeqtl.genetics.ucla.edu/ website, previously published (12).
Statistical analysis.
Results are reported as means ± SE. Statistical tests were done using JMP5.1.2, except for linkage and association analyses. Differences between three or more groups were analyzed by one-way ANOVA followed by post hoc comparisons of all means by Tukey-Kramer honestly significant difference test. The level of P < 0.05 was regarded as significant.
RESULTS
Hemodynamic traits in the C3HeB×SJL backcross.
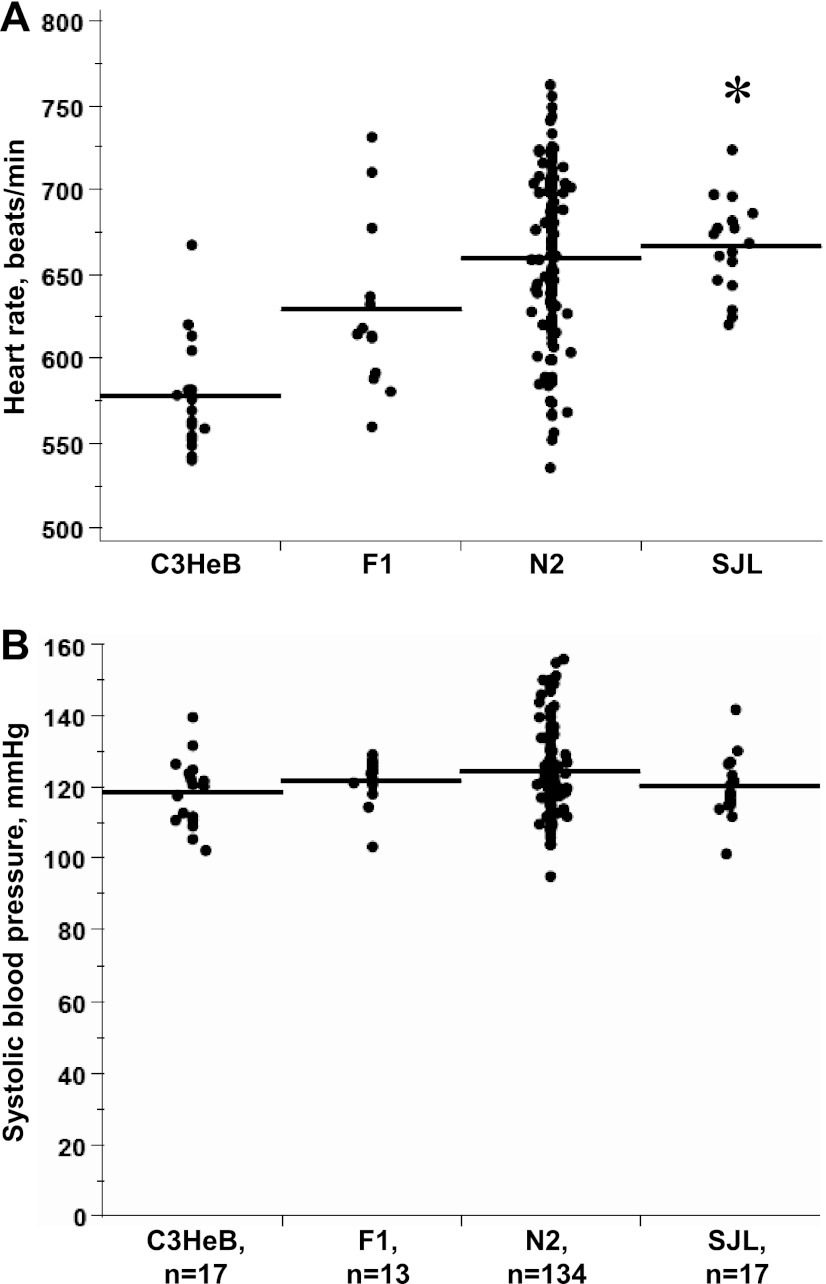
As previously reported by our group (20), HR was ∼80 beats/min higher in SJL compared with C3HeB (Fig. 1A). This 14% elevation of HR was similar between males and females in N2 progeny (not shown). However, there were no differences in SBP between C3HeB and SJL mice (Fig. 1B), which we previously observed (20). Mean SBP was not statistically different among C3HeB, SJL, F1, and N2 progeny (Fig. 1B). As expected, N2 progeny exhibited a wide range of HR consistent with the polygenic regulation of HR (Fig. 1A).
Fig. 1.
Hemodynamic phenotypes in C3HeB×SJL backcross. A: heart rate (HR), beats/min. B: systolic blood pressure (BP), mmHg. Individual values from each genetic progeny are plotted. Line shows mean values. n, Number of mice in each group. *P < 0.05 vs. C3HeB.
Linkage analyses in the C3HeB×SJL backcross.
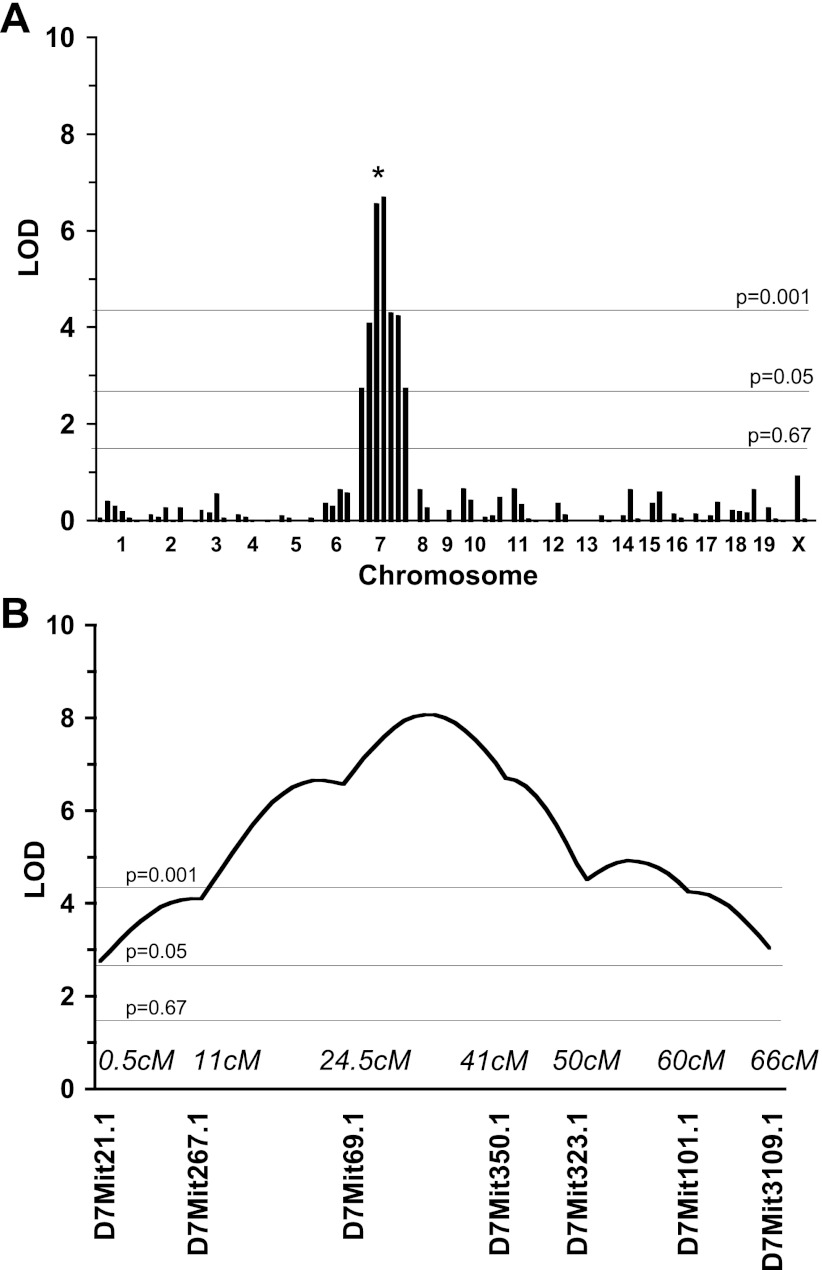
Using interval mapping we identified a significant locus on chr7 [logarithm of odds (LOD) = 6.7, P < 0.001; Fig. 2, Table 2]. It should be noted that HR was greater in the N2 progeny with SJL/SJL compared with C3HeB at D7Mit69.1 and D7Mit350.1 (not shown). Independent analysis of males and females revealed the absence of sex effect at this locus (not shown). We also found a suggestive QTL on the proximal part of chr7 for SBP in the backcross (Table 2).
Fig. 2.
Quantitative trait loci (QTLs) linked to HR in N2 mice in C3HeB×SJL backcross. A: whole genome scan. Each bar represents a single microsatellite marker. Thresholds are indicated by thin lines that were established by permutation tests. B: interval mapping of the HR on chromosome 7.
Table 2.
Significant and suggestive hemodynamic QTLs in the C3HeB×SJL backcross
| Chr | Trait | Locus (cM) | LOD | CI, cM | Variance, % |
|---|---|---|---|---|---|
| 7 | heart rate | D7Mit21.1 (0.5) | 2.8 | 44 | 9 |
| 7 | heart rate | D7Mit267.1 (11) | 4.1 | 30 | 13 |
| 7 | heart rate | D7Mit69.1 (24.5) | 6.6 | 20 | 20 |
| 7 | heart rate | D7Mit350.1 (41) | 6.7 | 19 | 21 |
| 7 | heart rate | D7Mit323.1 (50) | 4.3 | 29 | 14 |
| 7 | heart rate | D7Mit101.1 (60) | 4.3 | 29 | 14 |
| 7 | heart rate | D7Mit109.1 (66) | 2.8 | 44 | 9 |
| 7 | systolic blood pressure | D7Mit21.1 (0.5) | 1.7 | 69 | 6 |
QTL, quantitative trait locus; Chr, chromosome; LOD, logarithm of odds; CI, confidence interval. Thresholds: highly significant at P = 0.001; significant at P = 0.05; suggestive at P = 0.67. Variance (%), a percentage of the total phenotypic variance detected in the N2 with each locus shown linkage was associated. Significant markers are shown in boldface.
Variation of hemodynamic parameters in the HMDP.
To map the elevated HR locus on chr7 with improved resolution, we examined the HMDP for variability in SBP and HR (Figs. 3 and 4). Hemodynamic traits were normally distributed in the HMDP (insets, Figs. 3 and 4). Analyses of variances showed significant differences for SBP (F ratio = 14.1, P < 0.0001) and HR (F ratio = 13.4, P < 0.0001) across 58 inbred strains. The post hoc comparisons of hemodynamic parameters are shown in Tables 3 and 4 for SBP and HR, respectively. Levels not connected by the same reference letter (in boldface; referred to as “A”, “B,” and “C”) are significantly different. For example, in SBP the mean (± SE) values measured in mmHg ranged from 99 ± 6 in I to 151 ± 4 in BXA4 strain (Fig. 3, Table 3). HR values in SJL were statistically different from 22 strains (Table 4). This phenotypic characterization of HMDP mice revealed that HR (± SE), measured by beats/min, ranged from 546 ± 12 in C58 to 717 ± 7 in MA mice (Fig. 4).
Fig. 3.
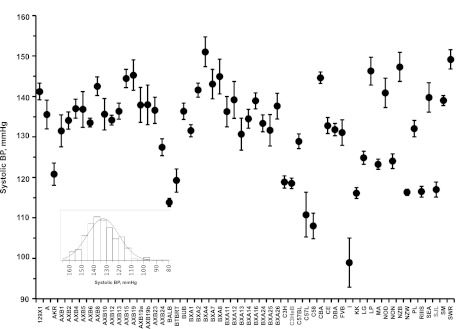
Variations in systolic BP in the Hybrid Mouse Diversity Panel (HMDP). Strains in gray color were used for the backcross. Inset: data distribution. Statistical comparisons across 58 strains are shown in Table 3.
Fig. 4.
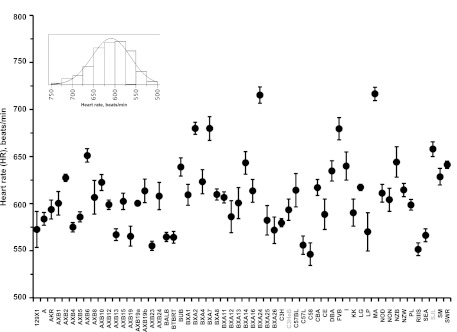
Variations in HR in the HMDP. Strains in gray color were used for the backcross. Inset: data distribution. Statistical comparisons across 58 strains are shown in Table 4.
Table 3.
Statistical comparisons of systolic blood pressure across 58 inbred strains
| Level | Mean | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BXA4 | A | 151.2 | |||||||||||||||
| SWR | A | B | 149.2 | ||||||||||||||
| AXB1 | A | B | 149.0 | ||||||||||||||
| NZB | A | B | C | 147.2 | |||||||||||||
| AXB19 | A | B | C | D | 145.4 | ||||||||||||
| BXA8 | A | B | C | D | 144.8 | ||||||||||||
| CBA | A | B | C | D | 144.7 | ||||||||||||
| LP | A | B | C | D | 144.6 | ||||||||||||
| AXB15 | A | B | C | D | 144.5 | ||||||||||||
| BXA7 | A | B | C | D | 142.8 | ||||||||||||
| AXB8 | A | B | C | D | 142.7 | ||||||||||||
| BXA2 | A | B | C | D | E | 141.5 | |||||||||||
| 129X1 | A | B | C | D | E | F | 141.4 | ||||||||||
| NOD | A | B | C | D | E | F | 140.8 | ||||||||||
| SEA | A | B | C | D | E | F | G | 139.8 | |||||||||
| BXA12 | A | B | C | D | E | F | G | 139.2 | |||||||||
| SM | A | B | C | D | E | F | G | 139.0 | |||||||||
| BXA16 | A | B | C | D | E | F | G | 139.0 | |||||||||
| AXB19b | A | B | C | D | E | F | G | H | 137.8 | ||||||||
| AXB19a | A | B | C | D | E | F | G | H | 137.8 | ||||||||
| BXA26 | A | B | C | D | E | F | G | H | 137.5 | ||||||||
| AXB5 | A | B | C | D | E | F | G | H | 137.0 | ||||||||
| AXB4 | A | B | C | D | E | F | G | H | 137.0 | ||||||||
| AXB23 | A | B | C | D | E | F | G | H | I | J | 136.8 | ||||||
| AXB13 | A | B | C | D | E | F | G | H | J | 136.3 | |||||||
| BUB | A | B | C | D | E | F | G | H | J | 136.3 | |||||||
| BXA11 | A | B | C | D | E | F | G | H | I | J | 136.2 | ||||||
| AXB10 | A | B | C | D | E | F | G | H | I | J | 135.7 | ||||||
| A | A | B | C | D | E | F | G | H | I | J | 135.3 | ||||||
| BXA14 | A | B | C | D | E | F | G | H | I | J | 134.5 | ||||||
| AXB2 | A | B | C | D | E | F | G | H | I | J | 134.0 | ||||||
| AXB12 | A | B | C | D | E | F | G | H | I | J | K | 134.0 | |||||
| AXB6 | B | C | D | E | F | G | H | I | J | 133.5 | |||||||
| BXA24 | B | C | D | E | F | G | H | I | J | K | 133.5 | ||||||
| CE | B | C | D | E | F | G | H | I | J | K | L | 132.8 | |||||
| PL | B | C | D | E | F | G | H | I | J | K | L | 132.2 | |||||
| DBA | B | C | D | E | F | G | H | I | J | K | L | M | 131.8 | ||||
| BXA25 | C | D | E | F | G | H | I | J | K | L | 131.5 | ||||||
| BXA1 | C | D | E | F | G | H | I | J | K | L | M | 131.3 | |||||
| FVB | C | D | E | F | G | H | I | J | K | L | M | 131.0 | |||||
| BXA13 | C | D | E | F | G | H | I | J | K | L | M | 130.7 | |||||
| C57BL | D | E | F | G | H | I | J | K | L | M | N | 128.8 | |||||
| AXB24 | D | E | F | G | H | I | J | K | L | M | N | 127.5 | |||||
| LG | E | F | G | H | I | J | K | L | M | N | O | 124.8 | |||||
| NON | F | G | H | I | J | K | L | M | N | O | 124.0 | ||||||
| MA | G | H | I | J | K | L | M | N | O | 123.3 | |||||||
| AKR | H | I | J | K | L | M | N | O | 120.6 | ||||||||
| BTBRT | I | K | L | M | N | O | 119.3 | ||||||||||
| C3H | I | J | K | L | M | N | O | 119.2 | |||||||||
| C3HeB | I | K | L | M | N | O | 118.4 | ||||||||||
| SJL | K | L | M | N | O | 117.0 | |||||||||||
| RIIIS | L | M | N | O | P | 116.5 | |||||||||||
| NZW | L | M | N | O | P | 116.3 | |||||||||||
| KK | L | M | N | O | P | 116.2 | |||||||||||
| BALB | M | N | O | P | 114.0 | ||||||||||||
| C57L | N | O | P | 110.8 | |||||||||||||
| C58 | O | P | 108.2 | ||||||||||||||
| I | P | 99.2 |
Strains not connected by the same letter are significantly different.
Table 4.
Statistical comparisons of heart rate across 58 mouse inbred strains
| Level | Mean | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MA | A | 716.7 | ||||||||||||||
| BXA24 | A | 715.5 | ||||||||||||||
| BXA2 | A | B | 679.8 | |||||||||||||
| BXA7 | A | B | 679.7 | |||||||||||||
| FVB | A | B | 679.5 | |||||||||||||
| SJL | A | B | C | 658.0 | ||||||||||||
| AXB | B | C | D | 651.3 | ||||||||||||
| NZB | B | C | D | E | F | 644.4 | ||||||||||
| BXA14 | B | C | D | E | 643.3 | |||||||||||
| SWR | B | C | D | E | F | 641.3 | ||||||||||
| I | B | C | D | E | F | 640.0 | ||||||||||
| BUB | B | C | D | E | F | 639.0 | ||||||||||
| DBA | B | C | D | E | F | G | 634.6 | |||||||||
| SM | B | C | D | E | F | G | H | I | 628.6 | |||||||
| AXB2 | B | C | D | E | F | G | I | 627.3 | ||||||||
| BXA4 | B | C | D | E | F | G | H | I | J | 623.4 | ||||||
| AXB10 | B | C | D | E | F | G | H | I | J | 622.3 | ||||||
| LG | B | C | D | E | F | G | H | I | J | K | L | 617.4 | ||||
| CBA | C | D | E | F | G | H | I | J | K | 617.2 | ||||||
| NZW | C | D | E | F | G | H | I | J | K | L | 614.7 | |||||
| C57BL | C | D | E | F | G | H | I | J | K | L | M | 614.0 | ||||
| AXB13 | C | D | E | F | G | H | I | J | K | L | 613.7 | |||||
| BXA16 | C | D | E | F | G | H | I | J | K | L | 613.5 | |||||
| NOD | C | D | E | F | G | H | I | J | K | L | M | 611.2 | ||||
| BXA8 | C | D | E | F | G | H | I | J | K | L | M | N | 609.6 | |||
| BXA1 | C | D | E | F | G | H | I | J | K | L | M | N | 609.3 | |||
| AXB24 | C | D | E | F | G | H | I | J | K | L | M | N | O | 608.0 | ||
| BXA11 | C | D | E | F | G | H | I | J | K | L | M | N | O | 606.8 | ||
| AXB8 | C | D | E | F | G | H | I | J | K | L | M | N | 606.5 | |||
| NON | C | D | E | F | G | H | I | J | K | L | M | N | O | 604.2 | ||
| AXB12 | C | D | E | F | G | H | I | J | K | L | M | N | O | 602.4 | ||
| AXB23 | C | D | E | F | G | H | I | J | K | L | M | N | O | 600.5 | ||
| BXA13 | C | D | E | F | G | H | I | J | K | L | M | N | O | 600.5 | ||
| AXB1 | C | D | E | F | G | H | I | J | K | L | M | N | O | 600.3 | ||
| AXB15 | C | D | E | F | G | H | I | J | K | L | M | N | O | 598.8 | ||
| PL | C | D | E | F | G | H | I | J | K | L | M | N | O | 598.6 | ||
| C3HeB | D | E | F | G | H | I | J | K | L | M | N | O | 593.6 | |||
| AKR | D | E | F | G | H | I | J | K | L | M | N | O | 593.6 | |||
| KK | E | F | G | H | I | J | K | L | M | N | O | 590.7 | ||||
| CE | D | E | F | G | H | I | J | K | L | M | N | O | 589.0 | |||
| BXA12 | E | F | G | H | I | J | K | L | M | N | O | 586.2 | ||||
| AXB5 | E | F | G | H | I | J | K | L | M | N | O | 585.8 | ||||
| A | E | F | G | H | I | J | K | L | M | N | O | 583.8 | ||||
| BXA25 | F | G | H | I | J | K | L | M | N | O | 582.7 | |||||
| C3H | F | G | H | I | J | K | L | M | N | O | 579.8 | |||||
| AXB4 | G | H | I | J | K | L | M | N | O | 574.8 | ||||||
| 129X1 | G | H | I | J | K | L | M | N | O | 572.8 | ||||||
| BXA26 | G | H | I | J | K | L | M | N | O | 571.7 | ||||||
| AXB19b | H | J | K | L | M | N | O | 567.2 | ||||||||
| SEA | H | I | J | K | L | M | N | O | 566.6 | |||||||
| AXB19 | H | I | J | K | L | M | N | O | 565.6 | |||||||
| BALB | H | I | J | K | L | M | N | O | 564.6 | |||||||
| BTBRT | J | K | L | M | N | O | 564.5 | |||||||||
| C57L | K | L | M | N | O | 556.0 | ||||||||||
| AXB19a | L | M | N | O | 555.3 | |||||||||||
| RIIIS | M | N | O | 551.8 | ||||||||||||
| LP | N | O | 546.6 | |||||||||||||
| C58 | O | 546.3 |
Strains not connected by the same letter are significantly different.
GWA analysis HR trait in the HMDP.
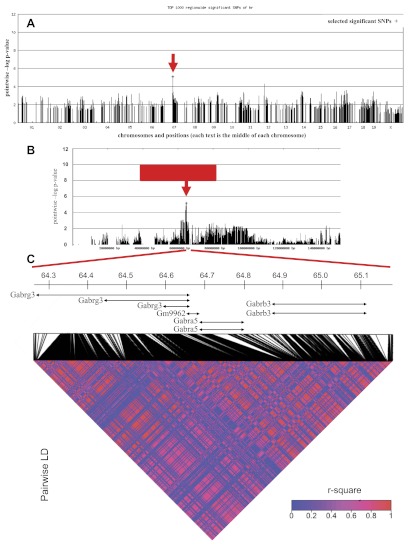
To validate the chr7 locus and further identify genomic loci contributing to HR, we performed GWA analysis in the subset of HMDP inbred mouse strains. The initial GWA analysis identified several significant loci for HR at the genome-wide cutoff of 1-e06 (not shown). After we corrected for population structure, however, none of the identified loci passed the stringent genome-wide threshold mainly due to lack of statistical power associated with limited number of strains used in our study. Despite the limited power in GWA study, however, the locus identified in the backcross on chr7 exhibited evidence for association (P < 0.001) with the peak marker D7Mit350.1 at 41 cM (Fig. 5A). This finding further validated the linkage results on chr7 (Figs. 5B and 2A).
Fig. 5.
Genome-wide association mapping of hemodynamic parameters in the HDMP. A: genome-wide association for HR. SNP, single nucleotide polymorphism. B: chromosome 7 that contains elevated HR locus. Red block represents the QTL, and the red arrow indicates the peak within the QTL for elevated HR in the C3HeB×SJL backcross. C: linkage disequilibrium (LD) heat map for SNPs from 64.2 to 65.1 Mb.
LD of SNPs in the HMDP panel.
The GWA peak marker on the chr7 locus was located at 64.9 Mb. To identify the potential candidate genes, we followed two complementary approaches to define the boundaries of the region and to fine-map the locus to a smaller region. First, we defined the confidence interval for the region based on the extent of regional LD in the HMDP panel. In the full HMDP panel consisting of 150 inbred and recombinant inbred strains, the locus on chr7 is located within an LD block extending from 64.2 to 65.1 Mb (not shown). To investigate whether this 0.9 Mb LD block were also present in the subset of HMDP strains utilized in our study, we calculated pair-wise r-square for every pair of SNPs in the region using genotypes for 1,459 polymorphic SNPs obtained from the Perlegen website. This analysis revealed that the majority of the SNPs in the 0.9 Mb region are highly correlated with each other, leading to the conclusion that the extent of LD for the chr7 locus in the subset of 58 inbred strains in our study is similar to the LD block in the complete HMDP panel (Fig. 5C). Consequently, the high confidence interval for elevated HR locus is 0.9 Mb beginning at 64.2 Mb and ending at 65.1 Mb on chr7. This locus contains a cluster of three gamma-aminobutyric acid (GABA) receptor subunit genes, Gabrb3, Gabra5, and Gabrg3, encoding receptor subunits for the inhibitory neurotransmitter GABAA. We found 17 SNPs that are localized either near or within these three genes (Table 5). In particular, one SNP was located inside the Gabrg3 gene, one inside Gabra5, three inside Gabrb3, and 12 SNPs in intergenic regions (Table 5). We found strong correlation between SNP alleles and HR variation in HMDP in all 17 SNPs excluding rs32461823, which is located inside the Gabrg3 gene. Interestingly, there is a functional variant resulting in a nonsense mutation between C3HeB and SJL in the coding sequence of Gabrg3 (not shown). The presence of GABA genes, which primarily function in brain, as the only genes in this region, suggests that genetic regulation of HR by chr7 is mainly due to the cellular pathways operating in brain. Alternatively, GABA genes could have functions outside the brain that regulate HR. Consistent with this, we found a cis-eQTL (LOD 6.7) in female F2s from a genetic cross between C57BL and DBA strains for Gabrb3 in the liver (12), suggesting a role for Gabrb3 outside the central nervous system. To fine-map the locus, we searched the chr7 locus to identify regions that are IBD between C3HeB and SJL mouse strains. The presence of one or more IBD indicates common ancestral genotype between two strains and, as such, highlights regions with low probability of having variation between the two strains. For this, we utilized 1,753 SNP genotypes available in the Perlegen database encompassing the 0.9 Mb locus identified in both the backcross and the GWA analysis and calculated the number of SNPs within the 10 Kb region using 2 Kb sliding windows. These analyses revealed the absence of IBD between C3HeB and SJL in the locus, leaving the entire 0.9 Mb genomic region as the locus containing the causal variants for elevated HR (Fig. 6). The IBD analysis strengthens our SJL×C3HeB cross QTL data, in that it shows that SJL mice are genetically distinct from C3HeB in the 0.9 Mb locus on chr7 (Fig. 6).
Table 5.
Significant SNPs on chr7 associated with heart rate trait in the HMDP
| P Value | Position, bp | rsID | Variation | Gene Name | Location |
|---|---|---|---|---|---|
| 9.50E-06 | 64560058 | rs32461823 | C/T | Gabrg3 | intron |
| 9.50E-06 | 64743240 | rs32261123 | C/A | Gabra5 | intron |
| 9.50E-06 | 64793391 | rs37051088 | A/G | ||
| 9.50E-06 | 64793478 | rs38734523 | G/A | ||
| 9.50E-06 | 64806025 | rs37878797 | A/T | ||
| 9.50E-06 | 64806096 | rs37291782 | A/G | ||
| 9.50E-06 | 64809420 | rs36856753 | C/T | ||
| 9.50E-06 | 64816242 | rs37328406 | T/C | ||
| 9.50E-06 | 64829670 | rs31944466 | T/G | ||
| 9.50E-06 | 64834798 | rs32195065 | T/C | ||
| 9.50E-06 | 64835087 | rs31278322 | T/C | ||
| 9.50E-06 | 64835490 | rs32188009 | C/T | ||
| 9.50E-06 | 64837449 | rs31889005 | A/C | ||
| 9.50E-06 | 64843819 | rs31247999 | A/T | ||
| 9.50E-06 | 64844063 | rs31920657 | C/A | Gabrb3 | cSNP |
| 9.50E-06 | 64844605 | rs38033329 | G/A | Gabrb3 | cSNP |
| 7.80E-06 | 64926113 | rs38530916 | T/A | Gabrb3 | intron |
SNP, single nucleotide polymorphism; HMDP, Hybrid Mouse Diversity Panel.
Fig. 6.
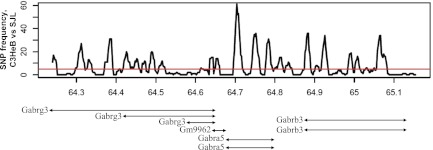
Analysis of QTL regions that are identical by descent. Peaks above the red line indicate high probability of SNP variation between strains. SJL mice are genetically distinct from C3HeB. A threshold of SNP frequency was 5.
DISCUSSION
The major finding of our study is that a novel genomic locus on chr7 regulates elevated HR in mice. We used two complementary approaches including linkage analysis in the C3HeB×SJL backcross and GWA analysis with the HMDP (30 inbred and 28 AXB/BXA RI strains). In the linkage analysis, we demonstrated that HR was significantly higher in the SJL compared with C3HeB mice and that HR exhibited a broad phenotypic spectrum in the N2 progeny from the C3HeB×SJL backcross (Fig. 1). We also found a highly significant QTL (LOD = 6.7, P < 0.0001) on chr7 (41 cM) for HR in the C3HeB×SJL backcross (Fig. 2). We observed a continuous distribution of both SBP and HR traits in the HMDP (Figs. 3 and 4 and Tables 3 and 4). The most important finding from GWA analysis was the locus on chr7, which colocalized with the QTL found in the C3HeB×SJL backcross (Fig. 5, A and B). Fine-mapping in the chr7 HR locus indicates that three GABAA receptor genes, Gabrb3, Gabra5, and Gabrg3, are strong candidates for regulation of HR (Figs. 5 and 6).
Several studies have found genetic loci that control HR in mice and humans (Table 6). Previously, eight loci had been reported to regulate HR in mice. A study using a cross of C57BL and DBA strains (5) and another study investigating a specific mutation attributed to conduction disease in FVB and 129P2 mice (28) each found HR loci localized on chr4. Significant QTLs were also identified on chr2 (hrq1) in the BALB×CBA backcross (32) and on chr6 (hr1) using AXA/BXA recombinant mouse inbred strains (14). A number of genes were identified on chr1 including corticotropin releasing factor 2 and neuropeptide Y (14). However, when increasing the number of strains to 58 in the HDMP we identified one locus on chr7, which coincided with a novel locus identified in the C3HeB×SJL cross.
Table 6.
Heart rate controling genetic loci in mice and humans
| Study | Strains | Chr | Peaks | LOD | QTL |
|---|---|---|---|---|---|
| Mouse | |||||
| Sugiyama et al., 2002 | BALB × CBA | 2 | 72 cM | 4.0 | Hrq1 |
| 15 | 25 cM | 3.1 | Hrq2 | ||
| Howden et al., 2008 | AXB and BXA recombinant inbred strains | 6 | 54 Mb | 3.8 | Hr1 |
| Blizard et al., 2009 | C57BL × DBA | 1 | 72 cM | 7.9 | Hrq4 |
| 4 | 2 cM | 3.2 | |||
| 5 | 54 cM | 8.5 | Hrq5 | ||
| 11 | 2 cM | 3.1 | Hrq6 | ||
| Scicluna et al., 2011 | FVB/NJ and 129P2; mutant mice for gene Scn5a-1798insD/+ | 4 | 136–151 Mb | 4.2 | |
| Human | |||||
| Singh et al., 2002 | Framingham Heart Study | 2 | 62 cM | 1.8 | |
| 15 | 153 cM | 1.8 | |||
| Martin et al., 2004 | Metabolic Risk Complications of Genes Obesity Project | 4 | 128 cM | 3.9 | |
| Spielmann et al., 2007 | HERITAGE Family Study | 3 | 103 Mb | 1.7 | HR50 |
| 9 | 118 Mb | 1.9 | HR50 | ||
| 10 | 102 Mb | 1.5 | HR50 | ||
| 111 Mb | 1.6 | HR50 | |||
| 111 Mb | 2.1 | HR60 | |||
| 113 Mb | 2.0 | HR60 | |||
| 116 Mb | 1.9 | HR60 | |||
| 116 Mb | 1.9 | HR60 | |||
| 117 Mb | 1.8 | HR60 | |||
| 18 | 42 Mb | 1.7 | HR50 | ||
| Melton et al., 2010 | Strong Heart Family Study in American Indians | 9 | 4.8 | ||
| Eijgelsheim et al., 2010 | RRGEN Study | 1 | |||
| 6 | |||||
| 7 | |||||
| 11 | |||||
| 12 | |||||
| 14 | |||||
Increased SBP, particularly in hypertension, is commonly accompanied by increases in HR (26). However, SBP and HR traits can be regulated independently of each other. Sugiyama et al. (32) found two QTLs for each of these traits in a mouse cross between CBA and BALB mice. It is likely that these QTLs harbor a number of genes that may interact to influence both hemodynamic traits. Blizard et al. (5) phenotyped mice prior to the onset of hypertension and found different loci that control SBP (chr4 and chr14) vs. HR (chr1 and chr5). Similarly, linkage analysis in a population of normotensive humans found a locus for HR on chr4 and identified two candidate genes, ankyrin-B and myozenin 2, that potentially influence resting HR (24). Our study provides additional evidence for genomic control of resting HR as we identified a novel QTL on chr7 for HR but no significant QTL for SBP using linkage analysis. This was strengthened by the GWA analysis, which confirmed that HR is regulated by a genetic locus on mouse chr7. GWA analysis, however, did reveal loci for SBP variation on several chrs (none on chr7) with the HMDP (not shown). This indicates that SBP is regulated by far more complex genetic mechanisms with a likely contribution from multiple loci, whereas elevated HR was linked to only one locus on chr7. Our study provides new insight into genomic control of resting HR while strongly supporting independent genetic regulation of HR and SBP traits.
SNP analysis of the QTL on chr7 identified polymorphisms in the GABA receptor family genes and one predicted gene Gm9962. Specifically, GABA receptors Gabrb3, Gabra5, and Gabrg3 localize in a 0.9 Mb region of chr7. LD analysis of this region indicated that at 64.9 Mb there was significant SNP correlation for Gabrg3. GABA is an inhibitory neurotransmitter and its multisubunit receptors are important for regulating neuronal excitability and muscle tone, including nonneuronal tissues (33). Sympathetic and parasympathetic signaling on the heart contributes largely to HR regulation. In particular ion-gated channels and G protein-coupled receptors are important for cardiac function. The GABA receptor family comprises both types of channels. Our candidate genes belongs to a small class of ion-gated channel GABAA receptors (34). Early studies have indicated that alterations in GABAergic signaling resulted in changes in cardiovascular function. Specifically, antagonism of the GABA inhibitory pathway resulted in decreased HR as well as blood pressure (1, 31). More recently it was reported that activation of GABAA receptors in cardiac vagal preganglionic neurons is necessary to set basal HR levels (13). Our study sheds light on this finding as we found a genomic locus and polymorphisms in specific GABAA receptors that may contribute to HR regulation. It is clear that GABAA receptor specificity on controlling HR warrants further investigation.
An elevated HR locus was identified in humans on chr15 (30) in the Framingham Heart Study (Table 6). Within this locus there is a region that contains genes encoding cholinergic receptors (CHRNB4 and CHRNA5) (27). Another recent human GWA study found a number of loci containing genes that are important in ion conductance (9). In particular, GNB4 (chr3) influences G protein-activated inwardly rectifying potassium channels, and CACNA1C (chr12) encodes a subunit in a voltage-gated calcium channel. Given that our study found SNPs in a class of receptors important in the autonomic nervous system, it is likely that this locus plays an important role in regulating autonomic tone in the heart, possibly by regulating ion conductance.
There are several limitations to our study. Measurements of the hemodynamic traits using tail-cuff plethysmography cannot exclude emotional stress as a confounding factor. Thus resting or basal HR may not be truly represented. However, we used a standard training procedure for each animal to reduce the effect of emotional stress in all experimental mice in both the genetic cross and the HMDP (18). Comparison of the data in our study in the HMDP and the data in the inbred strains obtained by telemetry techniques shows more similarities (∼2/3 of strains) than discrepancies in the HR trait (14). We understand that utilizing a traditional mapping approach by the generation of congenic mice carrying a single locus on chr7 could be more powerful mapping strategy, but it is time and labor consuming. Instead, we showed that an independent GWA not only identified the same locus but also provided higher mapping resolution and reduced the number of candidate genes from >500 to three genes.
Intriguingly, despite the limitations discussed here we found that the genetic locus on chr7 may harbor common genes controlling hemodynamic and vascular remodeling traits. In particular, we found a suggestive locus on chr7 that contributes to the carotid intima trait in the C3HeB×SJL backcross (19). These findings may link our previous observations of a correlation between HR and the extent of vascular remodeling in five inbred mouse strains (20). Additionally, our study may provide further knowledge into correlations between HR and other vascular diseases. For example, a strong relationship was found between HR and early atherosclerotic disease in humans (10) and a lower HR decreased atherosclerosis in cynomolgus monkeys (2, 3). Another study reported that there is a significant locus on distal part of mouse chr7 that controls vascular peripheral disease (8). In fact, our findings on HR and SBP may provide additional insights into genetic control of collateral circulation and ischemic injury in A and BALB strains (6).
In summary we used a combined genetic approach of genome-wide linkage and association analyses to identify regions on the mouse genome that control heart rate trait. A linkage analysis revealed a novel elevated HR locus on chr7 in the C3HeB×SJL backcross. For fine-mapping of the elevated HR locus, we used GWA with the HMDP of 58 mouse strains. Including the HMDP added genomic resolution to the QTL identified in the backcross. This combinatorial approach suggested novel candidate genes, Gabrb3, Gabra5, and Gabrg3, within the locus on chr7 that controls elevated HR in mice. Furthermore, we showed that the HR trait is regulated by a locus independent of SBP loci.
GRANTS
This study was supported in part by funds from University of Rochester, Russian Program “Scientific and scientific-pedagogical personnel of innovative Russia” for 2009-2013 GK no. 14.740.11.0923, and National Heart, Lung, and Blood Institute Grants HL-105623 (V. A. Korshunov) and HL-30568 and HL-42488 (A. J. Lusis).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: E.M.S., A.G., A.N.M., A.J.L., and V.A.K. conception and design of research; E.M.S., I.A.I., A.G., J.G., and V.A.K. performed experiments; E.M.S., I.A.I., A.G., J.G., and V.A.K. analyzed data; E.M.S., I.A.I., A.G., A.N.M., A.J.L., and V.A.K. interpreted results of experiments; E.M.S., I.A.I., A.G., and V.A.K. prepared figures; E.M.S. and I.A.I. drafted manuscript; E.M.S., I.A.I., A.G., J.G., A.N.M., A.J.L., and V.A.K. approved final version of manuscript; A.G., A.N.M., A.J.L., and V.A.K. edited and revised manuscript.
ACKNOWLEDGMENTS
The authors thank Drs. Brian Bennett and Renee LeBoeuf for thoughtful discussions of the manuscript.
REFERENCES
- 1. Antonaccio MJ, Taylor DG. Involvement of central GABA receptors in the regulation of blood pressure and heart rate of anesthetized cats. Eur J Pharmacol 46: 283–287, 1977 [DOI] [PubMed] [Google Scholar]
- 2. Beere PA, Glagov S, Zarins CK. Experimental atherosclerosis at the carotid bifurcation of the cynomolgus monkey. Localization, compensatory enlargement, and the sparing effect of lowered heart rate. Arterioscler Thromb 12: 1245–1253, 1992 [DOI] [PubMed] [Google Scholar]
- 3. Beere PA, Glagov S, Zarins CK. Retarding effect of lowered heart rate on coronary atherosclerosis. Science 226: 180–182, 1984 [DOI] [PubMed] [Google Scholar]
- 4. Bennett BJ, Farber CR, Orozco L, Kang HM, Ghazalpour A, Siemers N, Neubauer M, Neuhaus I, Yordanova R, Guan B, Truong A, Yang WP, He A, Kayne P, Gargalovic P, Kirchgessner T, Pan C, Castellani LW, Kostem E, Furlotte N, Drake TA, Eskin E, Lusis AJ. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res 20: 281–290, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blizard DA, Lionikas A, Vandenbergh DJ, Vasilopoulos T, Gerhard GS, Griffith JW, Klein LC, Stout JT, Mack HA, Lakoski JM, Larsson L, Spicer JM, Vogler GP, McClearn GE. Blood pressure and heart rate QTL in mice of the B6/D2 lineage: sex differences and environmental influences. Physiol Genomics 36: 158–166, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chalothorn D, Faber JE. Strain-dependent variation in collateral circulatory function in mouse hindlimb. Physiol Genomics 42: 469–479, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. DiPetrillo K, Tsaih SW, Sheehan S, Johns C, Kelmenson P, Gavras H, Churchill GA, Paigen B. Genetic analysis of blood pressure in C3H/HeJ and SWR/J mice. Physiol Genomics 17: 215–220, 2004 [DOI] [PubMed] [Google Scholar]
- 8. Dokun AO, Keum S, Hazarika S, Li Y, Lamonte GM, Wheeler F, Marchuk DA, Annex BH. A quantitative trait locus (LSq-1) on mouse chromosome 7 is linked to the absence of tissue loss after surgical hindlimb ischemia. Circulation 117: 1207–1215, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eijgelsheim M, Newton-Cheh C, Sotoodehnia N, de Bakker PI, Muller M, Morrison AC, Smith AV, Isaacs A, Sanna S, Dorr M, Navarro P, Fuchsberger C, Nolte IM, de Geus EJ, Estrada K, Hwang SJ, Bis JC, Ruckert IM, Alonso A, Launer LJ, Hottenga JJ, Rivadeneira F, Noseworthy PA, Rice KM, Perz S, Arking DE, Spector TD, Kors JA, Aulchenko YS, Tarasov KV, Homuth G, Wild SH, Marroni F, Gieger C, Licht CM, Prineas RJ, Hofman A, Rotter JI, Hicks AA, Ernst F, Najjar SS, Wright AF, Peters A, Fox ER, Oostra BA, Kroemer HK, Couper D, Volzke H, Campbell H, Meitinger T, Uda M, Witteman JC, Psaty BM, Wichmann HE, Harris TB, Kaab S, Siscovick DS, Jamshidi Y, Uitterlinden AG, Folsom AR, Larson MG, Wilson JF, Penninx BW, Snieder H, Pramstaller PP, van Duijn CM, Lakatta EG, Felix SB, Gudnason V, Pfeufer A, Heckbert SR, Stricker BH, Boerwinkle E, O'Donnell CJ. Genome-wide association analysis identifies multiple loci related to resting heart rate. Hum Mol Genet 19: 3885–3894, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Festa A, D'Agostino R, Jr, Rautaharju P, O'Leary DH, Rewers M, Mykkanen L, Haffner SM. Is QT interval a marker of subclinical atherosclerosis in nondiabetic subjects? The Insulin Resistance Atherosclerosis Study (IRAS). Stroke 30: 1566–1571, 1999 [DOI] [PubMed] [Google Scholar]
- 11. Flint J, Valdar W, Shifman S, Mott R. Strategies for mapping and cloning quantitative trait genes in rodents. Nat Rev Genet 6: 271–286, 2005 [DOI] [PubMed] [Google Scholar]
- 12. Ghazalpour A, Doss S, Sheth SS, Ingram-Drake LA, Schadt EE, Lusis AJ, Drake TA. Genomic analysis of metabolic pathway gene expression in mice. Genome Biol 6: R59, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hildreth CM, Goodchild AK. Role of ionotropic GABA, glutamate and glycine receptors in the tonic and reflex control of cardiac vagal outflow in the rat. BMC Neurosci 11: 128, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Howden R, Liu E, Miller-DeGraff L, Keener HL, Walker C, Clark JA, Myers PH, Rouse DC, Wiltshire T, Kleeberger SR. The genetic contribution to heart rate and heart rate variability in quiescent mice. Am J Physiol Heart Circ Physiol 295: H59–H68, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Julier C, Delepine M, Keavney B, Terwilliger J, Davis S, Weeks DE, Bui T, Jeunemaitre X, Velho G, Froguel P, Ratcliffe P, Corvol P, Soubrier F, Lathrop GM. Genetic susceptibility for human familial essential hypertension in a region of homology with blood pressure linkage on rat chromosome 10. Hum Mol Genet 6: 2077–2085, 1997 [DOI] [PubMed] [Google Scholar]
- 16. Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. King DE, Everett CJ, Mainous AG, 3rd, Liszka HA. Long-term prognostic value of resting heart rate in subjects with prehypertension. Am J Hypertens 19: 796–800, 2006 [DOI] [PubMed] [Google Scholar]
- 18. Korshunov VA, Berk BC. Flow-induced vascular remodeling in the mouse: a model for carotid intima-media thickening. Arterioscler Thromb Vasc Biol 23: 2185–2191, 2003 [DOI] [PubMed] [Google Scholar]
- 19. Korshunov VA, Berk BC. Genetic modifier loci linked to intima formation induced by low flow in the mouse carotid. Arterioscler Thromb Vasc Biol 29: 47–53, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Korshunov VA, Berk BC. Strain-dependent vascular remodeling: the “Glagov phenomenon” is genetically determined. Circulation 110: 220–226, 2004 [DOI] [PubMed] [Google Scholar]
- 21. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformat 9: 559, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lloyd-Jones DM. Cardiovascular risk prediction: basic concepts, current status, and future directions. Circulation 121: 1768–1777, 2010 [DOI] [PubMed] [Google Scholar]
- 23. Mansfield TA, Simon DB, Farfel Z, Bia M, Tucci JR, Lebel M, Gutkin M, Vialettes B, Christofilis MA, Kauppinen-Makelin R, Mayan H, Risch N, Lifton RP. Multilocus linkage of familial hyperkalaemia and hypertension, pseudohypoaldosteronism type II, to chromosomes 1q31–42 and 17p11-q21. Nat Genet 16: 202–205, 1997 [DOI] [PubMed] [Google Scholar]
- 24. Martin LJ, Comuzzie AG, Sonnenberg GE, Myklebust J, James R, Marks J, Blangero J, Kissebah AH. Major quantitative trait locus for resting heart rate maps to a region on chromosome 4. Hypertension 43: 1146–1151, 2004 [DOI] [PubMed] [Google Scholar]
- 25. Melton PE, Rutherford S, Voruganti VS, Goring HH, Laston S, Haack K, Comuzzie AG, Dyer TD, Johnson MP, Kent JW, Jr, Curran JE, Moses EK, Blangero J, Barac A, Lee ET, Best LG, Fabsitz RR, Devereux RB, Okin PM, Bella JN, Broeckel U, Howard BV, MacCluer JW, Cole SA, Almasy L. Bivariate genetic association of KIAA1797 with heart rate in American Indians: the Strong Heart Family Study. Hum Mol Genet 19: 3662–3671, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Palatini P. Elevated heart rate in cardiovascular diseases: a target for treatment? Prog Cardiovasc Dis 52: 46–60, 2009 [DOI] [PubMed] [Google Scholar]
- 27. Rempel N, Heyers S, Engels H, Sleegers E, Steinlein OK. The structures of the human neuronal nicotinic acetylcholine receptor beta2- and alpha3-subunit genes (CHRNB2 and CHRNA3). Hum Genet 103: 645–653, 1998 [DOI] [PubMed] [Google Scholar]
- 28. Scicluna BP, Tanck MW, Remme CA, Beekman L, Coronel R, Wilde AA, Bezzina CR. Quantitative trait loci for electrocardiographic parameters and arrhythmia in the mouse. J Mol Cell Cardiol 50: 380–389, 2011 [DOI] [PubMed] [Google Scholar]
- 29. Shin JH, Blay S, McNeney B, Graham J. LDheatmap: an R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J Stat Softw 16: 1–9, 2006 [Google Scholar]
- 30. Singh JP, Larson MG, O'Donnell CJ, Tsuji H, Corey D, Levy D. Genome scan linkage results for heart rate variability (the Framingham Heart Study). Am J Cardiol 90: 1290–1293, 2002 [DOI] [PubMed] [Google Scholar]
- 30a. Spielmann N, Leon AS, Rao DC, Rice T, Skinner JS, Rankinen T, Bouchard C. Genome-wide linkage scan for submaximal exercise heart rate in the HERITAGE family study. Am J Physiol Heart Circ Physiol 293: H3366–H3371, 2007 [DOI] [PubMed] [Google Scholar]
- 31. Stotz-Potter E, Benarroch E. Removal of GABAergic inhibition in the mediodorsal nucleus of the rat thalamus leads to increases in heart rate and blood pressure. Neurosci Lett 247: 127–130, 1998 [DOI] [PubMed] [Google Scholar]
- 32. Sugiyama F, Churchill GA, Li R, Libby LJ, Carver T, Yagami K, John SW, Paigen B. QTL associated with blood pressure, heart rate, and heart weight in CBA/CaJ and BALB/cJ mice. Physiol Genomics 10: 5–12, 2002 [DOI] [PubMed] [Google Scholar]
- 33. Watanabe M, Maemura K, Kanbara K, Tamayama T, Hayasaki H. GABA and GABA receptors in the central nervous system and other organs. Int Rev Cytol 213: 1–47, 2002 [DOI] [PubMed] [Google Scholar]
- 34. Whiting PJ. GABA-A receptor subtypes in the brain: a paradigm for CNS drug discovery? Drug Discov Today 8: 445–450, 2003 [DOI] [PubMed] [Google Scholar]
- 35. Woo DD, Kurtz I. Mapping blood pressure loci in (A/J × B6)F2 mice. Physiol Genomics 15: 236–242, 2003 [DOI] [PubMed] [Google Scholar]