Abstract
In fetal sheep during late gestation sulfoconjugated estrogens in plasma reach a concentration 40–100 times greater than unconjugated estrogens. The objective of the present study was to determine the genomics of estradiol-3-sulfate (E2S) action in the ovine fetal brain. The hypothesis was that E2S stimulates genes involved in the neuroendocrine pathways that direct or facilitate fetal development at the end of gestation. Four sets of chronically catheterized ovine twin fetuses were studied (gestational age: 120–127 days gestation) with one infused with E2S intracerebroventricularly (1 mg/day) and the other remaining untreated (control). After euthanasia, mRNA samples were extracted from fetal brains. Only hypothalamic samples were employed for this study given the important function of this brain region in the control of the hypothalamus-pituitary-adrenal axis. Microarray analysis was performed following the Agilent protocol for one-color 8 × 15 microarrays, designed for Ovis aries. A total of 363 known genes were significantly upregulated by the E2S treatment (P < 0.05). Network and enrichment analyses were performed using the Cytoscape/Bingo software, and the results validated by quantitative real-time PCR. The main overrepresented biological processes resulting from this analysis were feeding behavior, hypoxia response, and transforming growth factor signaling. Notably, the genes involved in the feeding behavior (neuropeptide Y and agouti-related protein) were the most strongly induced by the E2S treatment. In conclusion, E2S may be an important component of the mechanism for activating orexigenic, hypoxia responsiveness and neuroprotective pathways in the lamb as it approaches postnatal life.
Keywords: heart rate, estradiol, hypoxia, estrogen receptor, neuropeptides, neurosteroid
in fetal sheep and pregnant ewes, sulfoconjugated estrogens are far more abundant than unconjugated estrogens (9, 41, 46). High concentrations of estrone sulfate in uterine vein plasma (compared to peripheral vein plasma) of pregnant sheep suggested a high secretion rate for this steroid by placenta in late gestation (15). Plasma concentrations of estradiol-3-sulfate (E2S) are ∼40–100 times those of estradiol (E2) in the late-gestation fetal sheep. E2S is taken up by the fetal brain and stimulates responses that are both similar to and distinct from the responses to E2 (43, 46). Sulfoconjugation increases the half-life in the blood (33) and supplies a ready source of E2 in tissues (e.g., hypothalamus) that express steroid sulfatase (STS) (31, 43). E2S can bind estrogen receptor only after deconjugation by STS. We have proposed that, while the function of the sulfoconjugated estrogens in fetal and maternal sheep is unknown, deconjugation can increase estrogen action in specific tissues that express STS and estrogen receptor (43, 44, 46). The fetal brain expresses both STS and estrogen sulfotransferase (STF), allowing for the bidirectional interconversion of E2 and E2S (31, 32), although the ratio of expression for these enzymes favors deconjugation (43). The fetal brain also expresses transporters that have the capacity to transport the sulfoconjugated steroid across the blood-brain barrier (13).
We have recently observed that E2S has some actions that are similar to E2 and some actions that are distinct from E2 on the hypothalamus-pituitary-adrenal (HPA) (43, 45). Chronic infusions of E2 to fetal sheep produce sustained increases in both ACTH and cortisol, causing a potent stimulation of the fetal HPA axis (47). In contrast, long-term infusion (2–3 wk) of E2S inhibits the periparturient rise in ACTH and reduces HPA activity, even when plasma cortisol levels are not affected (45). Given the multifactorial response to E2S, it is probable that the response includes activation of genes that are not directly responsive to the estrogen receptor.
The present study was designed to reveal the genomics of E2S action in the fetal brain. Using a newly available ovine array, we tested the hypothesis that E2S both stimulates and inhibits genes involved in the neuroendocrine pathways that direct or facilitate fetal development at the end of gestation. We also hypothesized that E2S would significantly alter the activity of genes involved in late-gestation fetal development.
MATERIALS AND METHODS
Animal Procedures
A total of four sets of chronically catheterized ovine twin fetuses were studied with one infused with E2S intracerebroventricularly (1 mg/day) for 7–12 days, using an osmotic mini-pump implanted in the fetus, and the other served as an untreated control. The gestational age at the time of surgery was 120–127 days of gestation, a developmental window of time that is prior to the preparturient rise in ACTH. Twin fetuses were randomly assigned to the two groups at the time of surgery. All animals were housed in individual pens located in the Animal Resources Department at the University of Florida, and all of these experiments were approved by the University of Florida Institutional Animal Care and Use Committee. The rooms maintained controlled lighting and temperature, and sheep were given food and water ad libitum.
Food was withheld from the pregnant ewes for 24 h before surgery. Ewes were intubated and anesthetized with halothane (0.5–2%) in oxygen before and during surgical preparation as previously described (46). Surgery and catheter placement for all fetuses were performed using aseptic technique as previously described, with lateral cerebral ventricle, femoral arterial, and venous catheters as well as amniotic fluid catheters (35, 46). For placement of the catheter into the lateral cerebral ventricle, the scalp was retracted and a small catheter (outside diameter, 0.05 in.; inside diameter, 0.03 in.) attached to an osmotic mini-pump (size 2mL2; Alza, Palo Alto, CA; infusion rate 5 μl/h) was inserted through a hole made in the skull. This catheter was held in place using VetBond (3M, St. Paul, MN). The exposed catheter and osmotic mini-pump were placed subcutaneously before the incision on the head was closed. All minipumps in the treated fetuses were filled with E2SO4 (Sigma Aldrich, St. Louis, MO) in vehicle (water), and minipumps in the control fetuses were filled with vehicle only. The position of the catheters and the function of the pumps were verified by visual inspection at the time of death and tissue collection. At the end of the surgery, antibiotics (750 mg ampicillin) were administered into the amniotic cavity via direct injection. Vascular catheters were exteriorized through the flank of the ewe using a trochar, where they were maintained in a removable synthetic cloth pocket. Ewes were treated with 1 mg/kg flunixin meglumine (Webster Veterinary, Sterling, MA) for analgesia and returned to their pens where they were monitored until they could stand on their own. If needed, a second treatment with flunixin meglumine was administered 24–48 h after the first treatment with this drug. Twice daily during a 5-day recovery period ewes were treated with antibiotic (ampicillin, 750 mg im, Polyflex; Fort Dodge Laboratories, Fort Dodge, IA), and rectal temperatures were monitored for indication of postoperative infection. None of the animals in this study showed any signs of postoperative infection.
Blood Collection and Plasma Hormone Assays
Blood samples were collected to measure plasma hormone levels for both groups, to prove the efficacy of E2S treatment and to check for fetal-fetal transfer between treatment and control twin fetuses. Blood samples from treated and control fetuses were collected from the arterial catheter every other morning after the recovery period. Plasma from each sample was obtained by centrifugation at 3,000 g for 15 min at 4°C and stored at −20°C until analysis. Assays to measure E2S and E2 levels were performed as previously described (46). Mean values for E2S and E2 for these experiments have been reported previously (43).
Sample Collection
Pregnant ewes and twin fetuses of known gestational age (130–134 days) were euthanized with an overdose of pentobarbital sodium. Brains were rapidly removed, dissected into distinct regions, and snap-frozen in liquid nitrogen. Tissues were collected from hypothalamus, pituitary, hippocampus, medullary brain stem, cerebellum, and cerebral cortex and were stored at −80°C until processed for mRNA. These tissue samples were originally collected for other experiments (43).
In the present microarray experiment, the mRNA isolated from hypothalamus was analyzed. This region was selected for being a critical component of the HPA axis, a major feature of the fetal stress response and crucial for initiation of parturition in the sheep (27).
RNA Extraction and Preparation
RNA was extracted from the hypothalamus using TRIzol (Invitrogen, Carlsbad, CA) following the manufacturer's directions. The RNA was resuspended in RNAsecure and stored at −80°C in aliquots until use. For microarray analysis 20 μg of these RNAs, which had been stored for 7 yr, were DNase treated using the Turbo RNase-free DNase kit (Ambion, Foster City, CA), the concentration determined with a Nanodrop spectrophotometer (ND-1000; ThermoFisher, Wilmington DE), and the integrity of the RNA was measured using an Agilent Bioanalyzer, 2100 model. One RNA sample had an RNA integrity number (RIN) value of 5.1 and was excluded from further analysis; the remaining RNAs had RIN values of 6.9–8.0. DNase-treated RNA (1 μg) was labeled with cyanine 3 (Cy3) CTP with the Agilent Quick Amp kit (5190-0442, New Castle, DE) according to their methodology, purified with the Qiagen RNeasy kit (Valencia, CA) according to Agilent's revision of the Qiagen protocol as shown in the Quick Amp kit protocol except that the microcentrifugation spins were performed at room temperature instead of 4°C. The resulting labeled cRNA was analyzed with the NanoDrop spectrophotometer, and the specific activities and the yields of the cRNAs were calculated; these ranged from 10.41 to 19.73 pmol Cy3/μg RNA and from 5.0 to 12.8 μg, respectively. The labeled cRNA was stored at −80°C until use.
Microarray Hybridization
This was performed following protocols from Agilent; in brief, 600 ng of each labeled cRNA was fragmented and then mixed with hybridization buffer using the Agilent gene expression hybridization kit. These were applied to a sheep 8 X 15 K array slide (Agilent 019921), containing eight arrays with 15,744 oligomers with a length of 60 bases corresponding to 15,744 ovine genetic sequences published in the National Center for Biotechnology Information (NCBI) database and hybridized at 65°C for 17 h at 10 rpm. The arrays were washed, dried, stabilized, and scanned with an Agilent G2505B 2 dye scanner at the Interdisciplinary Center for Biotechnology Research at the University of Florida. Features were extracted with Agilent Feature extraction 9.1 software. Features flagged as Feature Nonuniform outliers were excluded from further analysis.
Statistical Analysis
Transcript levels were normalized to the chip median and log-transformed, to obtain more power in discovering differences between groups and compensate for systematic differences between the arrays. To identify the genes that are differentially regulated between the treated and control fetuses, we analyzed the normalized and transformed intensities by one-way ANOVA. To estimate whether microarray observations were able to predict the categorical outcome (treatment or control groups), we performed class prediction using Distance Scoring, a nonparametric discriminant method that bases predictions for an observation on distances between it and observations in a training set. Distances were computed from class centroids, and the statistical test employed was t-test (P < 0.05). All the statistical procedures were carried out using JMP Genomics 5 software (SAS Institute, Cary, NC).
A gene was considered to be significantly differentially expressed (over- or underexpressed) if both of the following conditions were met: 1) the ratio of the normalized intensity of the treatment fetus sample to normalized intensity in the control fetus sample was higher or lower than a twofold change (up- or downregulation, respectively); and 2) differences were considered statistically significant at P ≤ 0.05.
Functional Annotation
We used functional annotation of the genes as a useful tool to categorize the genes in functional classes, leading to a better understanding of the physiological relevance of altered gene expression. The most common method to access this analysis is to place each gene in the gene ontology (GO) hierarchy, developed at the GO Consortium (1). GO information for every gene is not available for the ovine genome. Thus, Blast2Go software (V 4.2.2) (12), a tool for functional annotation of (novel) sequences, was employed. This software uses the Basic Local Alignment Search Tool (BLAST) to find sequences similar to the queries. For each probe, the sequence of 60 nucleotides (as supplied by Agilent) was first compared with the nucleotide sequence database using blastn. The most congruent genes were selected, and their accession numbers were input into the NCBI site (EntrezBatch) to obtain their complete sequences. These sequences were blasted again using blastx, to compare the nucleotide query sequence translated in all reading frames against the known protein sequence database nr.
Clustering Analysis
The network inference and clustering analysis was performed using CytoScape version 2.7.0 (11), through the following plugins: GeneMania, ClusterONE, and BINGO. GeneMania was used to infer network data (42). The set of functional association data between genes was downloaded from the Homo sapiens database. The list of human official symbols for the genes of interest was input into the GeneMania plugin to retrieve the corresponding association network. The association data employed were protein-protein and protein-DNA interactions. The network was inferred for both upregulated and downregulated genes (treatment vs. control). ClusterONE was used to discover densely connected and possibly overlapping regions within the network (clusters) (2). These highly connected network regions can indicate protein complexes or fractions of them. A P value is calculated for each cluster, based on the one-sided Mann-Whitney U-test performed on the in-weights and out-weights of the vertices. An in-weights value significantly larger than the out-weights value would indicate a valid cluster and not the result of random fluctuations. Thus, a P value is assigned to the cluster. Only the clusters with a P value < 0.05 were considered in further analyses. BiNGO was run to determine which biological processes are statistically overrepresented in the set of genes corresponding to the identified cluster (25). The statistical test employed was the hypergeometric test (equivalent to the Fisher test). The threshold P value was 0.05, after correction by the Bonferroni method.
Quantitative real-time PCR Validation
The mRNA samples extracted from the hypothalami of the four sets of twin fetuses (E2S/control) were converted to cDNA with a High Capacity cDNA Archive kit using the methodology recommended by the kit manufacturer (Applied Biosystems, Foster City, CA). The newly synthesized cDNA was stored at −20°C until quantitative real-time (qRT)-PCR was performed.
A total of 11 selected genes from the significant clusters identified in the upregulated and downregulated networks were tested by qRT-PCR to validate the microarray results.
The upregulated selected genes were: agouti-related protein (AGRP); neuropeptide y (NPY); hypoxia inducible factor 1, alpha subunit (HIF1A); aryl-hydrocarbon receptor nuclear translocator 2 (ARNT2); coatomer protein complex, subunit alpha (COPA) and subunit beta 1 (COPB1); and transforming growth factor, beta 1 (TGFB1). The downregulated selected genes were: chemokine (C-C motif) ligand 3 (CCL3), interleukin 12B (IL12B), interleukin 18 (IL18), and tumor necrosis factor (TNF).
Relative expression of AGRP, HIF1A, TGFB1, and TNF were determined by qRT-PCR using FAM Taqman probes (HIF1A, TGFB1), VIC Taqman probes (AGRP, TNF), or MGB probe (NPY) and primers (Sigma-Aldrich, St. Louis, MO), and Taqman RT-PCR master mix (Applied Biosystems). Relative expression of ARNT2, COPA, COPB1, CCL3, IL12B, and IL18 was determined using primers (Sigma-Aldrich) and Syber Green PCR Master Mix (Applied Biosystems).
Probes and primers were designed with Primer Express software (Applied Biosystems). Primers for ARNT2, IL12B, and IL18 and primers and probes for NPY, AGRP, HIF1A, TGFB1, and TNF were designed from the corresponding ovine mRNA. Primers for COPA, COPB1, and CCL3 were designed from the bovine mRNA. Sequences and primer concentrations for primers and probes and accession numbers are reported in Table 1. All primer or probe and primer pairs had efficiencies >95%.
Table 1.
Sequences of primers and probes for real-time PCR analysis
| Gene | Forward Primer | Reverse Primer | Probe | Accession Number |
|---|---|---|---|---|
| ARNT2 | GGTCCACTGCACAGGCTACA | CGGCATCTTCTTCAGGTATAGTCA | CAAGGCTTGGCCGCCAGCA | NM_001009452 |
| IL12B | CATCAGGGACATCATCAAACCA | CCACCTGCCGAGAATTCTTTAG | Sybr | EU340264 |
| IL18 | AATCAACCTGTCTTTGAGGATATGC | CAGACCTCTAGTGAGGCTGTCCTTATA | Sybr | NM_001009438 |
| NPY | CGGAGGACTTGGCCAGATAC | TGCCTGGTGATGAGATTGATG | ACTCAGCGCTGCGAC | NM_001009263 |
| AGRP | GAGGTGCTAGATCCGGAAGGA | TGGTGTCCCAGACAGGATTCA | CCCACGTCGCTGCGTAAGGCTG | AY310396 |
| HIF1A | GCCACAACGTCACCATATAGTGA | TCTGTCTGTTCTATGACTCCTTTTCC | AAGTCGGACAGCCTCACCCAACAGAG | AY485676 |
| TGFB1 | CAGTAAGGATAACACGCTTCAAGTG | CCGGTTCATGCCGTGAAT | ACATCAACGGGTTCAGTTCCGGCC | NM_001009400 |
| TNF | CCCTTCCACCCCCTTGTT | ATGTTGACCTTGGTCTGGTAGGA | CCACACCATCAGCCGCATTGCA | NM_001024860 |
| COPA | GAAAAACCCCACAGATGCCTAT | CCGATAAGATGCAGCACAGATG | Sybr | NM_001105645 |
| COPB1 | GGTCTGTCATGCTAATCCATCAGA | AGCAGGGCTAGATGACTGTAGTAAGTT | Sybr | NM_001078007 |
| CCL3 | GGTGTCATCTTCCAGACCAAAAA | TCCTGGACCCAGTCCTCAGT | Sybr | NM_174511 |
The abundance of β-actin mRNA was determined in each sample, using primers and VIC Taqman probes designed from the ovine β-actin sequence and Taqman RT-PCR master mix (Applied Biosystems).
All samples were run in triplicate for each gene and for β-actin. Relative mRNA expression of each gene was calculated by determining change in threshold cycle (ΔCt) between the mean Ct for each gene and the mean Ct for β-actin mRNA from the same sample. The effect of E2S on each gene was compared by one-way ANOVA using the ΔCt values. Data are graphed as the mean fold change in mRNA relative to the control group; fold change in each sample was calculated as 2−ΔΔCt, where ΔΔCt is the difference between ΔCt in each sample and the mean ΔCt in the control group. For all statistical analyses, the criterion for achieving statistical significance was P < 0.05. A P value of 0.1 was considered a tendency.
RESULTS
Plasma Hormones
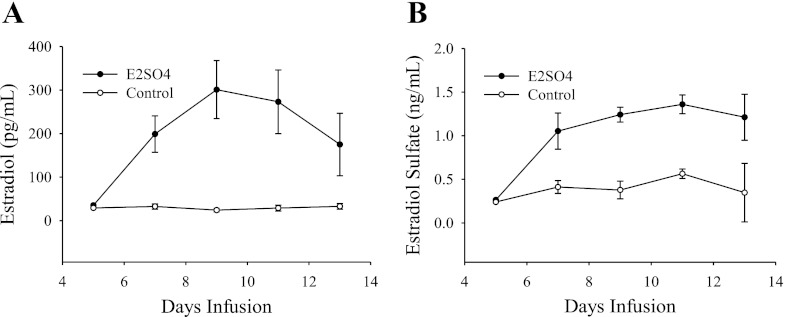
Treatment with E2S caused an increase in plasma levels of E2S and E2, with peaks of 1.36 ± 0.11 ng/ml and 301 ± 67 pg/ml respectively, compared with plasma levels of these hormones in the control fetuses (Fig. 1).
Fig. 1.
Plasma levels of estradiol (A) and estradiol-3-sulfate (B) during the infusion period in treated and control fetuses. The difference in hormone concentration between both groups was significant for both hormones (P < 0.001).
Microarray Results
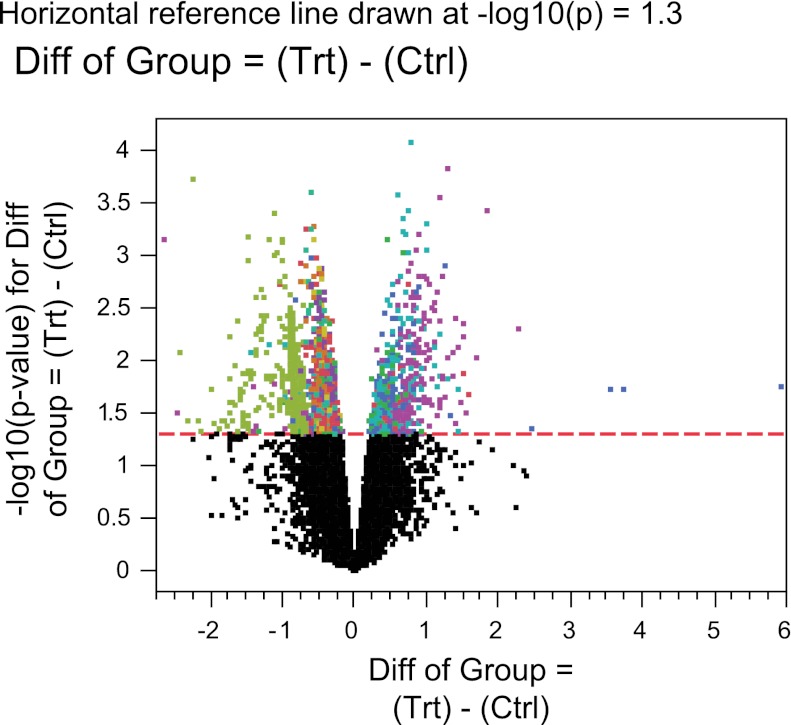
According to one-way ANOVA, the expression of a total of 2,442 genes was significantly regulated (P ≤ 0.05). Of these genes, expression of 526 had a fold change >2 (upregulated), and 1,916 had a fold change <2 (downregulated). The volcano plot generated from this analysis is shown in Fig. 2. Class prediction analysis showed that the probability for each observation of correctly predicting whether it belongs to the treatment or the control group was >91.18%, with a mean of 98.89% and a median of 99.99%.
Fig. 2.
Volcano plot showing the log-odds of differential expression vs. the log fold change in gene expression between estradiol-3-sulfate treatment and control arrays. Same color means same cluster of expression for both groups. Genes located above the significance cutoff (P value = 0.05, indicated by the dashed line) are those with statistically significant difference of expression between the treatment and control groups.
Functional Annotation
The gene represented by an oligomer on the microarray could be identified for 1,830 oligomers (out of 2,442 with significantly altered expression). After duplicate genes were removed 1,544 genes remained. The final gene lists that were submitted to the GeneMania plugin contained 363 genes in the upregulated category and 1,201 genes for the downregulated.
Clustering Analysis
Upregulated network.
The network inferred with the upregulated genes contained 218 nodes and 697 edges. The network as visualized using the BioLayout Express 3D software (38) is reported in Fig. 3. Nodes with similar color in this network represent the clusters selected by BioLayout Express 3D, that were consistent with the clusters found by ClusterOne plugin in Cytoscape. A total of nine significant clusters with overrepresented biological processes were identified on this network, as shown in Table 2 after redundant biological processes were simplified.
Fig. 3.
Network obtained from the list of upregulated genes, as visualized using BioLayout Express 3D software. Nodes with similar colors represent a cluster. Blue nodes denote those that do not belong to any cluster.
Table 2.
Statistically overrepresented biological processes found on the significant clusters of the treatment network (P value <0.05)
| Cl. | Biological Process | Genes |
|---|---|---|
| 1 | feeding behavior; positive regulation of response to stimulus | NPY - GHRL - AGRP |
| 2 | response to hypoxia; blood vessel development; positive regulation of cell proliferation; response to chemical stimulus | HIF1A - FLT1 - COL3A1 - EDN1 - ARNT2 - C-JUN |
| 3 | COPI coating of Golgi vesicle; vesicle targeting, to, from or within Golgi; Golgi vesicle budding; membrane budding; organelle localization | COPB2 - COPA - COPB1 - COPZ1 - PAFAH1B1 |
| 4 | transforming growth factor beta receptor signaling pathway; pathway-restricted SMAD protein phosphorylation; response to prostaglandin E stimulus; response to estrogen stimulus; response to hypoxia; immune system development; immune system process | CAV1 - TGFBR1 - TGFBR2 - TGFBR3 - TGFB1 |
| 5 | RNA splicing; nucleic acid metabolic process | PAPOLA - HNRNPH2 - SFRS5 - EFTUD2 - CDC40 - RBM5 - SRP75 - NFX1 |
| 6 | establishment of localization in cell | XPO1 - ARHGEF2 - DERL1 - HTATIP2 - CENPA - PAFAH1B1 - RANBP2 - CLTCL1 |
| 7 | translational initiation | EIF3A - EIF3G - EIF3-P36 |
| 8 | macromolecule catabolic process | ERCC5 - DCP2 - USP12 - USP46 |
| 9 | mitotic cell cycle | CDC6 - CDKN1A - NCAPD2 |
Boldfaced genes were validated by quantitative real-time PCR. Cl., cluster.
Downregulated network.
In this case, the network was composed of 756 nodes and 4,668 edges. There were nine significant clusters with overrepresented biological processes identified on this network (Table 3; redundant biological processes were simplified).
Table 3.
Statistically overrepresented biological processes found on the significant clusters of the control network (P value <0.05)
| Cl. | Biological Process | Genes |
|---|---|---|
| 1 | regulation of immune response; signaling pathway | FCER1A - LAT2 - CD36 - FYN - ITGB6 - FCER1G - MS4A2 - SKAP1 - SPN |
| 2 | regulation of leukocyte proliferation and activation | IL4 - IL2RA - SLA2 - IFNG - IL12B - CD28 - IL2–IL18 |
| 3 | immune system process; inflammatory response | CCL3 - CXCL9 - CCL15 - DPP4 - TNF |
| 4 | response to wounding | FGF7 - IL8 - CD46 - ITGA2 - SDC2 |
| 5 | nucleic acid metabolic process | POLR2F - SNAPC2 - SF3A1 - SF3B3 - POLR2A - SLBP - SC-35 - PHAX - TAF10 - GTF2E1 - GTF2F2 - SNRPB - LSM4 - GTF3C5 - LSM10 - SUPT4H1 - CPSF2 - NFIA - ERCC1 - GEMIN4 - SNRPG |
| 6 | protein metabolic process | COPS5 - UBA3 - NEDD8 - CUL4B - FBXO22 - NAE1 |
| 7 | lipoprotein catabolic process; cholesterol metabolic process | APOB - LDLR - APOE |
| 8 | regulation of macromolecule metabolic process | SREBF1 - MED30 - NR4A2 - MED12 - CDC23 - MED14 - K35 - MED13L - ERB |
| 9 | cellular protein localization | GRPEL1 - SEC24A - SRPR - SSR2 - SSR3 |
Boldfaced genes were validated by quantitative real-time PCR.
qRT-PCR
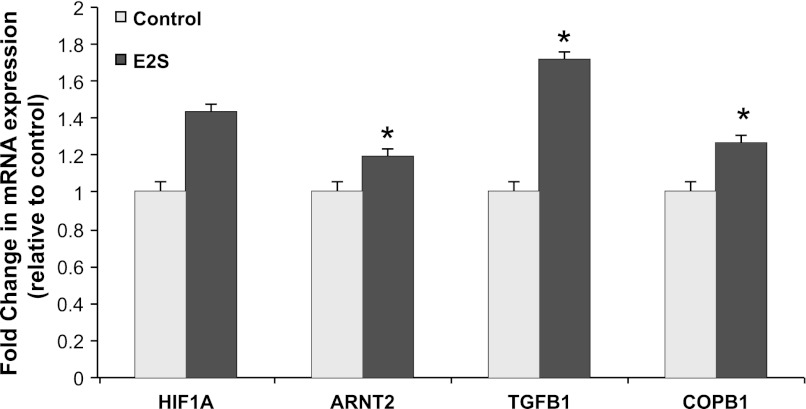
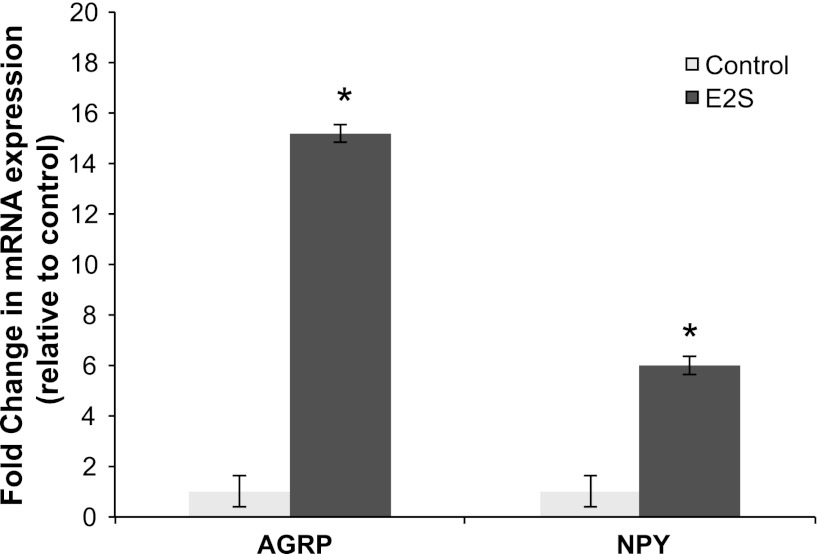
In concordance with the microarray results, expression of AGRP, NPY, ARNT2, COPB1, and TGFB1 mRNAs was significantly induced by the E2S treatment (P < 0.05), while the expression of HIF1A mRNA showed a tendency of induction by the E2S treatment (P = 0.1) (Fig. 4). The expression for NPY and AGRP genes showed the greatest increase (5- to 15-fold) in expression with E2S treatment compared with control twins (Fig. 5).
Fig. 4.
Estradiol-3-sulfate induced changes in mRNA expression of HIF1A (P = 0.1); ARNT2, TGFB1, and COPB1 (P < 0.05). Expression of each gene was normalized to β-actin expression in the same sample. Data are means ± SE fold differences relative to control group expression. *Statistically significant difference in mRNA expression between treatment and control groups.
Fig. 5.
Estradiol-3-sulfate induced strong changes in mRNA expression of AGRP and NPY (P < 0.05). Expression of each gene was normalized to β-actin expression in the same sample. Data are means ± SE fold differences relative to control group expression. *Statistically significant difference in mRNA expression between treatment and control groups.
There was no significant increase for COPA mRNA expression, and there were no statistically significant changes in the mRNA expression for the downregulated genes in the microarray analysis.
Overlap With ESR-1- and HIF1A-regulated Genes
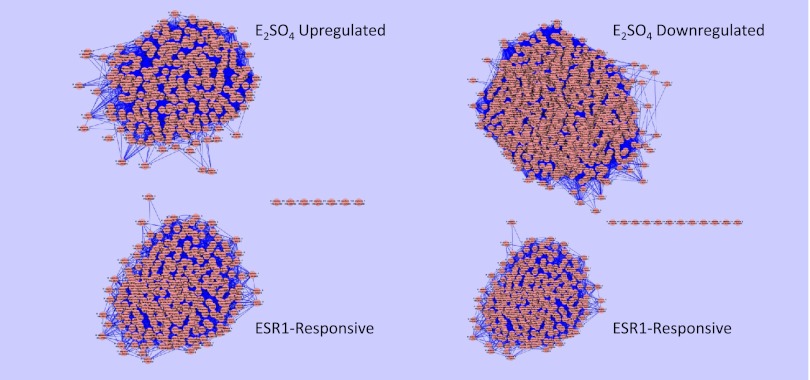
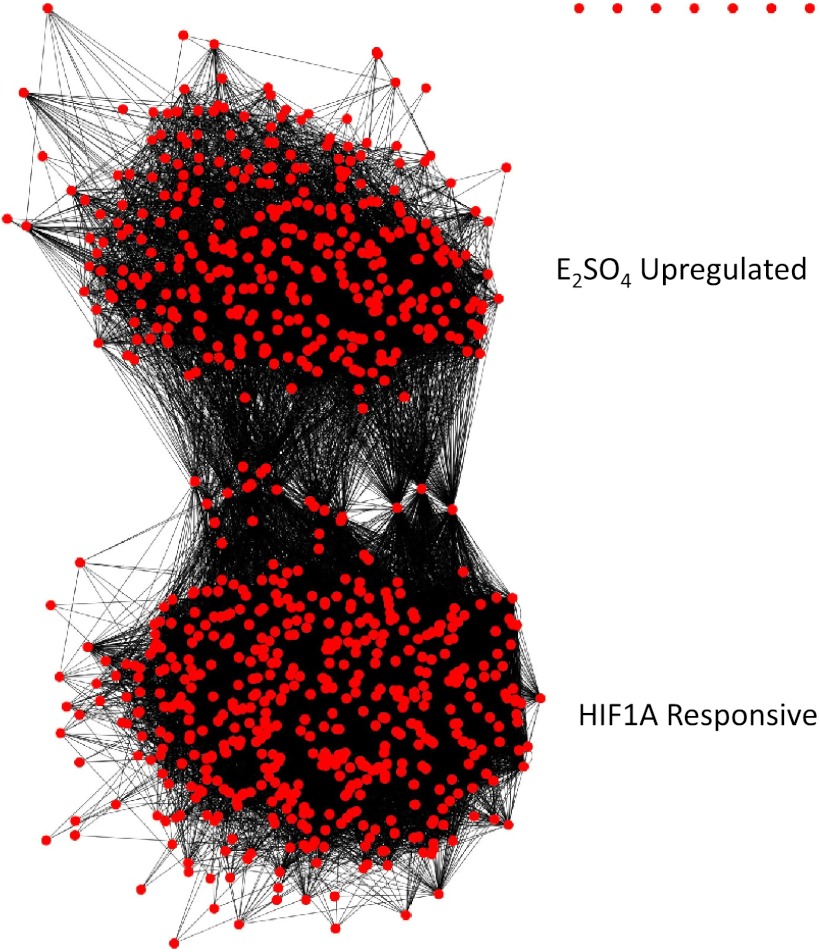
To identify genes in the up- and downregulated networks whose expression could be directly controlled by ERα (ESR-1), we tested for overlap of these networks with the known ESR-1 sensitive genes (23). There was no overlap of ESR-1 sensitive genes with genes whose expression were up- or downregulated by E2SO4 (Fig. 6). To identify genes whose expression could be controlled by HIF1A, we performed a similar analysis with genes whose expression is known to be regulated by HIF1A (4). In this case, we found significant overlap with the genes whose expression was upregulated by E2SO4 (Fig. 7).
Fig. 6.
Clusters of genes upregulated (left top) and downregulated (right top) by E2SO4, and (left and right bottom) clusters of genes known to be transcriptionally regulated by ERα (ESR1) (23). Note that there is no overlap between the known ESR1-sensitive genes and the genes differentially regulated by E2SO4.
Fig. 7.
Overlapping clusters of genes upregulated by E2SO4 (top) and of genes known to be transcriptionally regulated by HIF1A (bottom) (4).
DISCUSSION
Circulation of E2 in the sulfoconjugated form in the ovine fetus represents an important source of biologically active E2 after deconjugation by STS. The fetal hypothalamus expresses STS, and the ratio of STS to STF (the enzyme that catalyzes the reverse reaction) is high in the fetal hypothalamus (31, 43). The fetal hypothalamus also expresses both estrogen receptor alpha and beta (ERα and ERβ) throughout the latter half of gestation (34). Because E2S cannot bind ER directly and must be converted to E2 by STS, we have proposed that it functions as a precursor hormone. The increase of STS levels in the latter stages of gestation suggests an increasing capacity for converting E2S to E2 as the fetus approaches spontaneous parturition. Recent results from this laboratory have indicated that the actions of E2S are not identical to those of E2 (36). It is therefore possible that E2S exerts actions that are not ER-mediated. A limitation of the present study is that the design cannot distinguish ER-mediated from ER-independent mechanisms and cannot distinguish responses to E2 from possible direct responses to E2S (via a novel mechanism). Gene expression could be influenced by E2S, E2, and/or both since intracerebral administration of E2S in the treated animals increased the plasma levels of both E2S and E2 compared with the control (Fig. 1).
Interestingly, expression of genes known to be directly controlled by ER was not upregulated by E2S (Fig. 5). This suggests that some, if not all, of the fetal hypothalamic genomic response to E2S is mediated by other mechanisms. As discussed below, these mechanisms can include nongenomic actions of E2 (after deconjugation of E2S) or could include yet undiscovered signal transduction responses to E2S. The expression of many of the genes is likely to be influenced by the cellular responses to changes in neurotransmission that comprise a part of the downstream result of E2S administration. According to this logic, it is important to acknowledge that genomic responses, per se, are not necessarily direct responses to the infused hormone.
Neuropeptides Related to Feeding Behavior
A striking result in this experiment is the dramatic upregulation of NPY and AGRP gene expression in the E2S-treated fetuses, confirmed by qRT-PCR (Fig. 5). The neural network regulating appetite and energy balance in the adult sheep is established during fetal life: both NPY and AGRP, together with propiomelanocortin (POMC) and cocaine- and amphetamine-regulated transcript (CART), are expressed in the arcuate nucleus of the ovine fetal hypothalamus from at least 110 days of gestation, as well as the NPY projections from the arcuate nucleus to the paraventricular nucleus (29). Our data suggest that E2S treatment might stimulate alterations in appetite and/or energy balance in fetal sheep; however, the mechanism by which this might occur is unclear. Estrogen receptors are present on AGRP and NPY neurons in vivo (17) and in vitro (3, 39, 40) and can have a dual effect on NPY induction in the adult female. During proestrus, E2 stimulates NPY expression and release, which in turn contribute to the stimulation of preovulatory GnRH secretion into the hypophysial portal vessels (17). Despite this stimulatory action in these pathways, chronic administration of E2 has predominately anorectic properties (6, 48) in rats and monkeys that depend on the inhibition of NPY and AGRP expression and release (6, 14). In contrast to the present results with E2S, several reports have demonstrated inhibition of NPY by E2. E2 treatment of ovariectomized rats decreased both NPY and AGRP mRNA in the arcuate nucleus (36). Similarly, in rhesus monkeys E2 decreased AGRP and increased POMC secretion into cerebrospinal fluid (48). The upregulation of NPY and AGRP by E2S in the present study could be the consequence of an unknown stimulatory action of E2 in subpopulations of NPY-AGRP neurons or of non-ER-mediated E2S action, although differences of E2 action between fetal and adult life and of between species cannot be discounted. While the mechanism is unknown, E2S might be and important physiological stimulus for feeding behavior and regulation of energy balance necessary for the survival of the newborn.
Mediators of Vascularization and Hypoxia Response
E2S effects on genes related to the response to hypoxia more closely mirrored the expected effects of E2. E2 treatment in rat pituitary autografts increases the expression of proangiogenic factors, specially vascular endothelium growth factor (VEGF), its receptor, fms-related tyrosine kinase (FLT1), and HIF1A (24), and E2 increases the expression of mRNA for HIF1A (which encodes the protein HIF1α) in endometrium of ovariectomized ewes (19). Our microarray results showed an upregulation of FLT1 and HIF1A (PCR validation for HIF1A was not statistically significant, P = 0.1), but not VEGF, expression by E2S. It is therefore not clear whether E2S promotes angiogenesis in the fetal hypothalamus. Another upregulated gene from the same cluster was ARNT2, validated by qRT-PCR (Fig. 4). The encoded protein from the ARNT2 gene complexes with HIF1A in the nucleus, and this complex binds to hypoxia-response elements in enhancers and promoters of oxygen-responsive genes (37). HIF1A and ARNT2 are members of the bHLH-PAS family, together with ARNT and aryl hydrocarbon receptor (AHR) (18). ARNT and AHR can modulate ER-dependent transcription by protein-protein association (7, 30). Cho et al. (10) showed that both the NH2 terminus and COOH terminus of ER interact with the bHLH-PAS domain of HIF1α and suggested that under hypoxic conditions, HIF1α may target ERα to the proteasome, decreasing ERα abundance and providing a brake on the upregulation of HIF1A by E2. However, the physical association of HIF1A with ERα can be important in gene regulation: recruitment of ERα to the VEGF promoter (which has no ERE) by HIF1α is integral to the upregulation of VEGF expression in response to hypoxia (20). Why we did not observe an increase in VEGF expression in the present experiments is unclear but could be the result of competing responses to E2S.
The apparent upregulation of both HIF1A and ARNT2 by E2S in the present experiment suggests that during the last stage of gestation before spontaneous parturition, the fetal hypothalamus increases the expression of hypoxic-related genes in response to increased hypothalamic E2 and E2S concentrations or increased abundance of ERα (34), even when the fetus in not undergoing clinical hypoxia. Given that the genes of the bHLH-PAS family can interact with ER, the mediator of this effect might be E2 through ERα binding. In support to this notion, it was shown that injection of E2 increased the expression of ARNT2 mRNA in the hypothalamus of rats without affecting the expression of AHR and ARNT mRNA, possibly through interaction with ERα (23).
COPI System
COPI-coated vesicles are vesicular carriers that function in the early secretory pathway, especially the retrograde transport of luminal and membrane proteins in the ER-Golgi segment of the secretory pathway (5, 21). The COPI system is present in the CNS, but the effects of estrogen action on the expression of this system in the hypothalamus are unknown. The qRT-PCR validation showed a significant difference in mRNA expression between treatment and control only for COPB1 (Fig. 4) but not for COPA. Consequently, this action of E2S treatment cannot be confirmed in the present study.
TGFβ1
It has been previously documented that E2 treatment increases TGFβ1 release from hypothalamic astrocytes in vitro (8) and increases the expression of TGFB1 gene in the hypothalamus of ovariectomized female rats in vivo (22, 28). Increased TGB1 expression after E2 stimulation is mediated via an ER-dependent mechanism that involves the PI3K/Akt signaling pathway (16). The role of the estrogen-induced TGFβ1 in the hypothalamus may be that of a mediator of the ability of astroctytes to modulate GnRH release and as an intermediary of the neuroprotective effect of E2 (26). In support of this last action, the main biological processes that were downregulated by the E2S treatment in the present study were those related to inflammatory/immune response. However, changes in expression of none of the genes involved in these biological processes could be validated by qRT-PCR (CCL3, IL12B, IL18, and TNF).
We have suggested that the actions of E2S are likely to be mediated by E2, liberated after deconjugation of the E2S and binding of E2 to the estrogen receptor. While the present results reveal some genomic responses to E2S that are reminiscent of responses to E2, the NPY and AGRP response does not appear to mimic the response to E2 reported in the adult. Nevertheless, a limitation of this study is that we did not compare responses to those after treatment with E2 alone, and the question of whether E2S can act through a novel mechanism is still an open question.
In conclusion, E2S treatment of ovine fetuses near the end of gestation induces an upregulation of genes encoding factors involved in feeding behavior and response to hypoxia and possibly provides neuroprotection in the hypothalamus. The effect of E2S treatment on the orexigenic peptides observed in this study was not predicted by studies in adult animals. Our results demonstrate that E2S induced a strong increase in the expression of NPY and AGRP genes. The increased appetite induced by these neuropeptides could be an important component for the survival of the newborn and could have an effect on the regulation of energy balance regulation before and after birth.
GRANTS
This work was supported by National Institute of Child Health and Human Development Grant HD-057561 and by an ARRA supplement to this grant (HD-057561-01A2S1) to C. E. Wood.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: M.B.R., E.R., N.D., and C.E.W. analyzed data; M.B.R., E.R., N.D., M.K.-W., and C.E.W. interpreted results of experiments; M.B.R., E.R., and C.E.W. prepared figures; M.B.R., E.R., N.D., M.K.-W., and C.E.W. drafted manuscript; M.B.R., E.R., N.D., M.K.-W., and C.E.W. edited and revised manuscript; M.B.R., E.R., N.D., M.K.-W., and C.E.W. approved final version of manuscript; E.R. and C.E.W. performed experiments; M.K.-W. and C.E.W. conception and design of research.
ACKNOWLEDGMENTS
We thank Dr. Henry Baker for advice concerning the analysis and interpretation of the genomics data. We also thank Xiaoying (Lisa) Fang for help with RNA isolation. We thank Dr. Christine Schlaerth for work performed in the in vivo experiments. Finally, we thank the Interdisciplinary Center for Biotechnology Research at the University of Florida for the use of the Agilent Bioanalyzer and Agilent scanner.
REFERENCES
- 1. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4: 2, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belsham DD, Cai F, Cui H, Smukler SR, Salapatek AM, Shkreta L. Generation of a phenotypic array of hypothalamic neuronal cell models to study complex neuroendocrine disorders. Endocrinology 145: 393–400, 2004 [DOI] [PubMed] [Google Scholar]
- 4. Benita Y, Kikuchi H, Smith AD, Zhang MQ, Chung DC, Xavier RJ. An integrative genomics approach identifies Hypoxia Inducible Factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res 37: 4587–4602, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bethune J, Wieland F, Moelleken J. COPI-mediated transport. J Membr Biol 211: 65–79, 2006 [DOI] [PubMed] [Google Scholar]
- 6. Bonavera JJ, Dube MG, Kalra PS, Kalra SP. Anorectic effects of estrogen may be mediated by decreased neuropeptide-Y release in the hypothalamic paraventricular nucleus. Endocrinology 134: 2367–2370, 1994 [DOI] [PubMed] [Google Scholar]
- 7. Brunnberg S, Pettersson K, Rydin E, Matthews J, Hanberg A, Pongratz I. The basic helix-loop-helix-PAS protein ARNT functions as a potent coactivator of estrogen receptor-dependent transcription. Proc Natl Acad Sci USA 100: 6517–6522, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buchanan CD, Mahesh VB, Brann DW. Estrogen-astrocyte-luteinizing hormone-releasing hormone signaling: a role for transforming growth factor-beta(1). Biol Reprod 62: 1710–1721, 2000 [DOI] [PubMed] [Google Scholar]
- 9. Carnegie JA, Robertson HA. Conjugated and unconjugated estrogens in fetal and maternal fluids of the pregnant ewe: a possible role for estrone sulfate during early pregnancy. Biol Reprod 19: 202–211, 1978 [DOI] [PubMed] [Google Scholar]
- 10. Cho J, Kim D, Lee S, Lee Y. Cobalt chloride-induced estrogen receptor alpha down-regulation involves hypoxia-inducible factor-1alpha in MCF-7 human breast cancer cells. Mol Endocrinol 19: 1191–1199, 2005 [DOI] [PubMed] [Google Scholar]
- 11. Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2: 2366–2382, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676, 2005 [DOI] [PubMed] [Google Scholar]
- 13. Cousins R, Wood CE. Expression of organic anion transporters 1 and 3 in the ovine fetal brain during the latter half of gestation. Neurosci Lett 484: 22–25, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cowley MA, Pronchuk N, Fan W, Dinulescu DM, Colmers WF, Cone RD. Integration of NPY, AGRP, and melanocortin signals in the hypothalamic paraventricular nucleus: evidence of a cellular basis for the adipostat. Neuron 24: 155–163, 1999 [DOI] [PubMed] [Google Scholar]
- 15. Deayton JM, Young IR, Thorburn GD. Early hypophysectomy of sheep fetuses: effects on growth, placental steroidogenesis and prostaglandin production. J Reprod Fertil 97: 513–520, 1993 [DOI] [PubMed] [Google Scholar]
- 16. Dhandapani KM, Wade FM, Mahesh VB, Brann DW. Astrocyte-derived transforming growth factor-beta mediates the neuroprotective effects of 17beta-estradiol: involvement of nonclassical genomic signaling pathways. Endocrinology 146: 2749–2759, 2005 [DOI] [PubMed] [Google Scholar]
- 17. Estrada KM, Pompolo S, Morris MJ, Tilbrook AJ, Clarke IJ. Neuropeptide Y (NPY) delays the oestrogen-induced luteinizing hormone (LH) surge in the ovariectomized ewe: further evidence that NPY has a predominant negative effect on LH secretion in the ewe. J Neuroendocrinol 15: 1011–1020, 2003 [DOI] [PubMed] [Google Scholar]
- 18. Gu YZ, Hogenesch JB, Bradfield CA. The PAS superfamily: sensors of environmental and developmental signals. Annu Rev Pharmacol Toxicol 40: 519–561, 2000 [DOI] [PubMed] [Google Scholar]
- 19. Johnson ML, Grazul-Bilska AT, Redmer DA, Reynolds LP. Effects of estradiol-17beta on expression of mRNA for seven angiogenic factors and their receptors in the endometrium of ovariectomized (OVX) ewes. Endocrine 30: 333–342, 2006 [DOI] [PubMed] [Google Scholar]
- 20. Kazi AA, Jones JM, Koos RD. Chromatin immunoprecipitation analysis of gene expression in the rat uterus in vivo: estrogen-induced recruitment of both estrogen receptor alpha and hypoxia-inducible factor 1 to the vascular endothelial growth factor promoter. Mol Endocrinol 19: 2006–2019, 2005 [DOI] [PubMed] [Google Scholar]
- 21. Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol 20: 87–123, 2004 [DOI] [PubMed] [Google Scholar]
- 22. Levings MK, Sangregorio R, Galbiati F, Squadrone S, de Waal MR, Roncarolo MG. IFN-alpha and IL-10 induce the differentiation of human type 1 T regulatory cells. J Immunol 166: 5530–5539, 2001 [DOI] [PubMed] [Google Scholar]
- 23. Lin CY, Strom A, Vega VB, Kong SL, Yeo AL, Thomsen JS, Chan WC, Doray B, Bangarusamy DK, Ramasamy A, Vergara LA, Tang S, Chong A, Bajic VB, Miller LD, Gustafsson JA, Liu ET. Discovery of estrogen receptor alpha target genes and response elements in breast tumor cells. Genome Biol 5: R66, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lombardero M, Quintanar-Stephano A, Vidal S, Horvath E, Kovacs K, Lloyd RV, Scheithauer BW. Effect of estrogen on the blood supply of pituitary autografts in rats J Anat 214: 235–244, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449, 2005 [DOI] [PubMed] [Google Scholar]
- 26. Mahesh VB, Dhandapani KM, Brann DW. Role of astrocytes in reproduction and neuroprotection. Mol Cell Endocrinol 246: 1–9, 2006 [DOI] [PubMed] [Google Scholar]
- 27. McDonald TJ, Nathanielsz PW. Bilateral destruction of the fetal paraventricular nuclei prolongs gestation in sheep. Am J Obstet Gynecol 165: 764–770, 1991 [DOI] [PubMed] [Google Scholar]
- 28. Melcangi RC, Cavarretta I, Magnaghi V, Martini L, Galbiati M. Interactions between growth factors and steroids in the control of LHRH-secreting neurons. Brain Res Brain Res Rev 37: 223–234, 2001 [DOI] [PubMed] [Google Scholar]
- 29. Muhlhausler BS, McMillen IC, Rouzaud G, Findlay PA, Marrocco EM, Rhind SM, Adam CL. Appetite regulatory neuropeptides are expressed in the sheep hypothalamus before birth. J Neuroendocrinol 16: 502–507, 2004 [DOI] [PubMed] [Google Scholar]
- 30. Ohtake F, Takeyama K, Matsumoto T, Kitagawa H, Yamamoto Y, Nohara K, Tohyama C, Krust A, Mimura J, Chambon P, Yanagisawa J, Fujii-Kuriyama Y, Kato S. Modulation of oestrogen receptor signalling by association with the activated dioxin receptor. Nature 423: 545–550, 2003 [DOI] [PubMed] [Google Scholar]
- 31. Purinton SC, Newman H, Castro MI, Wood CE. Ontogeny of estrogen sulfatase activity in ovine fetal hypothalamus, hippocampus, and brain stem. Am J Physiol Regul Integr Comp Physiol 276: R1647–R1652, 1999 [DOI] [PubMed] [Google Scholar]
- 32. Purinton SC, Wood CE. Ovine fetal estrogen sulfotransferase in brain regions important for hypothalamus-pituitary-adrenal axis control. Neuroendocrinology 71: 237–242, 2000 [DOI] [PubMed] [Google Scholar]
- 33. Ruder HJ, Loriaux L, Lipsett MB. Estrone sulfate: production rate and metabolism in man. J Clin Invest 51: 1020–1033, 1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schaub CE, Gersting JA, Keller-Wood M, Wood CE. Development of ER-alpha and ER-beta expression in the developing ovine brain and pituitary. Gene Expr Patterns 8: 457–463, 2008 [DOI] [PubMed] [Google Scholar]
- 35. Schaub CE, Keller-Wood M, Wood CE. Blockade of estrogen receptors decreases CNS and pituitary prostaglandin synthase expression in fetal sheep. Neuroendocrinology 87: 121–128, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Silva LE, Castro M, Amaral FC, ntunes-Rodrigues J, Elias LL. Estradiol-induced hypophagia is associated with the differential mRNA expression of hypothalamic neuropeptides. Braz J Med Biol Res 43: 759–766, 2010 [DOI] [PubMed] [Google Scholar]
- 37. Stolze I, Berchner-Pfannschmidt U, Freitag P, Wotzlaw C, Rossler J, Frede S, Acker H, Fandrey J. Hypoxia-inducible erythropoietin gene expression in human neuroblastoma cells. Blood 100: 2623–2628, 2002 [DOI] [PubMed] [Google Scholar]
- 38. Theocharidis A, van Dongen S, Enright AJ, Freeman TC. Network visualization and analysis of gene expression data using BioLayout Express(3D). Nat Protoc 4: 1535–1550, 2009 [DOI] [PubMed] [Google Scholar]
- 39. Titolo D, Cai F, Belsham DD. Coordinate regulation of neuropeptide Y and agouti-related peptide gene expression by estrogen depends on the ratio of estrogen receptor (ER) alpha to ERbeta in clonal hypothalamic neurons. Mol Endocrinol 20: 2080–2092, 2006 [DOI] [PubMed] [Google Scholar]
- 40. Titolo D, Mayer CM, Dhillon SS, Cai F, Belsham DD. Estrogen facilitates both phosphatidylinositol 3-kinase/Akt and ERK1/2 mitogen-activated protein kinase membrane signaling required for long-term neuropeptide Y transcriptional regulation in clonal, immortalized neurons. J Neurosci 28: 6473–6482, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tsang CPW. Changes in plasma levels of estrone sulfate and estrone in the pregnant ewe around parturition. Steroids 23: 855–868, 1974 [DOI] [PubMed] [Google Scholar]
- 42. Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38: W214–W220, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Winikor J, Schlaerth C, Rabaglino MB, Cousins R, Sutherland M, Wood CE. Complex actions of estradiol-3-sulfate in late gestation fetal brain. Reprod Sci 18: 654–665, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wood CE. Estrogen/hypothalamus-pituitary-adrenal axis interactions in the fetus: the interplay between placenta and fetal brain. J Soc Gynecol Investig 12: 67–76, 2005 [DOI] [PubMed] [Google Scholar]
- 45. Wood CE. Fetal hypothalamus-pituitary-adrenal responses to estradiol sulfate. Endocrinology 152: 4966–4973, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wood CE, Gridley KE, Keller-Wood M. Biological activity of 17beta-estradiol-3-sulfate in ovine fetal plasma and uptake in fetal brain. Endocrinology 144: 599–604, 2003 [DOI] [PubMed] [Google Scholar]
- 47. Wood CE, Saoud CJ. Influence of estradiol and androstenedione on ACTH and cortisol secretion in the ovine fetus. J Soc Gynecol Investig 4: 279–283, 1997 [PubMed] [Google Scholar]
- 48. Xiao E, Kim AJ, Dutia R, Conwell I, Ferin M, Wardlaw SL. Effects of estradiol on cerebrospinal fluid levels of agouti-related protein in ovariectomized rhesus monkeys. Endocrinology 151: 1002–1009, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]