Abstract
Genomic studies on fetal alcohol spectrum disorders (FASD) have utilized either genome-wide microarrays/bioinformatics or targeted real-time PCR (RT-PCR). We utilized herein for the first time a novel digital approach with high throughput as well as the capability to focus on one physiological system. The aim of the present study was to investigate alcohol-induced alterations in uterine angiogenesis-related mRNA abundance using digital mRNA technology. Four biological and three technical replicates of uterine arterial endothelial cells from third-trimester ewes were fluorescence-activated cell sorted, validated, and treated without or with binge-like alcohol. A capture probe covalently bound to an oligonucleotide containing biotin and a color-coded reporter probe were designed for 85 angiogenesis-related genes and analyzed with the Nanostring nCounter system. Twenty genes were downregulated (↓) and two upregulated (↑), including angiogenic growth factors/receptors (↓placental growth factor), adhesion molecules (↓angiopoietin-like-3; ↓collagen-18A1; ↓endoglin), proteases/matrix proteins/inhibitors (↓alanyl aminopeptidase; ↓collagen-4A3; ↓heparanase; ↓plasminogen, ↑plasminogen activator urokinase; ↓platelet factor-4; ↓plexin domain containing-1; ↓tissue inhibitor of metalloproteinases-3), transcription/signaling molecules (↓heart and neural crest derivatives-2; ↓DNA-binding protein inhibitor; ↓NOTCH-4; ↓ribosomal protein-L13a1; ↓ribosomal protein large-P1), cytokines/chemokines (↓interleukin-1B), and miscellaneous growth factors (↓leptin; ↓platelet-derived growth factor-α); ↓transforming growth factor (TGF-α; ↑TGF-β receptor-1). These novel data show significant detrimental alcohol effects on genes controlling angiogenesis supporting a mechanistic role for abnormal uteroplacental vascular development in FASD. The tripartite digital gene expression system is therefore a valuable tool to answer many additional questions about FASD from both mechanistic as well as ameliorative perspectives.
Keywords: fetal alcohol spectrum disorders, pregnancy, alcohol, genomics
fetal alcohol spectrum disorders (FASD) refers to the set of deficits observed in the fetus or the offspring exposed to alcohol in utero (50). Since the recognition of prenatal alcohol-induced malformations in children (27), four decades of clinical, animal model, and cell culture system-based research has demonstrated alcohol effects on numerous physiological systems spanning neural development and function (1, 19, 49), cardiovascular (4, 8, 52), endocrine (10, 58), immune (60), nutrient homeostasis (47), and utero-placental (14, 16, 24, 44) systems. Multiple important mechanisms have been proposed for these deficits including altered fetal and uteroplacental angiogenesis, i.e., formation of new blood vessels from existing ones.
Gestational uterine angiogenesis is an important vascular adaptation that occurs to meet the nutrient and substrate requirement of the developing fetus and has important implications for fetal growth and development (34, 45, 61). The angiogenic process is initiated and regulated by the coordinated activity of numerous angiogenic growth factors, adhesion molecules, extracellular matrix proteins, transcription factors, signaling molecules, cytokines, and chemokines (61). Few reports exist on maternal alcohol effects on angiogenesis per se. For instance, a human prenatal alcohol study on embryos and fetuses from terminated pregnancies showed a significant decrease in many angiogenic indexes, including the mean brain vessel cross-sectional area and vessel perimeter by the 11th wk of gestation (51). In rats, one particular study showed a decrease in the capillary density of the dentate gyrus of the fetal hippocampal formation but not in the cerebellum (29). Alcohol is also reported to inhibit estrogen-induced ovine uterine artery endothelial proliferation (43). In a chick extraembryonic model, numerous angiogeneic genes were dose-dependently downregulated including basic fibroblast growth factor (FGF), vascular endothelial growth factor (VEGF), VEGF family (Fms-related tyrosine) kinase (FLT)-1, and Flk-1 with perturbation of angiogenic processes with moderate and heavy alcohol doses (54). However, none of these studies have utilized a global genomic approach to assess effects of alcohol on angiogenesis during gestation nor taken advantage of high-throughput methodologies like genomics and proteomics that have the potential to yield a wealth of information on the underlying mechanisms.
Genomic studies in FASD have utilized the microarray technology, computational bioinformatics or real-time PCR (RT-PCR) (20, 22, 46, 48). Microarrays and bioinformatics are global genome-wide approaches, whereas RT-PCR is an extremely targeted method of profiling with both platforms having little utility to assess specific physiological functions. Therefore, bridging the gap between microarrays and RT-PCR, we utilized a multiplexed tripartite high-throughput quantitative digital mRNA approach for simultaneously studying the expression of 85 genes but focusing on one physiological system, i.e., uterine angiogenesis. In the current study, we examined the effects of chronic binge-like alcohol on maternal uterine angiogenic mRNA transcriptome using the digital mRNA technology. In this method, the total RNA is hybridized with gene-specific capture probes that are covalently bound to biotin-containing oligonucleotides and color-coded reporter probes forming a tripartite complex, to directly provide the total number of transcripts (15). The digital mRNA method thus eliminates cDNA synthesis, enzymatic reactions, amplification, or bias and has a sensitivity greater than microarrays and similar to TaqMan-PCR (15, 41). In this study, we examined uterine angiogenesis during the third-trimester equivalent of human gestation, a period highly sensitive to alcohol exposure (9, 31, 59).
MATERIALS AND METHODS
Alcohol dosing.
The Animal Care and Use Committee of the University of Wisconsin-Madison approved procedures for obtaining uterine arteries from pregnant ewes (day 120–130, term = 147) for endothelial cells isolation using collagenase digestion procedures (2). Four biological replicates (cell lines derived from four different pregnant ewes) and three technical replicates were utilized. Thus, 12 Control and 12 Alcohol treatment group samples were utilized for the 24 hybridization reactions. Samples were not pooled. The procedure for alcohol binging has been described elsewhere (44). In brief, cells were purified by fluorescence-activated cell sorting, devoid of vascular smooth muscle cell contamination, and maintained in culture to passage 4. Cells were cultured to ∼70% confluence in the absence (0 mg/dl, Control) or presence of alcohol (300 mg/dl), a dose similar to the peak blood alcohol concentrations in previous ovine in vivo as well as cell culture system-based FASD studies (9, 43, 44, 59). Cells were exposed to a 2 wk binge-like paradigm of alcohol exposure in a sealed compensating system equilibrated with aqueous alcohol for 3 h on 3 consecutive days for 2 wk (13, 59), a pattern common among drinking women of childbearing age (5, 18, 35). Cell viability was validated prior to commencement of the study. At the end of the experiment, the endothelial cells were scraped and collected in RNeasy Mini lysis buffer (Qiagen, Valencia, CA).
Design of gene-specific probes.
We selected 85 angiogenic genes and five housekeeping genes based on previously published reports (25, 30), and gene-specific probes were constructed as described in the Supplementary Information.1 All targets were either RefSeq Ovis aries records or O. aries expressed sequence tag sequences that have sequence identity to Bos taurus orthologs. The gene identifier, accession number, targeted region, target sequence, melting temperatures, and the respective gene name are specified in the Supplementary Information.
Reporter and capture probe construction.
Details on the reporter and capture probe construction have been previously described (17). In brief, the reporter backbone consisted of a single-stranded DNA substrate with four 15-base repeats at the 5′ end. The backbone was then annealed to fluorescently labeled amino-allyl-labeled RNA transcripts coupled to a fluorophore (Alexa 488/594/647 or Cy3). Gene-specific probes annealed to a bridge oligonucleotide were then ligated to the reporter backbone to make a DNA/RNA double-stranded hybrid. This step was followed by affinity purification using magnetic beads coupled to oligonucleotides complementary to the 5′ end. Each labeled region was a 300 nm spot in an epifluorescent microscope. For capture probes, the gene-specific probes annealed to a bridge oligonucleotide were ligated to 3′ repeats (two 15-base repeats) linked to a biotin molecule. Each capture probe was then affinity purified with magnetic beads coupled to oligonucleotides complementary to the 3′ end. Selection and screening were performed to eliminate long direct and inverted repeats, high GC content, and long poly-C stretches followed by cross hybridization screening with the National Center for Biotechnology's basic local alignment search tool.
Tripartite hybridization.
We incubated 100 pM of capture probe, 25 pM reporter probe, 5× saline sodium phosphate EDTA buffer (pH, 7.5), 3 μl of uterine artery endothelial cell lysate (3 technical replicates, 4 pairs of biological replicates), and 6 positive and 8 negative control probe pairs at 65°C in a thermocycler block for 20 h (17). As previously described, unhybridized reporter and capture probes were removed via affinity purification with 3′ repeat complementary sequence followed by 5′ (17). Samples were driven through a channel in Nanostring fluidic device (has streptavidin) and washed with Tris-acetate EDTA. Reporters were then stretched using 160 V/cm for 1 min in the channel and immobilized, and Slowfade reagent was added. Imaging was performed using Nikon Eclipse TE2000E, 1.4 NA, Plan APO VC 60X, oil with a digital analyzer, and the mRNA transcripts were counted (17). The data were then normalized to the average counts for all control spikes in each sample and to the geometric mean of five housekeeper genes (actin-β, β2-microglobulin, glyceraldehyde-3-phosphate dehydrogenase, hypoxanthine phosphoribosyltransferase 1, and lactate dehydrogenase A).
Statistics.
The number of transcripts were log transformed and then compared between the Control (n = 4) and Alcohol groups (n = 4) using a paired Student's t-test. α-Level of significance was established a priori at P < 0.05.
RESULTS
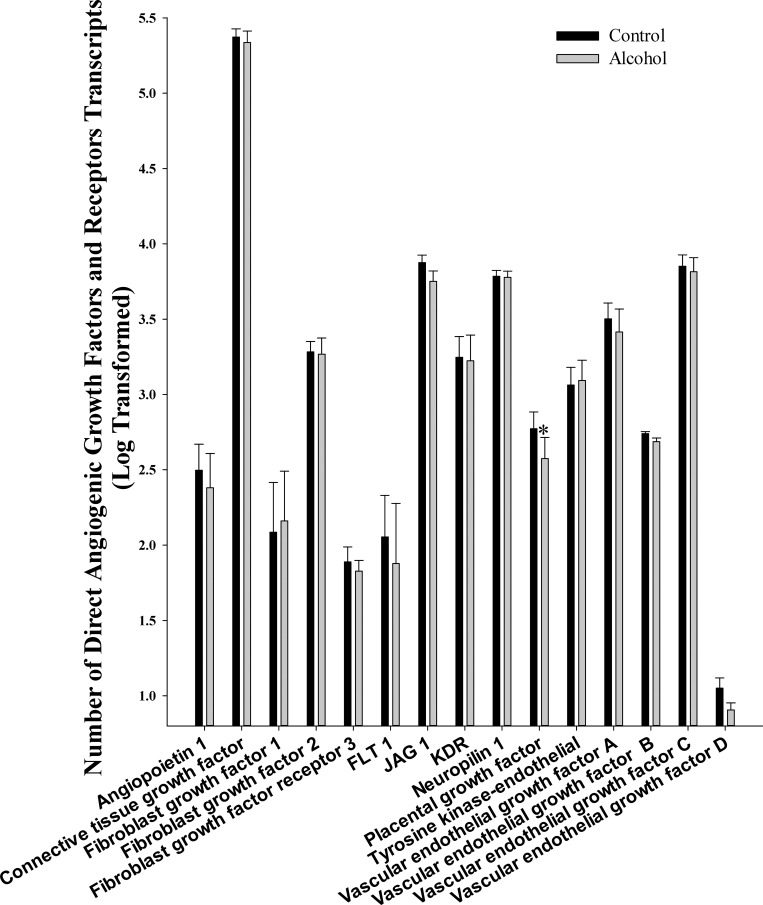
We analyzed 16 growth factors that directly promote angiogenesis including angiopoietin 1, connective tissue growth factor, FGF 1 and 2, FGF receptor 3, FLT-1, kinase insert domain receptor (KDR), VEGF A, B, C, and D, Jagged 1, neuropilin 1, placental growth factor, and tyrosine kinase-endothelial (Fig. 1). Of these, alcohol significantly decreased only placental growth factor (P = 0.036).
Fig. 1.
Effect of chronic binge-like alcohol on the number of direct angiogenic growth factor and receptor transcripts. Out of the 16 growth factors/receptors analyzed, alcohol decreased only placental growth factor (P = 0.036) mRNA expression. Data are log transformed and presented as means ± SE. *Significant difference between Control and Alcohol groups.
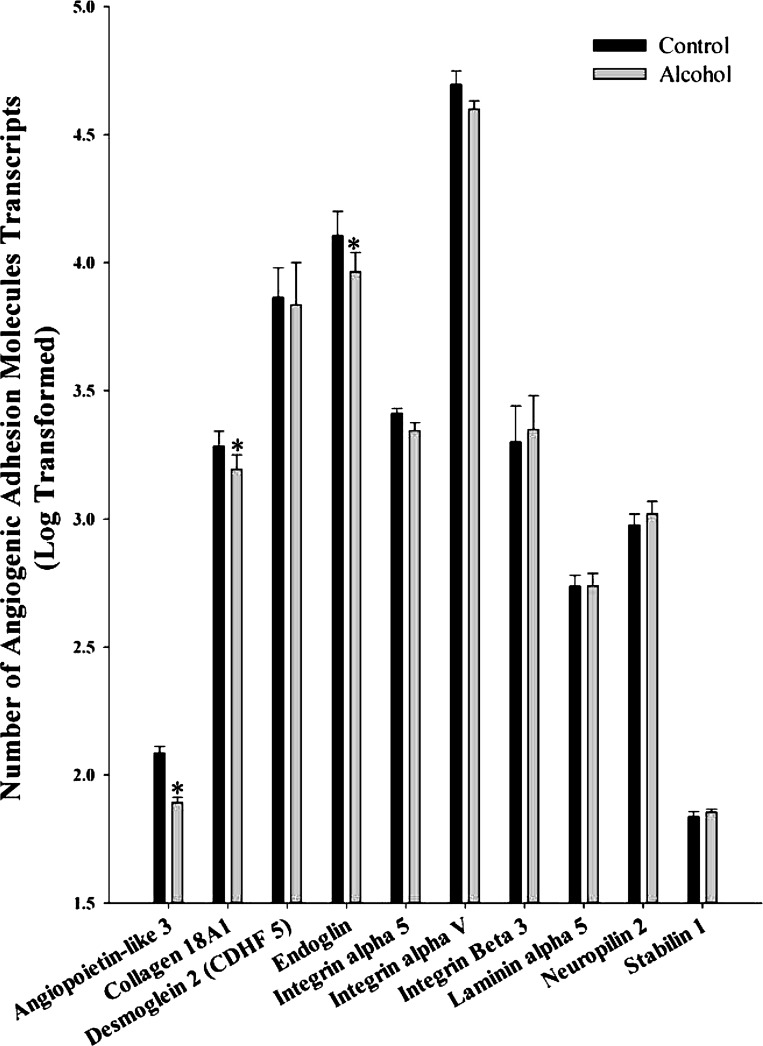
We analyzed 10 adhesion molecules associated with angiogenesis including angiopoietin-like 3, collagen 18A1, desmoglein 2 (CDHF 5), endoglin, integrin α5, integrin αV, integrin β3, laminin α5, neuropilin 2, and stabilin 1 (Fig. 2). Of these, three were significantly decreased in response to alcohol including angiopoietin-like 3 (P = 0.007), collagen 18A1 (P = 0.031), and endoglin (P = 0.032).
Fig. 2.
Effect of chronic binge-like alcohol on the number of adhesion molecule transcripts associated with angiogenesis. Out of the 10 genes analyzed, alcohol decreased angiopoietin-like 3 (P = 0.007), collagen 18A1 (P = 0.031), and endoglin (P = 0.032) mRNA expression. Data are log transformed and presented as means ± SE. *Significant difference between Control and Alcohol groups.
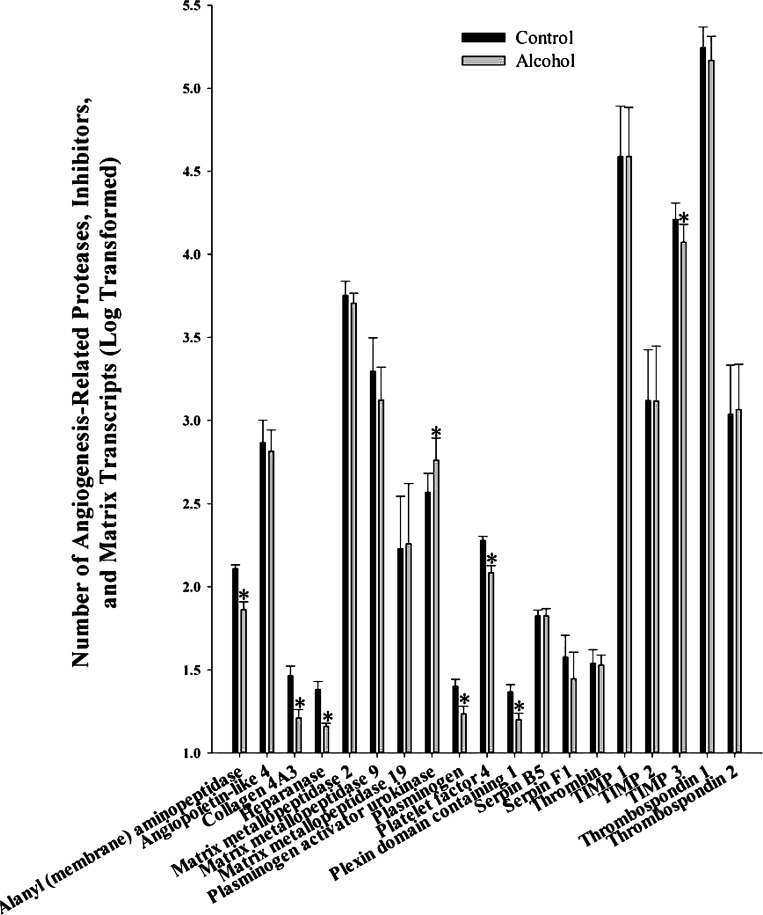
We analyzed 19 angiogenesis-related proteases, matrix proteins, and inhibitors including alanyl (membrane) aminopeptidase, angiopoietin-like 4, collagen 4A3, heparanase, matrix metallopeptidase 2, matrix metallopeptidase 9, matrix metallopeptidase 19, plasminogen activator urokinase, plasminogen, platelet factor 4, plexin domain containing 1, serpin B5, serpin F1, thrombin, tissue inhibitor of metalloproteinases (TIMP) 1, 2, and 3, thrombospondin 1 and 2 (Fig. 3). Of these, alcohol significantly decreased alanyl (membrane) aminopeptidase (P = 0.008), collagen 4A3 (P = 0.010), heparanase (P = 0.041), plasminogen (P = 0.002), platelet factor 4 (P = 0.008), plexin domain containing 1 (P = 0.037), and TIMP 3 (P = 0.014) and increased plasminogen activator urokinase (P = 0.019).
Fig. 3.
Effect of chronic binge-like alcohol on the number of angiogenesis-related proteases, matrix proteins, and inhibitor transcripts. Out of the 19 genes analyzed, alcohol decreased alanyl (membrane) aminopeptidase (P = 0.008), collagen 4A3 (P = 0.010), heparanase (P = 0.041), plasminogen (P = 0.002), platelet factor 4 (P = 0.008), plexin domain containing 1 (P = 0.037), and tissue inhibitor of metalloproteinases (TIMP) 3 (P = 0.014) and increased plasminogen activator urokinase (P = 0.019) mRNA expression. Data are log transformed and presented as means ± SE. *Significant difference between Control and Alcohol groups.
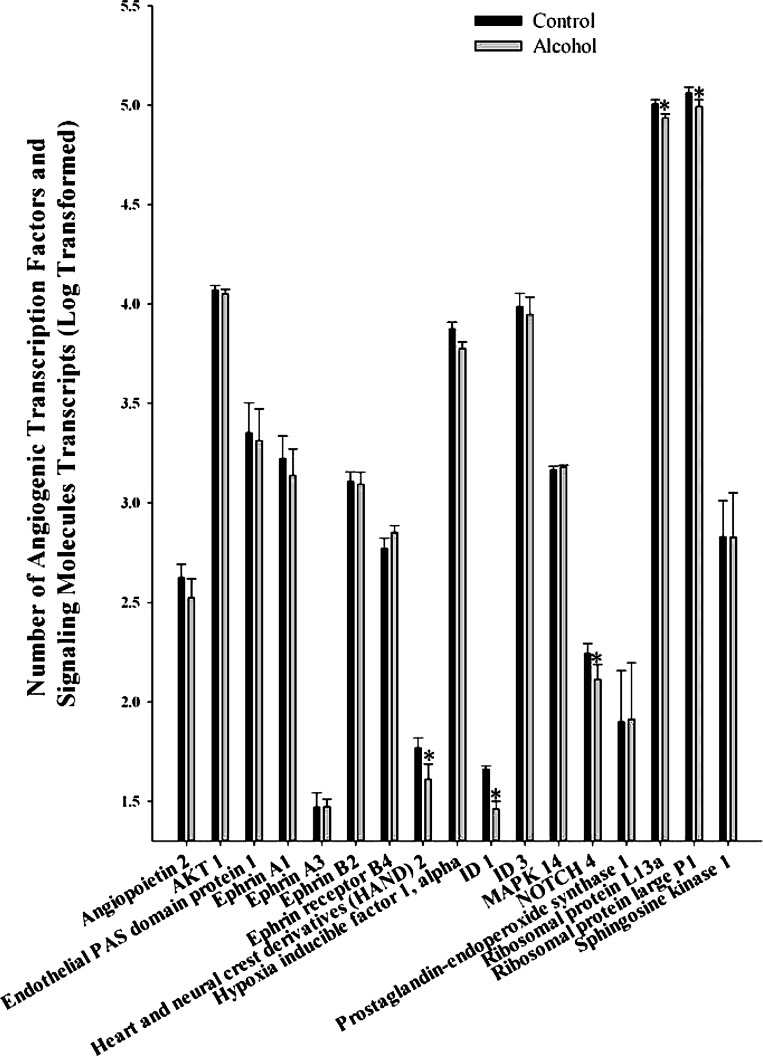
We studied 17 transcription factors and signaling molecules associated with angiogenesis including angiopoietin 2, AKT 1, endothelial PAS domain protein 1, ephrin A1, ephrin A3, ephrin B2, ephrin receptor B4, heart and neural crest derivatives (HAND) 2, hypoxia-inducible factor 1 alpha, DNA-binding protein inhibitor (ID) 1, ID 3, mitogen-activated protein kinase (MAPK) 14, NOTCH 4, prostaglandin-endoperoxide synthase 1, ribosomal protein L13a, ribosomal protein large P1, and sphingosine kinase 1 (Fig. 4). Of these, five were significantly decreased including HAND 2 (P = 0.033), ID 1 (P = 0.013), NOTCH 4 (P = 0.016), ribosomal protein L13a (P = 0.014), and ribosomal protein large P1 (P = 0.016).
Fig. 4.
Effect of chronic binge-like alcohol on the number of transcription factors and signaling molecule transcripts associated with angiogenesis. Out of the 17 genes analyzed, alcohol decreased heart and neural crest derivatives 2 (P = 0.033), DNA-binding protein inhibitor (ID) 1 (P = 0.013), NOTCH 4 (P = 0.016), ribosomal protein L13a (P = 0.014), and ribosomal protein large P1 (P = 0.016) mRNA expression. Data are log transformed and presented as means ± SE. *Significant difference between Control and Alcohol groups.
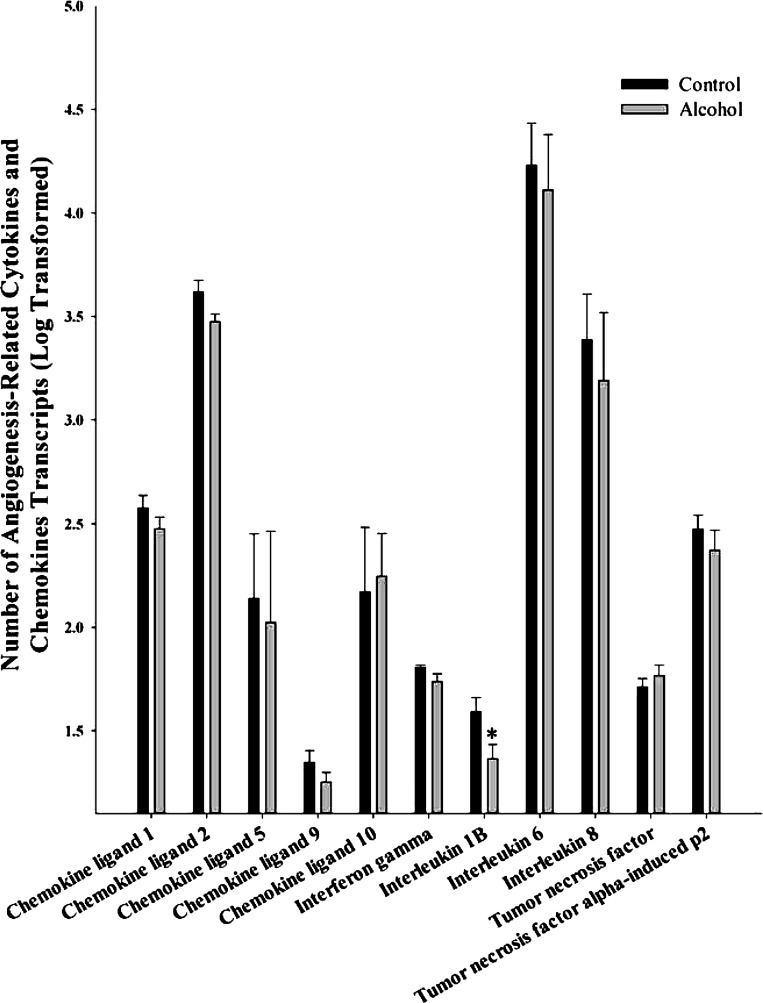
We examined 11 cytokines and chemokines related to angiogenesis including chemokine ligand 1, chemokine ligand 2, chemokine ligand 5, chemokine ligand 9, chemokine ligand 10, interferon gamma, interleukin 1B, interleukin 6, interleukin 8, tumor necrosis factor (TNF), and TNF α-induced protein (α-Ip)2 (Fig. 5). Of these, only interleukin 1B (P = 0.037) was significantly decreased by alcohol.
Fig. 5.
Effect of chronic binge-like alcohol on the number of cytokines and chemokine transcripts related to angiogenesis. Out of the 11 genes analyzed, interleukin 1B (P = 0.037) alone was significantly decreased by alcohol. Data are log transformed and presented as means ± SE. *Significant difference between Control and Alcohol groups.
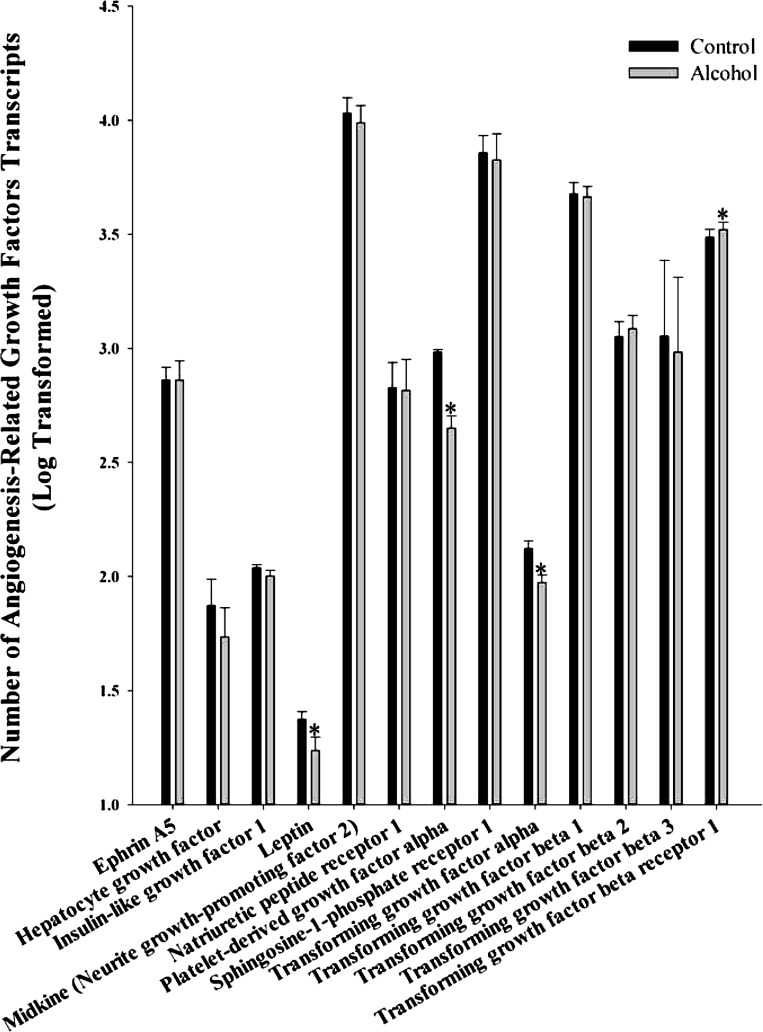
We also analyzed 13 miscellaneous angiogenic growth factors including ephrin A5, hepatocyte growth factor, insulin-like growth factor 1, leptin, midkine (neurite growth-promoting factor 2), natriuretic peptide receptor 1, platelet-derived growth factor (PDGF)-α, sphingosine-1-phosphate receptor 1, transforming growth factor (TGF)-α, TGF-β1, TGF-β2, TGF-β3, and TGF-β receptor 1 (Fig. 6). Of these, leptin (P = 0.016), PDGF-α (P = 0.011), and TGF-α (P = 0.045) were decreased, whereas TGF-β receptor 1 (P = 0.021) was increased.
Fig. 6.
Effect of chronic binge-like alcohol on the number of miscellaneous angiogenic growth factor transcripts. Out of the 13 genes analyzed, alcohol decreased leptin (P = 0.016), platelet-derived growth factor-α (P = 0.011), transforming growth factor (TGF)-α (P = 0.045) and increased TGF-β receptor 1 (P = 0.021) mRNA expression. Data are log transformed and presented as means ± SE. *Significant difference between Control and Alcohol groups.
DISCUSSION
This is the first study to utilize the technology of tripartite multiplexed digital mRNA in the field of FASD. Results from these data suggest that alcohol has detrimental effects on uterine angiogenesis, as overall 20 angiogenesis-related genes were downregulated and two genes upregulated. From a technical perspective, the current study is significant as tripartite multiplexed digital PCR technology is considered a state-of-the-art method that provides a digital count of the number of transcripts for multiple genes; at the same time we targeted one physiological system, thus bridging the gap between microarray technique and the highly specific RT-PCR (15). In addition, this technology has the capability to measure less abundant targets (as low as 0.5 fM of mRNA) (15, 41). The technique is also reported to have a high level of linearity and reproducibility (17).
So far in the field of FASD, few studies have utilized high-throughput techniques. Genomic studies in FASD have utilized the microarray technology, computational bioinformatics, or RT-PCR. In a mouse model of FASD, gene expression profiling utilizing Perkin-Elmer and Affymetrix platforms demonstrated substrain-specific metabolic and cellular reprogramming in the embryonic headfold (20). Hard and coworkers (22), again in mice, used microarray to identify 25 genes that were all downregulated in the developing brain. In rats, a microarray study of 28,000 genes was conducted; 304 placental genes were altered by greater than twofold by alcohol (46). Utilizing a computational gene expression mining approach, a study illustrated candidate pathways including TGF-β and MAPK signaling (32). Another study using mathematical modeling of microarray gene expression data from alcohol-treated mouse embryos identified candidate transcription factor and microRNA binding sites (56). RT-PCR has also been utilized; Shukla et al. (48) showed alteration in thyroid and glucocorticoid system-associated mRNAs in the placenta of rats exposed to alcohol during gestation. Thus, it can be noted that both these reports have utilized a global mining approach, like the microarrays and bioinformatics, or an extremely targeted approach, like RT-PCR. We herein have utilized the multiplexed high-throughput quantitative digital mRNA technology that is neither as nonspecific as the microarray platform nor as limiting as the RT-PCR (15). Thus, the current study is the first targeted report that utilizes a high-throughput methodology for global alterations in gene expression and to closely examine one particular physiological phenomenon, i.e., uterine angiogenesis.
Herein we show that alcohol potentially has detrimental effects on uterine angiogenesis during pregnancy. Our findings in general are in agreement with other reports on effects of prenatal alcohol exposure on placental and fetal angiogenesis. Human clinical studies show alteration in angiogeneic indexes such as mean vessel cross-sectional area and perimeter in the fetal brains (51). In rats, Kelly and coworkers (29) showed altered microvascularity in pups exposed to alcohol during the third-trimester equivalent of human gestation. Another study in rats showed that alcohol impairs placentation and prevented the conversion of uterine vessels and attributed these deficits to altered trophoblast aspartyl-(asparaginyl) β-hydroxylase levels (21). In sheep, it has been demonstrated that estradiol-17 β-induced uterine endothelial proliferation is decreased in response to alcohol (43). In a chick model of FASD, Tufan and Satiroglu-Tufan (54) demonstrated alcohol-induced deficits in extraembryonic vascular development and attributed these effects to oxidative stress, angiogenic growth factors, and their receptors. In summary, chronic binge-like alcohol exposure has significant negative effects on uterine angiogenic genes during pregnancy. As these genes control the regulation of angiogenesis including endothelial proliferation, migration, and extracellular matrix remodeling, these data provide important genomic information on alcohol-induced alterations to uteroplacental angiogenesis and gestational vascular adaptations.
Among direct angiogenic factors, only placental growth factor was significantly decreased in response to alcohol. Placental growth factor along with VEGF A and B acts via the VEGF receptor FLT 1, and amplifies the response to VEGF (7). VEGF A, C, and D, as well as Flt 1 and KDR were not altered in the current study. Though we did not consider statistical trends, it was interesting to note that VEGF B tended to be decreased (P = 0.054) in response to alcohol. Among adhesion-associated genes, angiopoietin-like 3, collagen 18A1, and endoglin were decreased. We show herein that angiopoietin-like 3, a factor that promotes angiogenesis with its fibrinogen-like domain, is expressed outside the liver (23), i.e., in the uterine vasculature. Collagen 18A1 may have a mixed role in angiogenesis, as it is important for cell polarity and differentiation while its COOH terminus produces endostatin, an inhibitor of angiogenesis (40). Endoglin is an essential angiogeneic member that has been implicated in vascular development diseases including pre-eclampsia (53). Among proteases, inhibitors, and matrix proteins that were decreased in response to alcohol in the present study, alanyl (membrane) aminopeptidase activity is known to be altered in adult male synaptosomes by alcohol (37); collagen 4A3 promotes nitric oxide-induced angiogenesis (57); heparanase induces angiogenesis via possibly basic FGF (12); plasminogen and urokinase are directly involved in extracellular matrix remodeling (39, 42); platelet factor 4 inhibits FGF 2 and VEGF binding to their respective receptors (28); plexin domain containing 1 is one of the plexin family members that directly promote angiogenesis (38); and TIMP 3 plays an important role in vascularization by affecting the extracellular matrix (26). Among transcription factors and signaling molecules decreased in response to alcohol in the current study, HAND 2 is one of the genes downregulated by alcohol in placenta (46); ID 1 is important for branching and sprouting of blood vessels (33); NOTCH 4 specifies arterial fate and suppresses venous differentiation (6); and ribosomal protein L13a1 and ribosomal protein large P1 are part of the translational machinery. Furthermore, these alcohol-induced alterations in transcription factors and signaling molecules might regulate the expression of other genes. Among cytokines and chemokines, interleukin 1B, a factor that increases VEGF expression, and its receptors were alone downregulated in the current study (36). Among miscellaneous angiogenesis-related growth factors that were decreased in this study, leptin generates a growth signal and thus promotes angiogenesis (3), PDGF-α is a mitogenic growth factor (11), and TGF-α is a cell survival growth factor through which TGF-β promotes angiogenesis (55). In contrast, TGF-β receptor 1 was significantly increased, and we reason that the increase was possibly a compensatory response to the decreases in the ligand gene expression levels.
We conclude that tripartite digital gene expression system provides a valuable tool to answer many additional questions of FASD from both mechanistic as well as ameliorative perspectives. Furthermore, these data clearly establish that chronic binge-like alcohol has negative effects on many genes related to uterine angiogenesis and thus may impair important gestational adaptations that are required for fetal growth and development.
GRANTS
Supported by National Institutes of Health Grants AA-19446 (to J. Ramadoss) and HL-49210, HD-38843, and HL-89144 (to R. R. Magness).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: J.R. and R.R.M. conception and design of research; J.R. and R.R.M. performed experiments; J.R. and R.R.M. analyzed data; J.R. and R.R.M. interpreted results of experiments; J.R. and R.R.M. prepared figures; J.R. and R.R.M. drafted manuscript; J.R. and R.R.M. edited and revised manuscript; J.R. and R.R.M. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Gladys Lopez for technical assistance at University of Wisconsin-Madison, and Nanostring Technologies, Seattle, Washington for nCounter system analyses.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1. Archibald SL, Fennema-Notestine C, Gamst A, Riley EP, Mattson SN, Jernigan TL. Brain dysmorphology in individuals with severe prenatal alcohol exposure. Dev Med Child Neurol 43: 148–154, 2001. [PubMed] [Google Scholar]
- 2. Bird IM, Sullivan JA, Di T, Cale JM, Zhang L, Zheng J, Magness RR. Pregnancy-dependent changes in cell signaling underlie changes in differential control of vasodilator production in uterine artery endothelial cells. Endocrinology 141: 1107–1117, 2000. [DOI] [PubMed] [Google Scholar]
- 3. Bouloumie A, Drexler HC, Lafontan M, Busse R. Leptin, the product of Ob gene, promotes angiogenesis. Circ res 83: 1059–1066, 1998. [DOI] [PubMed] [Google Scholar]
- 4. Burd L, Deal E, Rios R, Adickes E, Wynne J, Klug MG. Congenital heart defects and fetal alcohol spectrum disorders. Congen Heart Dis 2: 250–255, 2007. [DOI] [PubMed] [Google Scholar]
- 5. Caetano R, Ramisetty-Mikler S, Floyd LR, McGrath C. The epidemiology of drinking among women of child-bearing age. Alcohol Clin Exp Res 30: 1023–1030, 2006. [DOI] [PubMed] [Google Scholar]
- 6. Carmeliet P. Angiogenesis in health and disease. Nat Med 9: 653–660, 2003. [DOI] [PubMed] [Google Scholar]
- 7. Carmeliet P, Moons L, Luttun A, Vincenti V, Compernolle V, De Mol M, Wu Y, Bono F, Devy L, Beck H, Scholz D, Acker T, DiPalma T, Dewerchin M, Noel A, Stalmans I, Barra A, Blacher S, Vandendriessche T, Ponten A, Eriksson U, Plate KH, Foidart JM, Schaper W, Charnock-Jones DS, Hicklin DJ, Herbert JM, Collen D, Persico MG. Synergism between vascular endothelial growth factor and placental growth factor contributes to angiogenesis and plasma extravasation in pathological conditions. Nat Med 7: 575–583, 2001. [DOI] [PubMed] [Google Scholar]
- 8. Cook JL, Zhang Y, Davidge ST. Vascular function in alcohol-treated pregnant and nonpregnant mice. Am J Physiol Regul Integr Comp Physiol 281: R1449–R1455, 2001. [DOI] [PubMed] [Google Scholar]
- 9. Cudd TA, Chen WJ, Parnell SE, West JR. Third trimester binge ethanol exposure results in fetal hypercapnea and acidemia but not hypoxemia in pregnant sheep. Alcohol Clin Exp Res 25: 269–276, 2001. [PubMed] [Google Scholar]
- 10. Cudd TA, Chen WJ, West JR. Fetal and maternal thyroid hormone responses to ethanol exposure during the third trimester equivalent of gestation in sheep. Alcohol Clin Exp Res 26: 53–58, 2002. [PubMed] [Google Scholar]
- 11. Dunn IF, Heese O, Black PM. Growth factors in glioma angiogenesis: FGFs, PDGF, EGF, and TGFs. J Neuro-oncol 50: 121–137, 2000. [DOI] [PubMed] [Google Scholar]
- 12. Elkin M, Ilan N, Ishai-Michaeli R, Friedmann Y, Papo O, Pecker I, Vlodavsky I. Heparanase as mediator of angiogenesis: mode of action. FASEB J 15: 1661–1663, 2001. [DOI] [PubMed] [Google Scholar]
- 13. Eysseric H, Gonthier B, Soubeyran A, Bessard G, Saxod R, Barret L. There is not simple method to maintain a constant ethanol concentration in long-term cell culture: keys to a solution applied to the survey of astrocytic ethanol absorption. Alcohol 14: 111–115, 1997. [DOI] [PubMed] [Google Scholar]
- 14. Falconer J. The effect of maternal ethanol infusion on placental blood flow and fetal glucose metabolism in sheep. Alcohol Alcohol 25: 413–416, 1990. [PubMed] [Google Scholar]
- 15. Fortina P, Surrey S. Digital mRNA profiling. Nat Biotechnol 26: 293–294, 2008. [DOI] [PubMed] [Google Scholar]
- 16. Gabriel K, Hofmann C, Glavas M, Weinberg J. The hormonal effects of alcohol use on the mother and fetus. Alcohol Health Res World 22: 170–177, 1998. [PMC free article] [PubMed] [Google Scholar]
- 17. Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 26: 317–325, 2008. [DOI] [PubMed] [Google Scholar]
- 18. Gladstone J, Nulman I, Koren G. Reproductive risks of binge drinking during pregnancy. Reprod Toxicol 10: 3–13, 1996. [DOI] [PubMed] [Google Scholar]
- 19. Goodlett CR, Eilers AT. Alcohol-induced Purkinje cell loss with a single binge exposure in neonatal rats: a stereological study of temporal windows of vulnerability. Alcohol Clin Exp Res 21: 738–744, 1997. [PubMed] [Google Scholar]
- 20. Green ML, Singh AV, Zhang Y, Nemeth KA, Sulik KK, Knudsen TB. Reprogramming of genetic networks during initiation of the Fetal Alcohol Syndrome. Dev Dyn 236: 613–631, 2007. [DOI] [PubMed] [Google Scholar]
- 21. Gundogan F, Elwood G, Longato L, Tong M, Feijoo A, Carlson RI, Wands JR, de la Monte SM. Impaired placentation in fetal alcohol syndrome. Placenta 29: 148–157, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hard ML, Abdolell M, Robinson BH, Koren G. Gene-expression analysis after alcohol exposure in the developing mouse. J Lab Clin Med 145: 47–54, 2005. [DOI] [PubMed] [Google Scholar]
- 23. Hato T, Tabata M, Oike Y. The role of angiopoietin-like proteins in angiogenesis and metabolism. Trends Cardiovasc Med 18: 6–14, 2008. [DOI] [PubMed] [Google Scholar]
- 24. Helske S, Vuorela P, Carpen O, Hornig C, Weich H, Halmesmaki E. Expression of vascular endothelial growth factor receptors 1, 2 and 3 in placentas from normal and complicated pregnancies. Mol Hum Reprod 7: 205–210, 2001. [DOI] [PubMed] [Google Scholar]
- 25. Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res 71: 3792–3801, 2011. [DOI] [PubMed] [Google Scholar]
- 26. Janssen A, Hoellenriegel J, Fogarasi M, Schrewe H, Seeliger M, Tamm E, Ohlmann A, May CA, Weber BH, Stohr H. Abnormal vessel formation in the choroid of mice lacking tissue inhibitor of metalloprotease-3. Invest Ophthalmol Vis Sci 49: 2812–2822, 2008. [DOI] [PubMed] [Google Scholar]
- 27. Jones KL, Smith DW. Recognition of the fetal alcohol syndrome in early infancy. Lancet 302: 999–1001, 1973. [DOI] [PubMed] [Google Scholar]
- 28. Jouan V, Canron X, Alemany M, Caen JP, Quentin G, Plouet J, Bikfalvi A. Inhibition of in vitro angiogenesis by platelet factor-4-derived peptides and mechanism of action. Blood 94: 984–993, 1999. [PubMed] [Google Scholar]
- 29. Kelly SJ, Mahoney JC, West JR. Changes in brain microvasculature resulting from early postnatal alcohol exposure. Alcohol 7: 43–47, 1990. [DOI] [PubMed] [Google Scholar]
- 30. Ligi I, Simoncini S, Tellier E, Vassallo PF, Sabatier F, Guillet B, Lamy E, Sarlon G, Quemener C, Bikfalvi A, Marcelli M, Pascal A, Dizier B, Simeoni U, Dignat-George F, Anfosso F. A switch toward angiostatic gene expression impairs the angiogenic properties of endothelial progenitor cells in low birth weight preterm infants. Blood 118: 1699–1709, 2011. [DOI] [PubMed] [Google Scholar]
- 31. Livy DJ, Miller EK, Maier SE, West JR. Fetal alcohol exposure and temporal vulnerability: effects of binge-like alcohol exposure on the developing rat hippocampus. Neurotoxicol Teratol 25: 447–458, 2003. [DOI] [PubMed] [Google Scholar]
- 32. Lombard Z, Tiffin N, Hofmann O, Bajic VB, Hide W, Ramsay M. Computational selection and prioritization of candidate genes for fetal alcohol syndrome. BMC Genom 8: 389, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lyden D, Young AZ, Zagzag D, Yan W, Gerald W, O'Reilly R, Bader BL, Hynes RO, Zhuang Y, Manova K, Benezra R. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature 401: 670–677, 1999. [DOI] [PubMed] [Google Scholar]
- 34. Magness RR. Maternal cardiovascular and other physiologic responses to the endocrinology of pregnancy. In: The Endocrinology of Pregnancy, edited by Bazer FW. Totowa, NJ: Humana, 1998, p. 507–539. [Google Scholar]
- 35. Maier SE, West JR. Drinking patterns and alcohol-related birth defects. Alcohol Res Health 25: 168–174, 2001. [PMC free article] [PubMed] [Google Scholar]
- 36. Maruyama K, Mori Y, Murasawa S, Masaki H, Takahashi N, Tsutusmi Y, Moriguchi Y, Shibazaki Y, Tanaka Y, Shibuya M, Inada M, Matsubara H, Iwasaka T. Interleukin-1 beta upregulates cardiac expression of vascular endothelial growth factor and its receptor KDR/flk-1 via activation of protein tyrosine kinases. J Mol Cell Cardiol 31: 607–617, 1999. [DOI] [PubMed] [Google Scholar]
- 37. Mayas MD, Ramirez-Exposito MJ, Garcia MJ, Carrera P, Martinez-Martos JM. Ethanol modulates neuropeptide-degrading aminopeptidases at synapse level in calcium-dependent conditions. Alcohol Alcohol 39: 393–405, 2004. [DOI] [PubMed] [Google Scholar]
- 38. Neufeld G, Shraga-Heled N, Lange T, Kessler O. Semaphorins, plexins and neuropilins and their role in vasculogenesis and angiogenesis. In: Modern Concepts in Angiogenesis, edited by Simons M, Rubanyi GM. London: Imperial College Press, 2007, p. 1–25. [Google Scholar]
- 39. Oh CW, Hoover-Plow J, Plow EF. The role of plasminogen in angiogenesis in vivo. J Thromb Haemost 1: 1683–1687, 2003. [DOI] [PubMed] [Google Scholar]
- 40. Passos-Bueno MR, Suzuki OT, Armelin-Correa LM, Sertie AL, Errera FI, Bagatini K, Kok F, Leite KR. Mutations in collagen 18A1 and their relevance to the human phenotype. Anais da Academia Brasileira de Ciencias 78: 123–131, 2006. [DOI] [PubMed] [Google Scholar]
- 41. Payton JE, Grieselhuber NR, Chang LW, Murakami M, Geiss GK, Link DC, Nagarajan R, Watson MA, Ley TJ. High throughput digital quantification of mRNA abundance in primary human acute myeloid leukemia samples. J Clin Invest 119: 1714–1726, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rabbani SA. Metalloproteases and urokinase in angiogenesis and tumor progression. In Vivo 12: 135–142, 1998. [PubMed] [Google Scholar]
- 43. Ramadoss J, Jobe SO, Magness RR. Alcohol and maternal uterine vascular adaptations during pregnancy-part I: effects of chronic in vitro binge-like alcohol on uterine endothelial nitric oxide system and function. Alcohol Clin Exp Res 35: 1686–1693, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ramadoss J, Magness RR. 2-D DIGE uterine endothelial proteomic profile for maternal chronic binge-like alcohol exposure. J Proteomics 74: 2986–2994, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reynolds LP, Redmer DA. Angiogenesis in the placenta. Biol Reprod 64: 1033–1040, 2001. [DOI] [PubMed] [Google Scholar]
- 46. Rosenberg MJ, Wolff CR, El-Emawy A, Staples MC, Perrone-Bizzozero NI, Savage DD. Effects of moderate drinking during pregnancy on placental gene expression. Alcohol 44: 673–690, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shankar K, Ronis MJ, Badger TM. Effects of pregnancy and nutritional status on alcohol metabolism. Alcohol Res Health 30: 55–59, 2007. [PMC free article] [PubMed] [Google Scholar]
- 48. Shukla PK, Sittig LJ, Ullmann TM, Redei EE. Candidate placental biomarkers for intrauterine alcohol exposure. Alcohol Clin Exp Res 35: 559–565, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Simmons RW, Thomas JD, Levy SS, Riley EP. Motor response programming and movement time in children with heavy prenatal alcohol exposure. Alcohol 44: 371–378, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sokol RJ, Delaney-Black V, Nordstrom B. Fetal alcohol spectrum disorder. JAMA 290: 2996–2999, 2003. [DOI] [PubMed] [Google Scholar]
- 51. Solonskii AV, Logvinov SV, Kutepova NA. Development of brain vessels in human embryos and fetuses in conditions of prenatal exposure to alcohol. Neurosci Behav Physiol 38: 373–376, 2008. [DOI] [PubMed] [Google Scholar]
- 52. Steeg CN, Woolf P. Cardiovascular malformations in the fetal alcohol syndrome. Am Heart J 98: 635–637, 1979. [DOI] [PubMed] [Google Scholar]
- 53. ten Dijke P, Goumans MJ, Pardali E. Endoglin in angiogenesis and vascular diseases. Angiogenesis 11: 79–89, 2008. [DOI] [PubMed] [Google Scholar]
- 54. Tufan AC, Satiroglu-Tufan NL. The effect of ethanol exposure on extraembryonic vascular development in the chick area vasculosa. Cells Tiss Organs 175: 84–97, 2003. [DOI] [PubMed] [Google Scholar]
- 55. Vinals F, Pouyssegur J. Transforming growth factor beta1 (TGF-beta1) promotes endothelial cell survival during in vitro angiogenesis via an autocrine mechanism implicating TGF-alpha signaling. Mol Cell Biol 21: 7218–7230, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang G, Wang X, Wang Y, Yang JY, Li L, Nephew KP, Edenberg HJ, Zhou FC, Liu Y. Identification of transcription factor and microRNA binding sites in responsible to fetal alcohol syndrome. BMC Genom 9, Suppl 1: S19, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang H, Su Y. Collagen IV contributes to nitric oxide-induced angiogenesis of lung endothelial cells. Am J Physiol Cell Physiol 300: C979–C988, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Weinberg J, Sliwowska JH, Lan N, Hellemans KG. Prenatal alcohol exposure: foetal programming, the hypothalamic-pituitary-adrenal axis and sex differences in outcome. J Neuroendocrinol 20: 470–488, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. West JR, Parnell SE, Chen WJ, Cudd TA. Alcohol-mediated Purkinje cell loss in the absence of hypoxemia during the third trimester in an ovine model system. Alcohol Clin Exp Res 25: 1051–1057, 2001. [PubMed] [Google Scholar]
- 60. Zhang X, Sliwowska JH, Weinberg J. Prenatal alcohol exposure and fetal programming: effects on neuroendocrine and immune function. Exp Biol Med (Maywood) 230: 376–388, 2005. [DOI] [PubMed] [Google Scholar]
- 61. Zygmunt M, Herr F, Munstedt K, Lang U, Liang OD. Angiogenesis and vasculogenesis in pregnancy. Eur J Obstet Gynecol Reprod Biol 110, Suppl 1: S10–S18, 2003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.