Abstract
Our immune system is charged with the vital mission of identifying invading pathogens and mounting proper inflammatory responses. During the process of clearing infections, the immune system often causes considerable tissue damage. Conversely, if the target of immunity is a member of the resident microbiota, uncontrolled inflammation may lead to host pathology in the absence of infectious agents. Recent evidence suggests that several inflammatory disorders may be caused by specific bacterial species found in most healthy hosts. Although the mechanisms that mediate pathology remain largely unclear, it appears that genetic defects and/or environmental factors may predispose mammals to immune-mediated diseases triggered by potentially pathogenic symbionts of the microbiota. We have termed this class of microbes `pathobionts', to distinguish them from acquired infectious agents. Herein, we explore burgeoning hypotheses that the combination of an immunocompromised state with colonization by pathobionts together comprise a risk factor for certain inflammatory disorders and gastrointestinal cancer.
Introduction
Microbes dominate as the most abundant life form on Earth, occupying almost every terrestrial, aquatic, and biological ecosystem on our planet. Humans are no exception. Throughout our lives, we continuously encounter microorganisms that range from those essential for health to those causing disease [1]. The human body is permanently colonized by microbial organisms on virtually all environmentally exposed surfaces. The vast majority of these microbes are harbored in the gastrointestinal (GI) tract where commensal bacteria can outnumber host cells by 10-fold (thus, we are all 90% bacteria on a cellular level). Many vital host functions are provided by the microbiota, including the synthesis of vitamins, digestion of complex polysaccharides, maintenance of the intestinal epithelial barrier, and resistance to pathogen colonization [2]. Millions of years of co-evolution have interdependently linked the health of mammals to their microbiotas [3]. The Human Microbiome Project is currently underway to sequence the microbiota of various populations of people, with a goal of identifying microbial species implicated in health and disease [4]. What is already clear is that microbes have flourished inside us since time immemorial, and have diverged to take on many functional roles that are now being uncovered at the genetic and mechanistic levels. Several descriptions of an intimate link between the microbiota and the immune system have recently emerged [5–8]. However, not all host-microbiota interactions promote health, and particular species of resident bacteria appear to activate the immune system resulting in inflammatory diseases. Thus, our association with the microbial world is precarious.
It is now appreciated that some symbiotic microorganisms in the GI tract induce pathology under certain conditions, usually involving environmental and/or genetic alterations. The term `pathobionts' has been suggested to describe resident microbes with pathogenic potential [9]. Organisms proposed as pathobionts are associated with chronic inflammatory conditions, unlike opportunistic pathogens which often cause acute infections and are typically acquired from the environment or other parts of the body. In addition, pathobionts are innocuous to the host under normal conditions, distinct from traditional pathogens which may cause disease even in healthy hosts. In this review, we highlight experimental evidence mostly from animal models that support the classification of specific microbes as pathobionts (see Table 1). Furthermore, we explore the role of bacterial pathobionts on intestinal health and their resulting impact on inflammatory bowel disease (IBD) and gastrointestinal cancers.
Table 1.
PATHOBIONTS OF THE GASTROINTESTINAL TRACT
| Bacterial strain | Conditions Promoting Pathogenesis | Refs |
|---|---|---|
| Segmented Filamentous Bacteria | • leads to colitis in SCID mice reconstituted with CD4+CD45Rbhigh T cells | 15 |
| • promotes disease in experimental models of rheumatoid arthritis and multiple sclerosis in mono-associated gnotobiotic mice | 16,17 | |
|
| ||
| Helicobacter hepaticus | • induces colitis in C57BI/6 IL-10−/− mice | 24 |
| • initiates colitis and large bowel carcinoma in 129/SvEv Rag2−/− mice | 28 | |
|
| ||
| Helicobacter pylori | • genetic polymorphisms of IL1B associated with increased risk of gastric cancer | 40 |
|
| ||
| Proteus mirabilis Klebsiella pneumonia | • responsible for inducing colitis and colorectal cancer in T-bet−/−Rag2−/− (TRUC) mice | 48 |
|
| ||
| Prevotellaceae TM7 | • responsible for inducing colitis in mice with mutations in the inflammasome pathway | 57 |
|
| ||
| Clostridium difficile | • can lead to pseudomembranous colitis in humans, following long-term antibiotic treatment | 58 |
|
| ||
| Vancomycin-resistant Enterococcus | • capable of invading the bloodstream in humans treated with broad-spectrum antibiotics | 59,60 |
Segmented Filamentous Bacteria
Segmented filamentous bacteria (SFB) comprise a group of Gram-positive Clostridia-related bacteria that closely adhere to Peyer's patches in the mammalian small intestine and have been shown to potently stimulate immune responses including IgA induction and B cell activation [10]. Recently, much attention has been focused on SFB due to their ability to induce T-helper 17 (Th17) cells in the gut [7, 11]. Th17 cells, characterized by IL-17A, IL-17F, and IL-22 cytokine production, are an important contributor to adaptive immunity, conferring protection against enteric infection with extracellular pathogens. Specific-pathogen free (SPF) mice colonized with these non-culturable bacteria showed greater numbers of Th17 cells in the gut and heightened protection against Citrobacter rodentium infection compared to mice without SFB [7]. Germ-free (GF) mice, which have very few Th17 cells in the gut [12–14], exhibited no appreciable change in Th17 levels when reconstituted with a microbiota lacking SFB. Surprisingly, reconstitution with SFB alone was able to significantly increase the number of intestinal Th17 cells [7]. Considering the microbiota harbors a complex bacterial consortium of hundreds of species, these remarkable findings indicate the ability to induce Th17 cells in the gut may be uniquely possessed by only a small subset of the microbiota. Therefore, SFB colonization of healthy animals appears to play an important role in priming the adaptive immune system and potentially enhancing immunity against enteric pathogens.
However, the heightened immunity conferred by SFB colonization may also come at a cost to the host when inflammation is inappropriately triggered. Studies have demonstrated a pathogenic role for SFB in the gut. SCID (severe combined immunodeficiency) mice reconstituted with CD4+CD45Rbhigh T cells and colonized with SFB developed severe colitis and intestinal inflammation [15]. In this particular animal model of colitis, SFB may synergize with the surrounding microbiota to exert its immunomodulatory effects, as mice mono-colonized with SFB did not develop intestinal pathology. Furthermore, the impact of SFB colonization on the host immune system appears to extend beyond the gut, as SFB mono-colonization in GF mice has been shown to increase the susceptibility of disease in animal models of rheumatoid arthritis and multiple sclerosis [16–17]. GF or antibiotic treated animals display reduced Th17 cells outside the gut and do not develop disease; this suggests that SFB alone can substitute for a complex microbiota in terms of driving pathology through Th17 cells induction. The observation that gut bacteria affect extra-intestinal compartments highlights the profound impact of the microbiota in modulating the overall health of the host. Although the microbial molecules that drive immunity are unknown, these results illustrate that in the context of an autoimmune environment, SFB are pathobionts that promote diseases not observed in healthy hosts.
Helicobacter hepaticus
Helicobacter hepaticus belongs to the enterohepatic Helicobacter species (EHS), a diverse group of spiral bacteria that thrive on mucosal surfaces of the intestinal tract and/or the liver of humans and other animals [18]. H. hepaticus is a well-studied member of EHS and is prevalent in mice from commercial and academic institutions all around the world [19]. Pioneering work by Fox and co-workers led to the discovery of H. hepaticus and its role in hepatitis, hepatocellular carcinoma, typhlitis and colitis in several strains of immunodeficient mice [20–22]. Further studies provided evidence suggesting the involvement of H. hepaticus in the development of pathogenic inflammation and carcinogenesis in certain immunocompromised rodent models. Experimental infection with H. hepaticus induces IBD-like lesions in SCID mice reconstituted with naïve CD4+CD45RBhigh T cells, as well as in C57Bl/6IL-10−/− mice [23–24]. Colonization with H. hepaticus also initiates rapid development of colitis and large bowel carcinoma in 129/SvEv Rag2−/− mice [25]. However, in immunocompetent wild-type (WT) mice, H. hepaticus fails to induce significant disease, irrespective of the mouse strains [24–25]. Therefore, H. hepaticus acts as a pathobiont that is able to promote colitis and in some cases colon cancer only in mouse strains with disrupted immune function.
Further questions arise from the pathobiont definition. For example, how does H. hepaticus interact with the host immune system to maintain a balanced relationship? Why do certain susceptible mouse strains with compromised immune systems develop inflammatory responses after H. hepaticus colonization, whereas WT mice do not? H. hepaticus infection induces Th1 and Th17 associated intestinal inflammation in IL-10−/− mice [26–27]. In lymphocyte-deficient Rag−/− mice, experimental infection with H. hepaticus induces colitis and colorectal cancer through proinflammatory cytokines TNF-α, IL-17, and IL-23 [25, 28–29]. Rag−/− mice lacking MyD88 in the hematopoietic compartment are resistant to H. hepaticus-induced colitis, indicating an essential role for toll-like receptor (TLR) signaling in H. hepaticus-induced innate inflammation [30]. Therefore, H. hepaticus is capable of causing both T cell-dependent and -independent inflammatory responses.
H. hepaticus induces intestinal inflammation in IL-10−/− mice but not WT mice, whose mesenteric lymph node (MLN) cells produce IL-10 in response to soluble H. hepaticus antigen (SHelAg), indicating a crucial role for IL-10 in balancing the H. hepaticus-induced inflammatory responses [26–27]. Furthermore, anti-IL-10R treated MLN cells derived from H. hepaticus-infected mice produce higher levels of IL-17 and IFN-γ compared with WT MLNs following response to SHelAg [31]. Transferring H. hepaticus-induced CD4+CD45RBlow regulatory T cells suppresses H. hepaticus-induced colitis in Rag−/− mice [32]. Also, CD4+CD25+ regulatory T cells isolated from Helicobacter-free 129SvEv mice prevent both T cell-dependent and -independent intestinal inflammation in an IL-10-dependent manner [33]. Moreover, an intact NF-κB signaling pathway is required for IL-10-mediated inhibition of H. hepaticus-induced colitis [34]. Therefore, it appears that H. hepaticus colonization induces a tolerogenic IL-10-secreting regulatory T cell response, which may be important for maintaining immunologic `balance' with the host. In addition, H. hepaticus was found to suppress TLR4 and TLR5-mediated immune responses in intestinal epithelial cells [35]. Our results also showed that H. hepaticus suppressed the expression of TLR4 in the intestinal epithelial cell line MODE-K [36], suggesting another possible regulatory strategy in epithelial cells mediated by TLR signaling. We propose that H. hepaticus maintains symbiotic crosstalk with the host by directing both inflammatory and tolerogenic responses in the innate and adaptive immune system during long-term colonization. However, in genetically susceptible hosts with defects in tolerogenic immune function and/or regulatory mechanisms, H. hepaticus may trigger an imbalanced immune response leading to pathologic inflammation.
Most studies have focused on the host immune response to H. hepaticus colonization and genetic alterations in mice that lead to disease. Little is known about the bacterial components produced by H. hepaticus that mediate these outcomes. Our recent discovery revealed that H. hepaticus utilizes a type VI secretion system (T6SS) to balance host colonization and intestinal inflammation [36]. T6SS are multi-protein complexes assembled on the bacterial surface that function as a biological needle and syringe, injecting microbial molecules into eukaryotic cells. Deletion of the T6SS apparatus resulted in higher colonization levels of H. hepaticus during experimental colitis. Moreover, a H. hepaticus T6SS mutant elicited elevated inflammatory responses in the intestine of Rag1−/− mice reconstituted with CD4+CD45RBhigh T cells compared to WT bacteria. Meanwhile, T6SS directed an anti-inflammatory response in an intestinal epithelial cell line, characterized by suppressed expression of TLR4, NF-κB and IL-17R. However, whether T6SS mediated suppression of innate inflammatory signaling correlates with its regulatory roles is still unknown. Identification of T6SS substrates of H. hepaticus may define the molecular mechanisms by which this pathobiont `communicates' with its host to establish symbiosis. Disruption of this communication, through genetic polymorphisms or mutations in the host, may form the basis for why H. hepaticus causes disease in compromised animals.
Helicobacter pylori
In 2005, Barry Marshall and Robin Warren won the Nobel Prize in Medicine for demonstrating that Helicobacter pylori, a bacterium that intimately colonizes the mucosal lining of the stomach, could directly cause peptic ulcer disease and gastritis. Classified as a class I carcinogen, H. pylori has been shown to lead to gastric adenocarcinoma in 1% of infected individuals. While 50% of the human population is thought to be colonized with H. pylori, only a small percentage actually develop gastric disorders [37]. Colonization of humans with H. pylori is believed to have occurred since humans migrated out of Africa 58,000 years ago. The bacteria are thought to colonize during early childhood and can thrive in the stomach for a lifetime. In countries with higher socio-economic standards (involving increased antibiotic use and hygiene), colonization appears to be less prevalent compared in developing countries.
The mechanistic details of how H. pylori promotes inflammation have been investigated. Using a Type IV secretion system (T4SS), H. pylori translocates the bacterial protein CagA into gastric epithelial cells. CagA subsequently interacts with host signal transduction pathways involved in inflammation and oncogenesis [38]. The presence of CagA, along with other virulence factors such as a pathogenicity island and additional secreted toxins, correlate well with increased virulence in strains of H. pylori [39]. However, even virulent strains of H. pylori are found in asymptomatic individuals suggesting there are other factors contributing to the induction of disease. Genetic polymorphisms in the IL1β gene, which encodes for a pro-inflammatory cytokine important for enhancing the inflammatory response to H. pylori infection, have been shown to be associated with increased risk of gastric cancer [40].
Adding another layer of complexity is the observation that colonization with H. pylori inversely correlates with esophageal adenocarcinoma and childhood asthma [41–42]. Furthermore, individuals colonized with CagA deficient strains of H. pylori are at increased risk for disease [43]. Although H. pylori-mediated protection against these pathologies still remains to be convincingly demonstrated, these results suggest the intriguing concept that H. pylori may have evolved to protect its host against disease in order to promote a healthier environment for its long-term survival [44]. Finally, sequencing efforts have revealed highly diverse panmictic populations of H. pylori between geographically separated groups [45]. The extensive diversification of the H. pylori genome may have proven advantageous in surviving the changing immunological and environmental pressures of the stomach.
Implications for Inflammatory Bowel Disease and GI Cancers
Inflammatory bowel diseases (including Crohn's disease (CD) and Ulcerative colitis (UC)), afflict approximately 1.5 million people in the United States. Currently there is no cure for IBD, although immunosuppressive therapies and probiotics alleviate symptoms in some cases. The causes of IBD appear to be multifactorial, integrating the microbiota, host genetics, and the immune system as factors determining predisposition to disease.
Shifts in the intestinal microenvironment (due to diet, antibiotics, hygiene, etc) may lead to changes in the microbiota known as dysbiosis. Dysbiosis may increase susceptibility to intestinal inflammation [46–47]. In support of this hypothesis, T-bet−/−Rag2−/− (TRUC) mice spontaneously develop dysbiosis and colitis, which can eventually progress into colorectal cancer [48]; remarkably, microbiota transfer from these donors into wild-type mice can confer disease [5]. Subsequent studies identified two proteobacteria over-represented in TRUC mice, Proteus mirabilis and Klebsiella pneumonia, as the colitogenic microbes [49]. However, full induction of disease required the presence of a diverse microbiota, indicating that interactions with other microbes may define whether a pathobiont will display a pathogenic profile. Culture-independent 16S rDNA sequence analysis of the microbiotas of individuals with Crohn's disease revealed lower diversity and greater temporal instability compared to controls [50]. In patients with IBD, the number of commensals belonging to the phyla Firmicutes and Bacteroidetes were found to be decreased, while concomitant increases in Actinobacteria and Proteobacteria were observed [51]. These findings highlight an important link between changes to the composition of the microbiota and intestinal health in animal models and humans.
Evidence over several decades suggests that the gut microbiota is a key factor in the pathogenesis of IBD. Studies have shown increased antibody titers against gut bacteria in IBD patients compared to healthy individuals [52]. Furthermore, treatment with antibiotics can help alleviate symptoms [53]. It is well documented that in certain mouse models of experimental colitis, rederivation under germ-free conditions abolishes disease [54]. However, host genetics and their impact on the resulting immunological environment significantly determine the type of response (or lack thereof) to the microbiota. Numerous genetic variants have been identified in individuals with IBD and correlate strongly with an increased risk of disease. Many of these genes are involved in bacterial recognition (NOD2, TLR genes, IRGM, ATG16L1) and innate and adaptive immunity (IL-23R, IL-10) [55]. Balanced immune responses to the microbiota are critical for intestinal homeostasis, as the microbiota itself has been shown to coordinate intestinal immunity. Illustrating this concept, mice expressing the human defensin DEFA5 showed a reduction in SFB colonization and a corresponding decrease in lamina propria Th17 cells [56]. In addition, perturbations in the mouse NLRP6 inflammasome pathway led to overgrowth of intestinal Prevotellaceae and TM7 bacteria, resulting in increased susceptibility to chemically-induced colitis [57]. Thus, although certain symbionts are prominent species in the gut and typically non-pathogenic, specific host defects can trigger IBD as a result of inflammation directed to pathobionts.
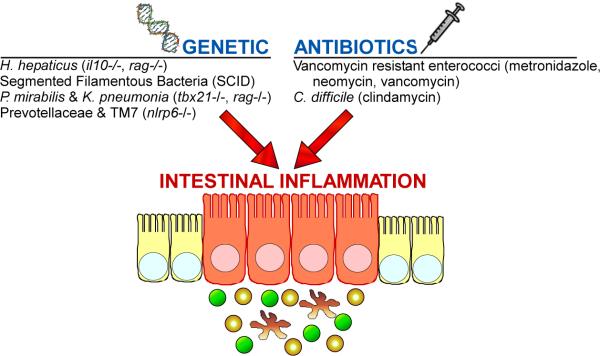
Although our understanding of the role of pathobionts on human health is still in its infancy, a few studies have highlighted the dangers of disrupting the human gut microbial community. Pseudomembranous colitis which results in severe diarrhea, fever and abdominal pain, is caused by overgrowth of Clostridium difficile following long-term antibiotic treatment [58]. Broad-spectrum antibiotics can also enhance vancomycin-resistant Enterococcus (VRE) survival and proliferation in the GI tract, which may subsequently lead to infection of the bloodstream [59–60]. As the source of C. difficile and Enterococcal infections is the microbiota, environmental factors may predispose patients to diseases caused by indigenous pathobionts. We predict that both genetic host alterations and/or environmental perturbations (such as antibiotic use) may lead to intestinal inflammation triggered by pathobionts (Figure 1).
Figure 1. Genetic and environmental alterations may synergize with pathobionts to cause intestinal inflammation and disease.
In addition to acquired pathogens which can cause gastroenteritis, resident gut bacteria trigger intestinal inflammation. However, unlike acute pathogens, pathobionts appear to require additional factors to cause disease. Based predominantly on animal models, certain symbionts of the microbiota can initiation gut inflammation and pathology when colonizing a genetically susceptible host (e.g., H. hepaticus, SFB, P. mirabilis & K. pneumonia, Prevotellaceae and TM7). In other cases, specific resident gut bacteria can expand following antibiotic use which clears competing symbionts to promote gastrointestinal disease (VRE, C. difficile). The associated genetic defects or antibiotics are denoted in parenthesis.
Recent studies have suggested that chronic inflammatory conditions can contribute to the development of some cancers by promoting cell proliferation, cell survival, and/or angiogenesis [61]. Individuals with IBD (in particular ulcerative colitis) have an increased risk of developing colorectal cancer [62]. In an experimental animal model of colitis-associated cancer, IL-10−/− mice treated with the chemical carcinogen azoxymethane, were devoid of tumors when raised under germ-free conditions, indicating the presence of intestinal bacteria is required for carcinogenesis [63]. Similar results were found in other animal models of spontaneous colon cancer. Germ-free rederivation of TCRβ−/−p53−/− mice and TGFβ1−/− mice eliminated the formation of intestinal tumors [64–65]. In addition, clinical studies have identified a higher incidence of adherent and invasive Escherichia coli (AIEC) in biopsies from carcinoma patients compared to controls [66–67]. Colorectal cancer is the second most common cause of malignant tumors in the United States [68], and often has life-threatening consequences. Moreover, epidemiologic and clinical data show that the incidence of colon cancer is dramatically increasing in Western countries. A genetic basis for cancer is well established; however it is being increasingly appreciated that non-genetic (environmental) factors are also crucial to the disease process. Whether there is a causal relationship between the microbiota, intestinal inflammation, and colon carcinogenesis will require further investigation.
Concluding Remarks
Recent ground-breaking studies of the interactions between humans and beneficial bacteria have marked a revolution in microbiology and immunology [3]. The human gastrointestinal tract harbors astounding multitudes of symbiotic bacterial species living in homeostasis with the immune system. However, some of these permanent residents appear to take on pathogenic properties during colonization of hosts with genetic and/or environmental alterations. Based on this rationale, we have speculated a category of indigenous gut bacteria termed pathobionts which cause disease only in susceptible hosts. This designation is based on recent data from animal models, with limited but growing support from clinical studies. The combination of a compromised host with colonization by pathobionts may be a risk factor in IBD, colon cancer and perhaps for diseases outside of the intestinal compartment. Identifying the molecular interactions between pathobionts and the mammalian immune system may be critical to understanding the etiology of certain diseases with a non-infectious microbial component. Finally, the design of drugs that inhibit the processes by which pathobionts promote inflammation may represent novel therapies for chronic human diseases.
Highlights
Research now shows that some members of the normal gut microbiota may promote disease
We term these microbes “Pathobionts” to distinguish them from acquired infections
Pathobionts appear to cause chronic inflammatory diseases
Understanding how Pathobionts induce disease may lead to anti-microbial therapies for IBD and colon cancer
Acknowledgments
J.C. is supported by a pre-doctoral training grant (NIH GM007616). S.K.M. is a Searle Scholar. Work in the authors' laboratory is supported by funding from the National Institutes of Health (DK078938, DK083633 & AI088626), Emerald Foundation, Damon Runyon Cancer Research Foundation and the Crohn's and Colitis Foundation of America to S.K.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest The authors declare no conflict of interest.
References
Papers of particular interest published within the period of review have been highlighted as:
* of special interest
** of outstanding interest
- 1.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–48. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 2.Backhed F, et al. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 3.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330(6012):1768–73. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson J, et al. The NIH Human Microbiome Project. Genome Res. 2009;19(12):2317–23. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131(1):33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107(27):12204–9. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **7.Ivanov, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]; Along with Ref. [11**], this paper showed that a specific group of bacteria is primarily responsible for inducing pro-inflammatory Th17 cells in the gut.
- 8.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–41. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–23. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Talham GL, et al. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67(4):1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **11.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31(4):677–89. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]; Along with Ref. [9**], this study showed that SFB are capable of modulating intestinal immune responses, particularly enhancing Th1 and Th17 cell proportions.
- 12.Ivanov, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4(4):337–49. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niess JH, et al. Commensal gut flora drives the expansion of proinflammatory CD4 T cells in the colonic lamina propria under normal and inflammatory conditions. J Immunol. 2008;180(1):559–68. doi: 10.4049/jimmunol.180.1.559. [DOI] [PubMed] [Google Scholar]
- 14.Atarashi K, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455(7214):808–12. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 15.Stepankova R, et al. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm Bowel Dis. 2007;13(10):1202–11. doi: 10.1002/ibd.20221. [DOI] [PubMed] [Google Scholar]
- *16.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32(6):815–27. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; Along with Ref. [17*], this study showed that a specific group of intestinal bacterial (namely Th17-inducing SFB) are capable of driving disease in extra-intestinal compartments.
- *17.Lee YK, et al. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4615–22. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]; Along with Ref. [16*], this paper showed that mono-associated GF mice colonized with SFB are more susceptible to disease in extra-intestinal compartments.
- 18.Solnick JV, Schauer DB. Emergence of diverse Helicobacter species in the pathogenesis of gastric and enterohepatic diseases. Clin Microbiol Rev. 2001;14(1):59–97. doi: 10.1128/CMR.14.1.59-97.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor NS, et al. Enterohepatic Helicobacter species are prevalent in mice from commercial and academic institutions in Asia, Europe, and North America. J Clin Microbiol. 2007;45(7):2166–72. doi: 10.1128/JCM.00137-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward JM, et al. Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J Natl Cancer Inst. 1994;86(16):1222–7. doi: 10.1093/jnci/86.16.1222. [DOI] [PubMed] [Google Scholar]
- 21.Ward JM, et al. Inflammatory large bowel disease in immunodeficient mice naturally infected with Helicobacter hepaticus. Lab Anim Sci. 1996;46(1):15–20. [PubMed] [Google Scholar]
- 22.Li X, et al. SCID/NCr mice naturally infected with Helicobacter hepaticus develop progressive hepatitis, proliferative typhlitis, and colitis. Infect Immun. 1998;66(11):5477–84. doi: 10.1128/iai.66.11.5477-5484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cahill RJ, et al. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect Immun. 1997;65(8):3126–31. doi: 10.1128/iai.65.8.3126-3131.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kullberg MC, et al. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect Immun. 1998;66(11):5157–66. doi: 10.1128/iai.66.11.5157-5166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erdman SE, et al. CD4+ CD25+ regulatory T lymphocytes inhibit microbially induced colon cancer in Rag2-deficient mice. Am J Pathol. 2003;162(2):691–702. doi: 10.1016/S0002-9440(10)63863-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kullberg MC, et al. Induction of colitis by a CD4+ T cell clone specific for a bacterial epitope. Proc Natl Acad Sci U S A. 2003;100(26):15830–5. doi: 10.1073/pnas.2534546100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matharu KS, et al. Toll-like receptor 4-mediated regulation of spontaneous Helicobacter-dependent colitis in IL-10-deficient mice. Gastroenterology. 2009;137(4):1380–90. e1–3. doi: 10.1053/j.gastro.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erdman SE, et al. Nitric oxide and TNF-alpha trigger colonic inflammation and carcinogenesis in Helicobacter hepaticus-infected, Rag2-deficient mice. Proc Natl Acad Sci U S A. 2009;106(4):1027–32. doi: 10.1073/pnas.0812347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hue S, et al. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J Exp Med. 2006;203(11):2473–83. doi: 10.1084/jem.20061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asquith MJ, et al. Pathogenic and protective roles of MyD88 in leukocytes and epithelial cells in mouse models of inflammatory bowel disease. Gastroenterology. 2010;139(2):519–29. 529, e1–2. doi: 10.1053/j.gastro.2010.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fox JG, et al. Helicobacter hepaticus infection in mice: models for understanding lower bowel inflammation and cancer. Mucosal Immunol. 2011;4(1):22–30. doi: 10.1038/mi.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kullberg MC, et al. Bacteria-triggered CD4(+) T regulatory cells suppress Helicobacter hepaticus-induced colitis. J Exp Med. 2002;196(4):505–15. doi: 10.1084/jem.20020556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maloy KJ, et al. CD4+CD25+ T(R) cells suppress innate immune pathology through cytokine-dependent mechanisms. J Exp Med. 2003;197(1):111–9. doi: 10.1084/jem.20021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomczak MF, et al. Inhibition of Helicobacter hepaticus-induced colitis by IL-10 requires the p50/p105 subunit of NF-kappa B. J Immunol. 2006;177(10):7332–9. doi: 10.4049/jimmunol.177.10.7332. [DOI] [PubMed] [Google Scholar]
- 35.Sterzenbach T, et al. Inhibitory effect of enterohepatic Helicobacter hepaticus on innate immune responses of mouse intestinal epithelial cells. Infect Immun. 2007;75(6):2717–28. doi: 10.1128/IAI.01935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **36.Chow J, Mazmanian SK. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe. 2010;7(4):265–76. doi: 10.1016/j.chom.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper revealed the first description of a molecular mechanism by which a pathobiont of the intestinal microbiota impacts host mucosal immunity.
- 37.Dorer MS, Talarico S, Salama NR. Helicobacter pylori's unconventional role in health and disease. PLoS Pathog. 2009;5(10):e1000544. doi: 10.1371/journal.ppat.1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Odenbreit S, et al. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287(5457):1497–500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 39.Polk DB, Peek RM., Jr. Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer. 2010;10(6):403–14. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El-Omar EM, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404(6776):398–402. doi: 10.1038/35006081. [DOI] [PubMed] [Google Scholar]
- 41.Sonnenberg A, Lash RH, Genta RM. A national study of Helicobactor pylori infection in gastric biopsy specimens. Gastroenterology. 2010;139(6):1894–1901. e2. doi: 10.1053/j.gastro.2010.08.018. quiz e12. [DOI] [PubMed] [Google Scholar]
- 42.Chen Y, Blaser MJ. Helicobacter pylori colonization is inversely associated with childhood asthma. J Infect Dis. 2008;198(4):553–60. doi: 10.1086/590158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chow WH, et al. An inverse relation between cagA+ strains of Helicobacter pylori infection and risk of esophageal and gastric cardia adenocarcinoma. Cancer Res. 1998;58(4):588–90. [PubMed] [Google Scholar]
- 44.Willyard C. A tough controversy to stomach. Nat Med. 2009;15(8):836–9. doi: 10.1038/nm0809-836. [DOI] [PubMed] [Google Scholar]
- 45.Gressmann H, et al. Gain and loss of multiple genes during the evolution of Helicobacter pylori. PLoS Genet. 2005;1(4):e43. doi: 10.1371/journal.pgen.0010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453(7195):620–5. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 47.Kinross JM, et al. The human gut microbiome: implications for future health care. Curr Gastroenterol Rep. 2008;10(4):396–403. doi: 10.1007/s11894-008-0075-y. [DOI] [PubMed] [Google Scholar]
- 48.Garrett WS, et al. Colitis-associated colorectal cancer driven by T-bet deficiency in dendritic cells. Cancer Cell. 2009;16(3):208–19. doi: 10.1016/j.ccr.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *49.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8(3):292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper showed that in T-bet−/−Rag2−/− (TRUC) mice, the presence of two Enterobacteriaceae species was responsible for inducing spontaneous colitis. Furthermore, transfer of the microbiota from TRUC mice was sufficient to cause disease in WT mice.
- 50.Scanlan PD, et al. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn's disease. J Clin Microbiol. 2006;44(11):3980–8. doi: 10.1128/JCM.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104(34):13780–5. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macpherson A, et al. Mucosal antibodies in inflammatory bowel disease are directed against intestinal bacteria. Gut. 1996;38(3):365–75. doi: 10.1136/gut.38.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sartor RB. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics, and prebiotics. Gastroenterology. 2004;126(6):1620–33. doi: 10.1053/j.gastro.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 54.Nell S, Suerbaum S, Josenhans C. The impact of the microbiota on the pathogenesis of IBD: lessons from mouse infection models. Nat Rev Microbiol. 2010;8(8):564–77. doi: 10.1038/nrmicro2403. [DOI] [PubMed] [Google Scholar]
- 55.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40(8):955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *56.Salzman NH, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11(1):76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study showed how DEFA5, an alpha-defensin secreted by Paneth cells in the gut, is capable of modulating the composition of the commensal microbiota. Specifically, DEFA5-expressing mice had a significant loss in SFB with a concomitant reduction in Th17 cells.
- **57.Elinav E, et al. NLRP6 Inflammasome Regulates Colonic Microbial Ecology and Risk for Colitis. Cell. 2011;145(5):745–57. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrated that NLRP6 −/− mice exhibit dysbiosis in the gut, specifically overgrowth of Prevotellaceae and TM7 bacteria that results in a transmissible increase in colitis susceptibility.
- 58.Palmer R. Fecal matters. Nat Med. 2011;17(2):150–2. doi: 10.1038/nm0211-150. [DOI] [PubMed] [Google Scholar]
- 59.Ubeda C, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest. 2010;120(12):4332–41. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *60.Brandl K, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455(7214):804–7. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study showed that extended antibiotic treatment can lead to a reduction in RegIIIγ, a secreted C-type lectin that kills Gram-positive bacteria, thus allowing overgrowth of VRE.
- 61.Mantovani A, et al. Cancer-related inflammation. Nature. 2008;454(7203):436–44. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 62.McConnell BB, Yang VW. The Role of Inflammation in the Pathogenesis of Colorectal Cancer. Curr Colorectal Cancer Rep. 2009;5(2):69–74. doi: 10.1007/s11888-009-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uronis JM, et al. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS One. 2009;4(6):e6026. doi: 10.1371/journal.pone.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Engle SJ, et al. Elimination of colon cancer in germ-free transforming growth factor beta 1-deficient mice. Cancer Res. 2002;62(22):6362–6. [PubMed] [Google Scholar]
- 65.Kado S, et al. Intestinal microflora are necessary for development of spontaneous adenocarcinoma of the large intestine in T-cell receptor beta chain and p53 double-knockout mice. Cancer Res. 2001;61(6):2395–8. [PubMed] [Google Scholar]
- 66.Swidsinski A, et al. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology. 1998;115(2):281–6. doi: 10.1016/s0016-5085(98)70194-5. [DOI] [PubMed] [Google Scholar]
- 67.Martin HM, et al. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology. 2004;127(1):80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 68.Wilmink AB. Overview of the epidemiology of colorectal cancer. Dis Colon Rectum. 1997;40(4):483–93. doi: 10.1007/BF02258397. [DOI] [PubMed] [Google Scholar]