Abstract
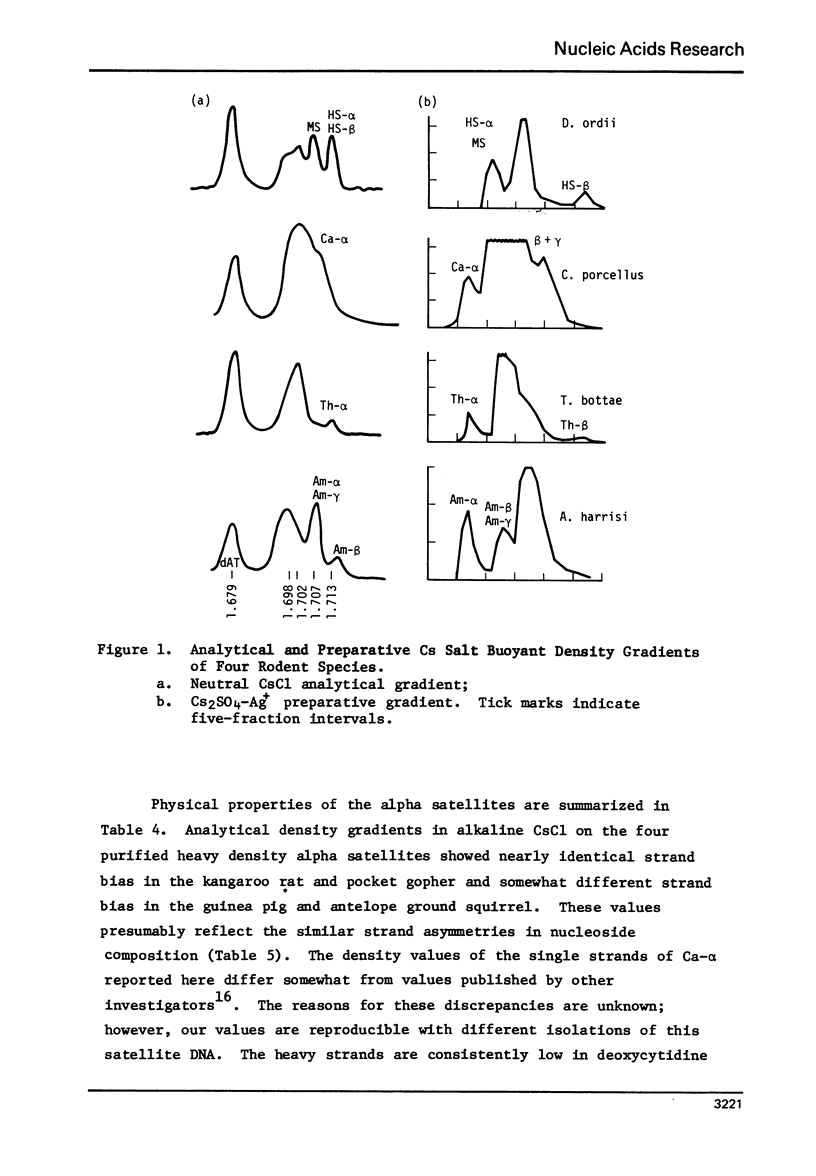
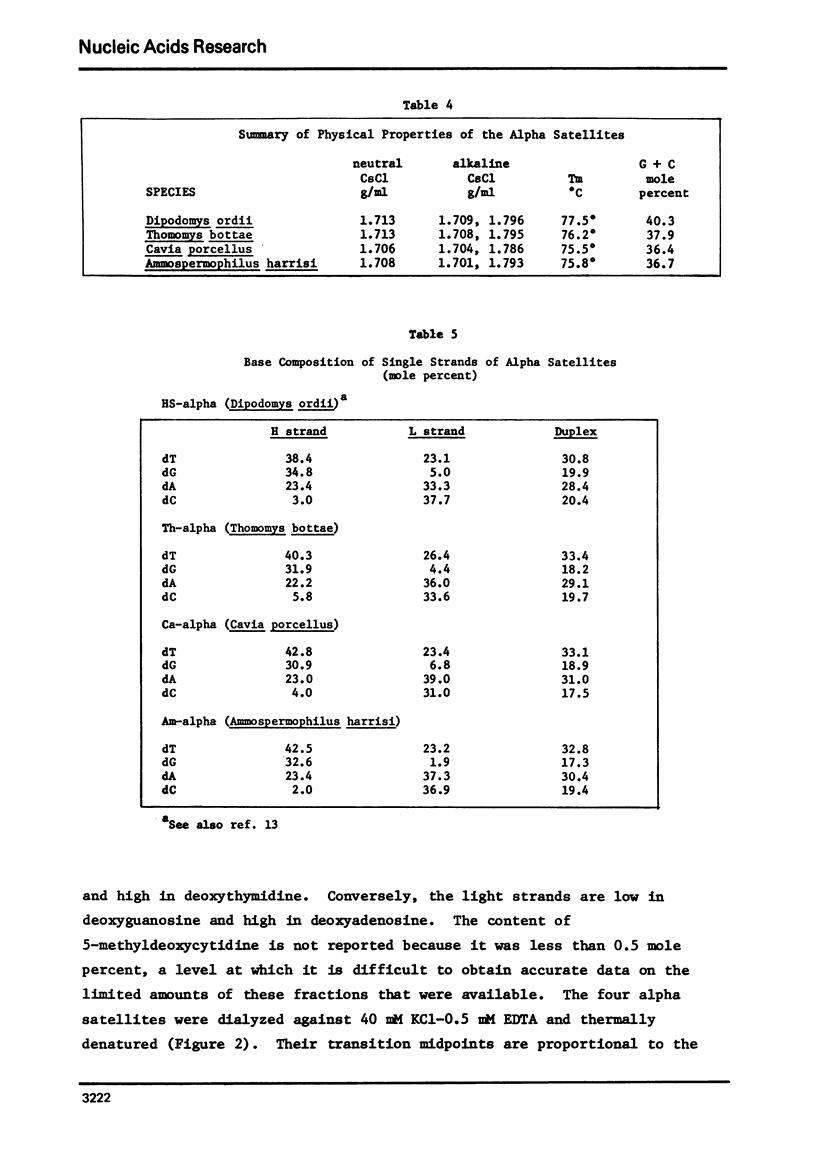
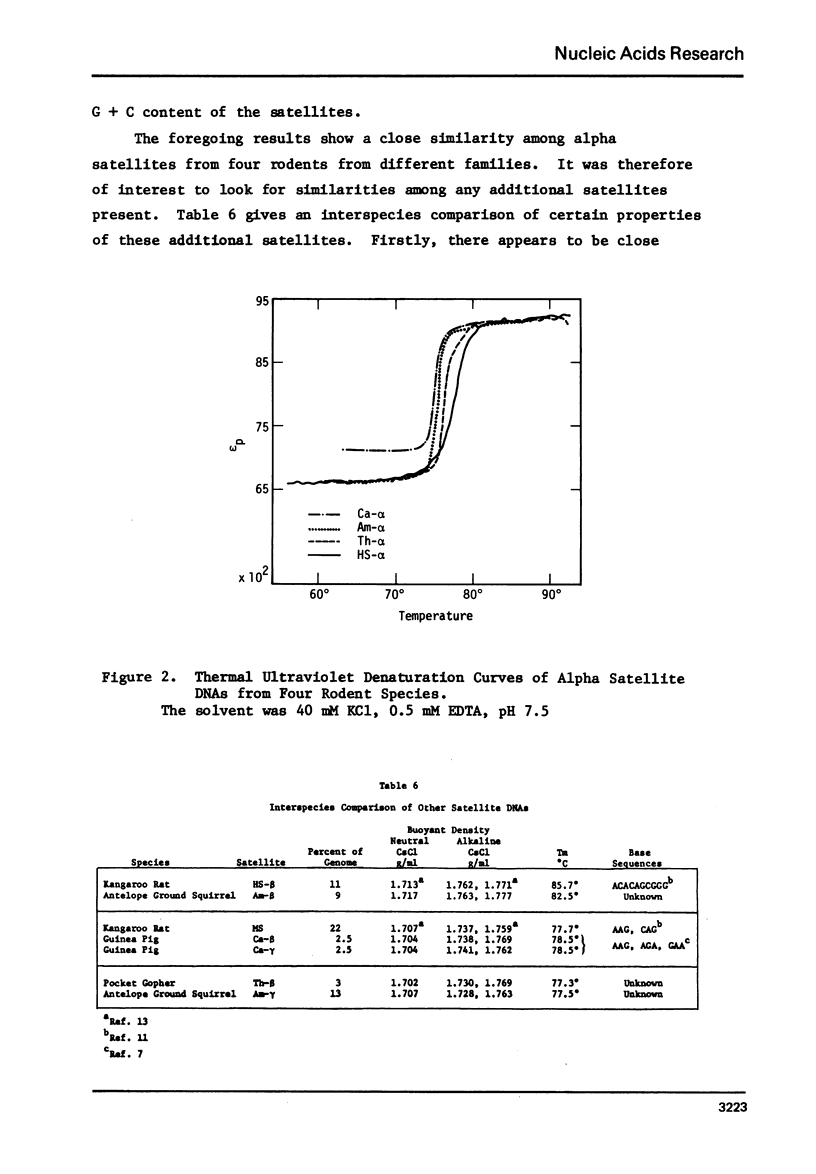
We have characterized satellite DNAs from 9 species of kangaroo rat (Dipodomys) and have shown that the HS-α and HS-β satellites, where present, are nearly identical in all species as to melting transition midpoint (Tm), and density in neutral CsCl, alkaline CsCl, and Cs2SO4-Ag+ gradients. However, the MS satellites exist in two internally similar classes. The satellite DNAs from three other rodents were characterized (densities listed are in neutral CsCl). The pocket gopher, Thomomysbottae, contains Th-α (1.713 g/ml) and Th-β (1.703 g/ml). The guinea pig (Caviaporcellus) contains Ca-α, Ca-β and Ca-γ at densities of 1.706 g/ml, 1.704 g/ml and 1.704 g/ml, respectively. The antelope ground squirrel (Ammospermophilusharrisi) contains Am-α, 1.708 g/ml, Am-β, 1.717 g/ml, and Am-γ, 1.707 g/ml. The physical and chemical properties of the alpha-satellites from the above four rodents representing four different families in two suborders of Rodentia were compared. They show nearly identical Tm, nucleoside composition of single strands, and single strand densities in alkaline CsCl. Similar comparisons on the second or third satellite DNAs from these rodents also indicate a close relationship to each other. Thus the high degree of similarity of satellite sequences found in such a diverse group of rodents suggests a cellular function that is subject to natural selection, and implies that these sequences have been conserved over a considerable span of evolutionary time since the divergence of these rodents about 50 million years ago.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bostock C. J., Christie S. Chromosome banding and DNA replication studies on a cell line of Dipodomys merriami. Chromosoma. 1974;48(1):73–87. doi: 10.1007/BF00284868. [DOI] [PubMed] [Google Scholar]
- Bostock C. J., Christie S. Chromosomes of a cell line of Dipodomys panamintinus (kangaroo rat). A banding and autoradiographic study. Chromosoma. 1975;51(1):25–34. doi: 10.1007/BF00285804. [DOI] [PubMed] [Google Scholar]
- Bostock C. J., Christie S., Lauder I. J., Hatch F. T., Mazrimas J. A. S phase patterns of replication of different satellite DNAs in three species of Dipodomys (kangaroo rat). J Mol Biol. 1976 Dec;108(2):417–433. doi: 10.1016/s0022-2836(76)80128-3. [DOI] [PubMed] [Google Scholar]
- Brown P. R., Bobick S., Hanley F. L. The analysis of purine and pyrimidine bases and their nucleosides by high-pressure liquid chromatography. J Chromatogr. 1974 Nov 6;99(0):586–595. [PubMed] [Google Scholar]
- Gall J. G., Atherton D. D. Satellite DNA sequences in Drosophila virilis. J Mol Biol. 1974 Jan 5;85(4):633–664. doi: 10.1016/0022-2836(74)90321-0. [DOI] [PubMed] [Google Scholar]
- Hatch F. T., Bodner A. J., Mazrimas J. A., Moore D. H. Satellite DNA and cytogenetic evolution. DNA quantity, satellite DNA and karyotypic variations in kangaroo rats (genus Dipodomys). Chromosoma. 1976 Oct 28;58(2):155–168. doi: 10.1007/BF00701356. [DOI] [PubMed] [Google Scholar]
- Hatch F. T., Mazrimas J. A. Fractionation and characterization of satellite DNAs of the kangaroo rat (Dipodomys ordii). Nucleic Acids Res. 1974 Apr;1(4):559–575. doi: 10.1093/nar/1.4.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig W., Walker P. M. Variations in the DNA from two rodent families (Cricetidae and Muridae). Nature. 1970 Mar 7;225(5236):915–919. doi: 10.1038/225915a0. [DOI] [PubMed] [Google Scholar]
- Mazrimas J. A., Hatch F. T. A possible relationship between satellite DNA and the evolution of kangaroo rat species (genus Dipodomys). Nat New Biol. 1972 Nov 22;240(99):102–105. doi: 10.1038/newbio240102a0. [DOI] [PubMed] [Google Scholar]
- Peacock W. J., Brutlag D., Goldring E., Appels R., Hinton C. W., Lindsley D. L. The organization of highly repeated DNA sequences in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol. 1974;38:405–416. doi: 10.1101/sqb.1974.038.01.043. [DOI] [PubMed] [Google Scholar]
- Prescott D. M., Bostock C. J., Hatch F. T., Mazrimas J. A. Location of satellite DNAs in the chromosomes of the kangaroo rat (Dipodomys ordii). Chromosoma. 1973;42(2):205–213. doi: 10.1007/BF00320941. [DOI] [PubMed] [Google Scholar]
- Prosser J., Moar M., Bobrow M., Jones K. W. Satellite sequences in chimpanzee (Pan troglodytes). Biochim Biophys Acta. 1973 Aug 24;319(2):122–134. doi: 10.1016/0005-2787(73)90003-8. [DOI] [PubMed] [Google Scholar]
- Rice N. R. Change in repeated DNA in evolution. Brookhaven Symp Biol. 1972;23:44–79. [PubMed] [Google Scholar]
- SIMPSON G. G. The nature and origin of supraspecific taxa. Cold Spring Harb Symp Quant Biol. 1959;24:255–271. doi: 10.1101/sqb.1959.024.01.025. [DOI] [PubMed] [Google Scholar]
- Salser W., Bowen S., Browne D., el-Adli F., Fedoroff N., Fry K., Heindell H., Paddock G., Poon R., Wallace B. Investigation of the organization of mammalian chromosomes at the DNA sequence level. Fed Proc. 1976 Jan;35(1):23–35. [PubMed] [Google Scholar]
- Smith G. P. Evolution of repeated DNA sequences by unequal crossover. Science. 1976 Feb 13;191(4227):528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Base sequence and evolution of guinea-pig alpha-satellite DNA. Nature. 1970 Aug 22;227(5260):794–798. doi: 10.1038/227794a0. [DOI] [PubMed] [Google Scholar]
- Sutton W. D., McCallum M. Related satellite DNA's in the genus Mus. J Mol Biol. 1972 Nov 28;71(3):633–652. doi: 10.1016/s0022-2836(72)80028-7. [DOI] [PubMed] [Google Scholar]
- Walker P. M. How different are the DNAs from related animals? Nature. 1968 Jul 20;219(5151):228–232. doi: 10.1038/219228a0. [DOI] [PubMed] [Google Scholar]



