Abstract
KEOPS is an important cellular complex conserved in Eukarya, with some subunits conserved in Archaea and Bacteria. This complex was recently found to play an essential role in formation of the tRNA modification threonylcarbamoyladenosine (t6A), and was previously associated with telomere length maintenance and transcription. KEOPS subunits are conserved in Archaea, especially in the Euryarchaea, where they had been studied in vitro. Here we attempted to delete the genes encoding the four conserved subunits of the KEOPS complex in the euryarchaeote Haloferax volcanii and study their phenotypes in vivo. The fused kae1-bud32 gene was shown to be essential as was cgi121, which is dispensable in yeast. In contrast, pcc1 (encoding the putative dimerizing unit of KEOPS) was not essential in H. volcanii. Deletion of pcc1 led to pleiotropic phenotypes, including decreased growth rate, reduced levels of t6A modification, and elevated levels of intra-cellular glycation products.
Introduction
Kae1 (also referred to as Gcp or YgjD, and recently renamed TsaD [1]) is one of about 60 proteins conserved in over 99% of organisms with sequenced genomes. Despite this impressive ubiquity, its function has remained elusive until recently, and is yet not fully understood. Kae1 therefore remains a top priority target for experimental study [2]. This protein is part of a complex, named KEOPS or EKC in Eukarya and Archaea [3], [4], [5]. The KEOPS (Kinase, Endopeptidase and Other Proteins of small Size) complex was first characterized in yeast and shown to be involved in the elongation and uncapping of telomeres, as well as a transcription factor. More recently, it was shown to be involved in the synthesis of the universal tRNA modification threonylcarbamoyladenosine or (t6A) [6], [7]. The KEOPS complex is composed of Kae1; the serine/threonine kinase-like Bud32; Cgi121; Pcc1, and Gon7 (unique to fungi) [5]. Kae1 is essential in Gram-positive and Gram-negative bacteria [8], [9], [10], in yeast, kae1- deficient mutants are very sick [4], [5], [6], [7], [11]. Saccharomyces cerevisiae strains carrying deletions of any of the other KEOPS subunit- encoding genes are viable, albeit with reduced growth rates [5], [12].
The bacterial Kae1 homologue TsaD, initially recognized as a secreted glycopeptidase in Mannheimia haemolytica [13], was later shown to be involved in the metabolism of Advanced Glycated End Products (AGEs). Depletion of TsaD resulted in cellular accumulation of AGEs followed by growth arrest in Escherichia coli cells [10]. AGEs are highly stable, toxic compounds which were found to be involved in several aspects of cell physiology. AGEs accumulate in bacteria and are actively secreted into the growth medium [14]. Several additional phenotypes resulting from TsaD depletion in E. coli include deficiencies in DNA maintenance, membrane and cell shape homeostasis, and cell division [15], [16], [17]. Although at least some of these pleiotropic phenotypes could be linked to glycation, a simpler explanation was offered by recent studies that link Kae1/TsaD to the synthesis of t6A. This universal modification is found at position 37, 3′ of the anti-codon, in all tRNAs that pair with ANN codons [6], [7]. tRNA extracted from yeast Δkae1 mutants lack t6A [6], [7], [18], and the TsaD protein from E. coli, in combination with TsaB (YrdC), TsaC (YjeE), and TsaE (YeaZ) is required to synthesize t6A in vitro [1]. As the absence of t6A leads to mis-initiation and frameshifts [7], [19], the phenotypes observed could be indirectly linked due to mistranslation of specific proteins, in accordance with the absence of other modifications of the anticodon such as cm5U derivatives in yeast [20], [21]. The eukaryotic and bacterial t6A machinery must have diverged as both TsaD/Kae1 and YrdC/Sua5 are universal, but TsaE and TsaB are bacterial-specific. Sua5 and Kae1 alone are not sufficient to synthesize t6A in the cytosol [6]. One possibility is that the other subunits of the KEOPS complex found in Eukaryotes and Archaea are required, as it was previously shown that mutations in pcc1 and bud32 in yeast affect t6A levels [6], [22].
Although archaeal proteins served as a structural model for the eukaryotic KEOPS, no in vivo studies of the KEOPS complex in Archaea have been reported. Previous studies on KEOPS combined in vitro analysis of archaeal proteins with in vivo genetic analysis in yeast, and were useful for determining some of the functional roles of the eukaryotic KEOPS. An in vitro study of the Kae1 homolog of the hyperthermophilic archaeon Pyrococcus abyssi showed it to be an endonuclease of apurinated nucleotides [23], hinting that it may be involved in DNA repair stability or processes. However, a similar study of the Methanococcus jannaschii homolog concluded that the latter Kae1 homolog does not possess such activity and cannot bind DNA [4]. Reconstruction and structural analysis of the entire KEOPS complex using proteins from Methanococcus jannaschii and Pyrococcus furiosus expressed in E. coli, suggested that Cgi121 activates Bud32 which in turn regulates Kae1 activity, while Pcc1 is a dimerizing module [4].
However, since in vivo experiments have not been carried out in Archaea, and attempts to complement the yeast or bacterial kae1/tsaD deletions with archaeal homologs failed [7], no physiological role could be assigned to the archaeal KEOPS. Here we perform a genetic analysis in the model archaeon Haloferax volcanii and establish the essentiality of this complex in the third domain of life.
Results
The Kae1-Bud32 fusion protein is essential in Haloferax volcanii
Four components of the KEOPS complex– Kae1, Bud32, Cgi121 and Pcc1 have homologs in Archaea [4]. Homologs of these four components were identified in the genome of H. volcanii using BLASTP [24]. The genes encoding Kae1 and Bud32 are fused to a single open reading frame in haloarchaea and methanogens [23].
To determine the essentiality of the Kae1-Bud32 fusion encoding gene in H. volcanii (gene number HVO_1895), we employed the “pop-in/pop-out” strategy for gene deletion [25], [26] see materials and methods. We attempted to delete the kae1-bud32 gene, using plasmid pAN1 transformed into H. volcanii strain H26 creating the pop-in strain H-AN2 (For plasmids and primers used see tables S1 and S2, respectively). Following counter selection (“pop-out”), 30 colonies were screened, but no kae1-bud32 deletions were obtained, suggesting that this fusion gene is probably essential. We then proceeded to perform a gene replacement experiment, replacing kae1-bud32 with a trpA selectable marker (using plasmid pAN2). pAN2 was transformed into H. volcanii strain H133 (ΔpyrE, ΔtrpA), creating the pop-in strain H-AN1. Following counter selection (“pop-out”), no colonies were obtained, supporting the essentiality of kae1-bud32 in H. volcanii. To validate this conclusion, a plasmid carrying the complete kae1-bud32 gene was cloned into the pRV1-Ptna-bgaH plasmid [27] placing kae1-bud32 under the control of the tryptophanase promoter (pAN4). This plasmid was subsequently transformed into the “pop-in” strain, HAN2. In this genetic background, where an exogenous gene is provided, it was possible to knock out the chromosomal gene and obtain strain HAN4 (Figure S1A).
A similar approach was used to examine the essentiality of both components of the fusion gene, kae1 and bud32. Fragments encoding each of the putative functional domains were cloned into the pRV plasmid and transformed into strain HAN1. The fusion gene was split to contain the individual domains, as previously done for the Methanocaldococcus jannaschii homolog [4] (for the exact positions of the fragment inserted, see table S1). If only one of the genes/domains is essential, one should be able to replace the fusion gene region with a trpA selectable marker, provided that the other portion is supplied exogenously (using plasmids pAN 12 and pAN 19). However, this was also unsuccessful, showing that both the Kae1 and Bud32 domains are likely to be essential.
Cgi121 is essential in Haloferax volcanii
The “pop-in/pop-out” methodology described above was also used in an attempt to delete cgi121 (gene number HVO_0013) using plasmid pAN23. When attempting to delete cgi121 with no selectable marker, 50 “pop-out” colonies were examined, and were found to have reverted back to wild-type. Moreover, when attempting to replace cgi121 with the trpA selectable marker (pAN24), as described above for kae1-bud32, it proved impossible to delete cgi121. When cgi121 was provided on a complementing plasmid, pAN25, we were able to delete cgi121 from its chromosomal location. This verifies that failure to knock out the gene is due to its essentiality and not a technical artifact (Figure S1B).
A Haloferax volcanii Δpcc1 mutant displays reduced growth
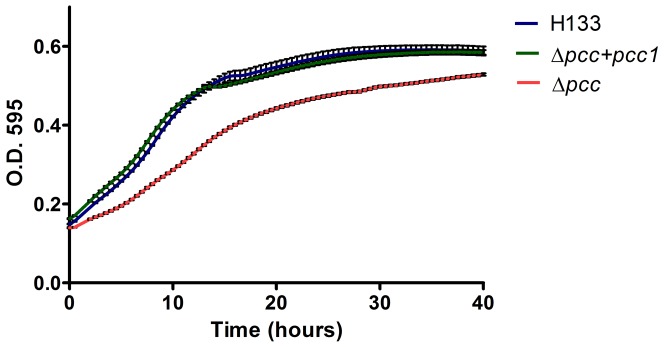
Our attempts to delete pcc1 (HVO_0652) by “pop-in/pop-out” (using plasmid pAN21) without a selectable marker were unsuccessful, indicating much reduced fitness. The 50 “pop-out” colonies that were scanned were found to have reverted back to the wild-type genotype. However, when replacing the gene with the trpA selectable marker in an H133 (trp −) background (using plasmid pAN22), some colonies were obtained following counter-selection. Since the “pop-out” colonies were smaller in size (Figure S1C). We therefore tested the growth rate of Δpcc1. As seen in Figure 1, the growth rate of Δpcc1 was significantly lower when compared to the H133 (wild-type) strain. To confirm that this significant decrease in growth rate is not due to other factors, we introduced a complementing plasmid (pAN26) containing pcc1 in trans. The growth rate of the complemented strain is similar to that of the wild type, indicating that the loss of pcc1 causes a reduced growth rate.
Figure 1. Growth of wild type Haloferax volcanii and the Δpcc1 mutant cells.
Growth curves comparing the growth rate of H133 (wild-type), Δpcc1 (HAN16) and Δpcc1 containing the complementing plasmid (HAN19). Results are a mean of 10 replicates, error bars indicates standard error of the mean.
A Haloferax volcanii Δpcc1 mutant contains more AGEs
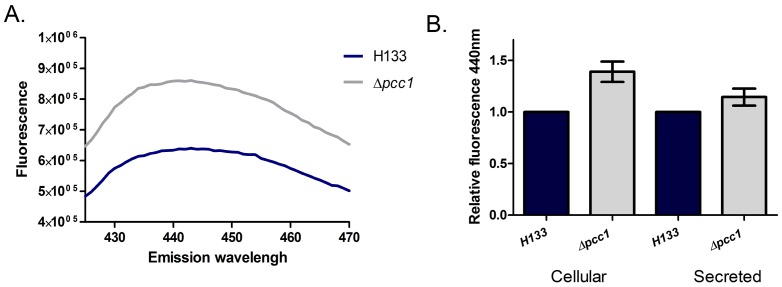
Since the bacterial homolog of Kae1 was shown to be involved in the accumulation of cellular AGEs, [10] we compared the levels of AGEs in wild-type H. volcanii cells to HAN16 Δpcc1 cells. The presence of AGEs can be determined by AGE-specific fluorescence, measured by emission at 440 nm upon excitation at 370 nm (see materials and methods). As shown in Figure 2A, lysates of wild-type cells (H133) showed a flattened peak at about 440 nm, indicating that AGEs were formed. Comparing the level of cellular AGEs in Δpcc1 cells to wild-type, we observed a 37% increase of cellular levels of AGEs (Figure 2A). The differences in the level of secreted AGEs were more subtle - about 14% more AGEs were secreted in the Δpcc1 cells. Thus, pcc1-null cells contain more AGEs, and AGEs secretion probably cannot keep up with the increased accumulation of AGEs.
Figure 2. Advanced glycation products in wild type Haloferax volcanii and the Δpcc1 mutant cells.
A. Measurement of AGEs-specific fluorescence in the H133 (wild-type) and in Δpcc1 (HAN16). The emission spectrum from 400 nm to 480 nm upon excitation at 370 nm is presented. A representative sample is shown. B. Accumulation and secretion of AGEs. AGE-specific fluorescence is shown relative to the wild-type. The results represent three independent experiments, error bars indicates standard error of mean.
A Haloferax volcanii Δpcc1 mutant has higher nucleic acid content
In order to assess the nucleic acid content and distribution in the Δpcc1 cells, we examined the cells in a flow cytometer (see materials and methods). The cell sizes of wild-type and Δpcc1 were similar (Figure 3A). Cell morphology was also similar under light microscopy (data not shown). Although cell size was normal, nucleic acid content was substantially higher with a broader distribution across cells in the Δpcc1 mutants (Figure 3B).
Figure 3. Flow cytometry analysis of Haloferax volcanii H195 and H133 Δpcc1 mutant cells.
H195 (based on H133, with a bgaHa-Bb deletion, and a leuB- Ag1 allele, which has flow cytometry profiles highly similar to H133– data not shown) A. Cell size as determined by forward light scatter of H195, and of Δpcc1 (HAN16). B. Nucleic acid content as determined by acridine orange fluorescence of H195 (wild-type) and in Δpcc1 (HAN16).
A Haloferax volcanii Δpcc1 mutant contains less t6A
tRNAs were extracted from the Δpcc1 and H133 (wild-type) strains, hydrolyzed to liberated nucleosides, and analyzed by both HPLC and LC/MS/MS. t6A was present in Δpcc1, but the mutant strain contained approximately 15–20% less t6A relative to H133 (Figure 4). This result is similar to the reduction seen in a pcc1–4 point mutant in yeast [22]. These results suggest that Pcc1 is not essential for t6A formation in Archaea or Eukarya, but contributes to the efficiency of its biosynthesis.
Figure 4. Nucleoside analysis analysis in wild type Haloferax volcanii and the Δpcc1 mutant cells.
tRNAs were extracted from each strain and hydrolyzed to nucleosides. The nucleoside content was determined by HPLC with detection by UV/Vis at 254 nm. Analyses were performed in triplicate from independent cultures. A. HPLC chromatographs of nucleosides from wild-type, wild-type with synthesized t6A added, or Δpcc1. t6A elutes at 24 minutes. B. Comparison of the t6A peak area of wild-type and Δpcc1. The ratios of Ψ-modified base/m2 2G were used to normalize tRNA concentrations across samples with t6A peak area of wild-type set at 100%. Δpcc1 contains approximately 19% less t6A than wild-type (P = 0.02).
Discussion
Whether the KEOPS complex is involved in one biological pathway or process that has pleiotropic effects or whether it is directly involved in several pathways is still unclear. Four of the five KEOPS proteins show extraordinary conservation from Archaea to mammals. Notably, one of the proteins, Kae1, is one of the very few proteins encoded by nearly all genomes sequenced in the three domains of life.
This work represents the first in vivo investigation of the KEOPS complex in Archaea. We have shown the critical importance of all four KEOPS components for growth of H. volcanii. Viability of the pcc1 deletion mutants was somewhat expected, since Pcc1 is thought to function as a dimerizing module connecting two KEOPS complexes together. Absence of Pcc1 should result in reduced KEOPS activity rather than a complete loss of KEOPS function [4]. Nevertheless, pcc1 deletion mutants are slow growing, and show several other defects that were previously shown to be associated with disruption of this complex. These phenotypes include an elevated level of AGEs, as was seen in E. coli TsaD depletion [10], reduced t6A modification, as seen in E. coli and S. cerevisiae, and an aberrant distribution of nucleic acid content in mutant cells [6], [18]. Curiously, a cgi121 deletion, which had little effect in yeast, proved to be essential in H. volcanii. In yeast, cgi121 null mutants show no altered t6A modification levels [6], perhaps indicating that the KEOPS complex is involved in more than one key cellular functions. This conclusion is further supported by the relative mildness of the t6A phenotype of the pcc1 mutant compared to the substantial growth impairment of that mutant in H. volcanii.
The fact that both AGEs-related and t6A-related phenotypes were observed in H. volcanii raises an interesting question - is the KEOPS complex involved in both protection from glycation damages and the carbamoylation reaction independently, or are these processes linked? One possibility that we are exploring is, that in the absence of a functional KEOPS complex, the TsaC/Sua5 enzyme generates a glycation product. Alternatively, translation errors caused by insufficient t6A modification of tRNAs can result in mistranslated proteins that have more exposed lysine residues, providing more abundant substrates for Amadori product formation and results in higher levels of AGEs. Regardless of the exact biochemical cross-talk between Amadori product formation and tRNA t6A modification, it is clear that defects in these processes are likely to have pleiotropic phenotypes, such as the differences in nucleic acid content observed in pcc1-null mutants.
The question of the essentiality of t6A in Archaea remains unanswered. Yeast mutants that lack t6A are viable, however E. coli mutants are not [6], [7], [28]. Naturally, one wonders whether essentiality is because of the absence of the t6A modification itself, or because t6A synthesis proteins have additional roles in E. coli. The fact that all four E. coli t6A biosynthesis proteins are essential strongly suggests that it is the modification itself that is essential. Members of the archaeal Sua5 family have been shown to complement both the t6A deficiency of the sua5Δ yeast mutant and the essentiality phenotype of E. coli ΔtsaC [28]. Like E. coli and unlike yeast, the H. volcanii Sua5 homolog HVO_0253 is essential [18]. One possibility explaining the essentiality of t6A in prokaryotes and not in eukaryotes is that C34 in tRNAIle CAU, decoding AUA codons, has to be modified to lysidine (k2C) in Bacteria and agmatidine (agm2C) in Archaea to function in decoding [29], [30], [31]. Both the lysidine synthase (TilS) encoding gene and the agmitidine synthase (TiaS) encoding genes are essential [18], [32]. If t6A is required for TilS and TiaS activity then t6A would be essential in prokaryotes and not in eukaryotes. We are currently investigating this scenario.
Materials and Methods
Strains
Strains used in this work and their genotype are listed in Table 1.
Table 1. Strains used in this study.
| strain | Description | Source |
| H26 | ΔpyrE2 | [25] |
| H133 | ΔpyrE2 ΔtrpA ΔleuB ΔhdrB | [25] |
| H195 | ΔpyrE2 ΔtrpA ΔhdrB bgaHa-Bb leuB- Ag1 | [37] |
| HAN1 | H133 with pAN2 pop in. | This work |
| HAN2 | H26 with pAN1 pop in. | This work |
| HAN4 | Deletion of kae1-bud32 (from pop in strain HAN2) with the pAN4 complementing plasmid | This work |
| HAN16 | Replacement of pcc1 with a trpA cassette (in a H133 background). | This work |
| HAN19 | HAN 16 containing the pAN26 complementing plasmid. | This work |
Culture conditions
H. volcanii was routinely grown in rich (HY) medium, or on a selective CA medium (see [33]. For counter-selection of uracil auxotrophs, 5-fluoroorotic acid (5-FOA) (United States Biological) was added to the medium at a final concentration of 100 µg/ml. When required, uracil was added to a final concentration of 50 µg/ml. When needed, novobiocin (Sigma-Aldrich) was added to the medium at a final concentration of 2 µg/ml.
Transformation
Transformation of H. volcanii was carried out using the PEG method as described in [34].
Gene knockouts
The gene knockouts were performed according to the protocol described in [25], [26]. In this method, the upstream and downstream flanking regions of the sequence to be exchanged are amplified by PCR and cloned together into the ‘suicide plasmid’ pTA131 that carries the pyrE2 selectable genetic marker and cannot replicate autonomously in H. volcanii. The plasmids are then transformed into a H. volcanii ΔpyrE mutant, and transformants, in which the plasmids have been integrated into the chromosome, are selected for on plates that lack uracil (‘pop-in’). Upon counter-selection on plates containing uracil and 5-fluoroorotic acid (5FOA), the only cells that survive are those in which the integrated plasmids have been excised by spontaneous intra-chromosomal homologous recombination (‘pop-out’), either restoring the wild-type gene or resulting in allele exchange. Gene replacements were performed as described in [25]. CA medium was used as a uracil- and tryptophan-minus medium for trpA cassette selection.
Growth curves
The growth curves were performed in 96-well plates at 42°C with continuous shaking, using the Biotek ELX808IU-PC microplate reader. Optical density was measured every 30 minutes at a wavelength of 595 nm.
Flow cytometry
Exponential phase samples (A600∼0.5) were stained with acridine orange (10 µg/ml, Sigma-Aldrich) for two minutes. Flow cytometry was immediately performed using an Apogee A40 equipped with a 50 mW 488 nm solid state laser (Coherent) and a 510–580 nm bandpass filter. Settings LS1: 420 V, FL1: 495 V, 50000 cells. Doublet signals were removed by gating on peak/area plots for LS1 and FL1. Data analyzed using FlowJo (TreeStar Inc).
Determination of AGEs
Advanced Glycation End-products (AGEs) were quantified using the natural AGE-specific fluorescence (Ex. 370 nm, Em. 440 nm) by scanning emission ranging from 400 nm to 500 nm upon excitation at 370 nm at 37°C, in a HORIBA scientific FluoroLog-3 Spectrofluorometer. Data represent either the full range spectrum, or the 440 nm emission peak, as indicated in the legends. In order to quantify AGEs, the culture was grown to exponential phase (OD600∼0.5), and washed once with 18% NaCl solution. When quantifying secreted AGEs, cells were incubated in the salt solution for 3 hours, precipitated by centrifugation and the solution was used for measurement. For the cellular AGEs quantification, cells were washed, lysed by sonication and the lysate was used for measurement.
Preparation of Bulk tRNA
H. volcanii H133 (wild-type) and Δpcc1 were grown in YPC [25]. Bulk tRNA was prepared by double phenol/chloroform extraction previously described [35]. Nucleoside preparations were prepared by incubating 100 µg of linearized bulk tRNA with 10 units of Nuclease P1 (Sigma) in 10 mM ammonium acetate (pH 5.3) overnight at 37°C. The next day, the digestion was adjusted to a final concentration of 100 mM ammonium bicarbonate, 0.01 units of Phosphodiesterase I (Sigma), 0.05 units E. coli alkaline phosphatase (Sigma), and incubated for an additional 2 hours at 37°C. The hydrolyzed nucleosides were further purified by filtering through a 5 Kd MCO filter (to remove enzymes), dried, and suspended in 20 µL of water prior to analysis by HPLC or LC-MS/MS. All tRNA extractions were performed in triplicate from independent cultures.
HPLC and LC-MS/MS Analysis
t6A was detected by HPLC as described by [36] and also by LC–MS/MS as described in [28]. Levels of t6A were measured by integrating the area under the peak from the extraction ion chromatograms. The ratios of Ψ-modified base/m2 2G were used to normalize for tRNA concentrations across samples. t6A levels for Δpcc1 were compared to wild-type levels, with wild-type set at 100%. The MS/MS fragmentation data as well as a synthesized t6A standard provided by D. Davis were used to confirm the presence of t6A.
Supporting Information
Confirmation of genetic mutations. Oligonucleotides anneal outside of the open reading frame, but within the region of homology used for “pop-in/pop-out” deletions. A. PCR analysis of kae1-bud31 fusion using primers AP26 and AP27. Lane 1 - DNA ladder, lane 2 - wild type. Lane 3- the “pop in” strain (HAN1). Lane 4 – “pop out” of kae1-bud32 with kae1-bud32 supplied in trans (HAN4). B. PCR analysis of cgi121 using primers AP229 and AP233. Lane 1- DNA Ladder. Lane 2 – “pop in” of plasmid pAN24 in H133. Lane 3 - DNA Ladder. Lane 4- wild-type. Lane 5 – “pop out” of cgi121 with cgi121 supplied in trans (pAN25). C. PCR analysis of pcc1 using primers AP295 and AP296. Lane 1- wild-type. Lane 2 – “pop in” of plasmid pAN22 in H133. Lane 3 and 4 “pop out” HAN16. Lane 5- DNA Ladder.
(TIF)
Plasmids used in this study.
(DOCX)
Oligonucleotides used in this study.
(DOCX)
Acknowledgments
The Authors would like to thank Moshe Mevareh, Eliora Ron and Martin Kupiec, for helpful advice and discussion. We would like to thank Rachel Sheriber and Ella Shtifman-Segal for technical assistance, Basma El Yacoubi for stimulating discussions and Darrell Davis for proving chemically synthesized t6A standard.
Funding Statement
UG was supported by the Israel Science Foundation and the German-Israeli Project Cooperation (DIP), AN was funded by the Federation of the Societies of Biochemistry and Molecular Biology short term fellowship grant, and VdC by National Institutes of Health (grant no. R01 GM70641). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Deutsch C, El Yacoubi B, de Crecy-Lagard V, Iwata-Reuyl D (2012) The biosynthesis of threonylcarbamoyl adenosine (t6A), a universal tRNA nucleoside. Journal of Biological Chemistry doi: 10.1074/jbc.M112.344028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Galperin MY, Koonin EV (2004) ‘Conserved hypothetical’ proteins: prioritization of targets for experimental study. Nucleic Acids Res 32: 5452–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Downey M, Houlsworth R, Maringele L, Rollie A, Brehme M, et al. (2006) A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell 124: 1155–1168. [DOI] [PubMed] [Google Scholar]
- 4. Mao DY, Neculai D, Downey M, Orlicky S, Haffani YZ, et al. (2008) Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine. Mol Cell 32: 259–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kisseleva-Romanova E, Lopreiato R, Baudin-Baillieu A, Rousselle JC, Ilan L, et al. (2006) Yeast homolog of a cancer-testis antigen defines a new transcription complex. Embo J 25: 3576–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Srinivasan M, Mehta P, Yu Y, Prugar E, Koonin EV, et al. (2011) The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J 30: 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. El Yacoubi B, Hatin I, Deutsch C, Kahveci T, Rousset JP, et al. (2011) A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J 30: 882–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arigoni F, Talabot F, Peitsch M, Edgerton MD, Meldrum E, et al. (1998) A genome-based approach for the identification of essential bacterial genes. Nat Biotechnol 16: 851–856. [DOI] [PubMed] [Google Scholar]
- 9. Zalacain M, Biswas S, Ingraham KA, Ambrad J, Bryant A, et al. (2003) A global approach to identify novel broad-spectrum antibacterial targets among proteins of unknown function. J Mol Microbiol Biotechnol 6: 109–126. [DOI] [PubMed] [Google Scholar]
- 10. Katz C, Cohen-Or I, Gophna U, Ron EZ (2010) The ubiquitous conserved glycopeptidase gcp prevents accumulation of toxic glycated proteins. MBio 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hecker A, Lopreiato R, Graille M, Collinet B, Forterre P, et al. (2008) Structure of the archaeal Kae1/Bud32 fusion protein MJ1130: a model for the eukaryotic EKC/KEOPS subcomplex. Embo J 27: 2340–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peggion C, Lopreiato R, Casanova E, Ruzzene M, Facchin S, et al. (2008) Phosphorylation of the Saccharomyces cerevisiae Grx4p glutaredoxin by the Bud32p kinase unveils a novel signaling pathway involving Sch9p, a yeast member of the Akt/PKB subfamily. Febs J 275: 5919–5933. [DOI] [PubMed] [Google Scholar]
- 13. Abdullah KM, Lo RY, Mellors A (1991) Cloning, nucleotide sequence, and expression of the Pasteurella haemolytica A1 glycoprotease gene. J Bacteriol 173: 5597–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cohen-Or I, Katz C, Ron EZ (2011) AGEs secreted by bacteria are involved in the inflammatory response. PLoS One 6: e17974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oberto J, Breuil N, Hecker A, Farina F, Brochier-Armanet C, et al. (2009) Qri7/OSGEPL, the mitochondrial version of the universal Kae1/YgjD protein, is essential for mitochondrial genome maintenance. Nucleic Acids Res 37: 5343–5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bergmiller T, Pena-Miller R, Boehm A, Ackermann M (2011) Single-cell time-lapse analysis of depletion of the universally conserved essential protein YgjD. BMC Microbiol 11: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hashimoto C, Sakaguchi K, Taniguchi Y, Honda H, Oshima T, et al. (2011) Effects on transcription of mutations in ygjD, yeaZ, and yjeE genes, which are involved in a universal tRNA modification in Escherichia coli . J Bacteriol 193: 6075–6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blaby IK, Phillips G, Blaby-Haas CE, Gulig KS, El Yacoubi B, et al. (2010) Towards a systems approach in the genetic analysis of archaea: Accelerating mutant construction and phenotypic analysis in Haloferax volcanii . Archaea 2010: 426239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin CA, Ellis SR, True HL (2010) The Sua5 protein is essential for normal translational regulation in yeast. Mol Cell Biol 30: 354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mehlgarten C, Jablonowski D, Wrackmeyer U, Tschitschmann S, Sondermann D, et al. (2010) Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol Microbiol 76: 1082–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen C, Huang B, Eliasson M, Ryden P, Bystrom AS (2011) Elongator complex influences telomeric gene silencing and DNA damage response by its role in wobble uridine tRNA modification. PLoS Genet 7: e1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Daugeron MC, Lenstra TL, Frizzarin M, El Yacoubi B, Liu X, et al. (2011) Gcn4 misregulation reveals a direct role for the evolutionary conserved EKC/KEOPS in the t6A modification of tRNAs. Nucleic Acids Res [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hecker A, Leulliot N, Gadelle D, Graille M, Justome A, et al. (2007) An archaeal orthologue of the universal protein Kae1 is an iron metalloprotein which exhibits atypical DNA-binding properties and apurinic-endonuclease activity in vitro. Nucleic Acids Res 35: 6042–6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 25. Allers T, Ngo HP, Mevarech M, Lloyd RG (2004) Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl Environ Microbiol 70: 943–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bitan-Banin G, Ortenberg R, Mevarech M (2003) Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J Bacteriol 185: 772–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Large A, Stamme C, Lange C, Duan Z, Allers T, et al. (2007) Characterization of a tightly controlled promoter of the halophilic archaeon Haloferax volcanii and its use in the analysis of the essential cct1 gene. Mol Microbiol 66: 1092–1106. [DOI] [PubMed] [Google Scholar]
- 28. El Yacoubi B, Lyons B, Cruz Y, Reddy R, Nordin B, et al. (2009) The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res 37: 2894–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Soma A, Ikeuchi Y, Kanemasa S, Kobayashi K, Ogasawara N, et al. (2003) An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol Cell 12: 689–698. [DOI] [PubMed] [Google Scholar]
- 30. Mandal D, Kohrer C, Su D, Russell SP, Krivos K, et al. (2010) Agmatidine, a modified cytidine in the anticodon of archaeal tRNA(Ile), base pairs with adenosine but not with guanosine. Proc Natl Acad Sci U S A 107: 2872–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ikeuchi Y, Kimura S, Numata T, Nakamura D, Yokogawa T, et al. (2010) Agmatine-conjugated cytidine in a tRNA anticodon is essential for AUA decoding in Archaea. Nat Chem Biol 6: 277–282. [DOI] [PubMed] [Google Scholar]
- 32. Soma A, Ikeuchi Y, Kanemasa S, Kobayashi K, Ogasawara N, et al. (2003) An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol Cell 12: 689–698. [DOI] [PubMed] [Google Scholar]
- 33. Naor A, Lazary R, Barzel A, Papke RT, Gophna U (2011) In vivo characterization of the homing endonuclease within the polB gene in the halophilic archaeon Haloferax volcanii . PLoS One 6: e15833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cline SW, Lam WL, Charlebois RL, Schalkwyk LC, Doolittle WF (1989) Transformation methods for halophilic archaebacteria. Can J Microbiol 35: 148–152. [DOI] [PubMed] [Google Scholar]
- 35. de Crecy-Lagard V, Brochier-Armanet C, Urbonavicius J, Fernandez B, Phillips G, et al. (2010) Biosynthesis of wyosine derivatives in tRNA: an ancient and highly diverse pathway in Archaea. Mol Biol Evol 27: 2062–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pomerantz SC, McCloskey JA (1990) Analysis of RNA hydrolyzates by liquid chromatography-mass spectrometry. Methods Enzymol 193: 796–824. [DOI] [PubMed] [Google Scholar]
- 37. Guy CP, Haldenby S, Brindley A, Walsh DA, Briggs GS, et al. (2006) Interactions of RadB, a DNA repair protein in archaea, with DNA and ATP. J Mol Biol 358: 46–56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Confirmation of genetic mutations. Oligonucleotides anneal outside of the open reading frame, but within the region of homology used for “pop-in/pop-out” deletions. A. PCR analysis of kae1-bud31 fusion using primers AP26 and AP27. Lane 1 - DNA ladder, lane 2 - wild type. Lane 3- the “pop in” strain (HAN1). Lane 4 – “pop out” of kae1-bud32 with kae1-bud32 supplied in trans (HAN4). B. PCR analysis of cgi121 using primers AP229 and AP233. Lane 1- DNA Ladder. Lane 2 – “pop in” of plasmid pAN24 in H133. Lane 3 - DNA Ladder. Lane 4- wild-type. Lane 5 – “pop out” of cgi121 with cgi121 supplied in trans (pAN25). C. PCR analysis of pcc1 using primers AP295 and AP296. Lane 1- wild-type. Lane 2 – “pop in” of plasmid pAN22 in H133. Lane 3 and 4 “pop out” HAN16. Lane 5- DNA Ladder.
(TIF)
Plasmids used in this study.
(DOCX)
Oligonucleotides used in this study.
(DOCX)