Abstract
Microdeletion of the Azoospermia Factor (AZF) regions in Y chromosome is a well-known genetic cause of male infertility resulting from spermatogenetic impairment. However, the partial deletions of AZFc region related to spermatogenetic impairment are controversial. In this study, we characterized partial deletion of AZFc region in Korean patients with spermatogenetic impairment and assessed whether the DAZ and CDY1 contributes to the phenotype in patients with gr/gr deletions. Total of 377 patients with azoo-/oligozoospermia and 217controls were analyzed using multiplex polymerase chain reaction (PCR), analysis of DAZ-CDY1 sequence family variants (SFVs), and quantitative fluorescent (QF)-PCR. Of the 377 men with impaired spermatogenesis, 59 cases (15.6%) had partial AZFc deletions, including 32 gr/gr (8.5%), 22 b2/b3 (5.8%), four b1/b3 (1.1%) and one b3/b4 (0.3%) deletion. In comparison, 14 of 217 normozoospermic controls (6.5%) had partial AZFc deletions, including five gr/gr (2.3%) and nine b2/b3 (4.1%) deletions. The frequency of gr/gr deletions was significantly higher in the azoo-/oligozoospermic group than in the normozoospermic control group (p = 0.003; OR = 3.933; 95% CI = 1.509–10.250). Concerning Y haplogroup, we observed no significant differences in the frequency of gr/gr deletions between the case and the control groups in the YAP+ lineages, while gr/gr deletion were significantly higher in azoo-/oligozoospermia than normozoospermia in the YAP− lineage (p = 0.004; OR = 6.341; 95% CI = 1.472–27.312). Our data suggested that gr/gr deletion is associated with impaired spermatogenesis in Koreans with YAP− lineage, regardless of the gr/gr subtypes.
Introduction
The azoospermia factor (AZF) locus has been mapped to the long arm of the human Y chromosome that is associated with spermatogenetic failure [1], [2]. Four recurrent microdeletions of the AZF region related to azoospermia or oligozoospermia have been identified to date: AZFa, P5/proximal-P1 (AZFb), P5/distal-P1 and AZFc (b2/b4) deletions [2], [3]. In addition to these deletions, several partial AZFc deletions (gr/gr, b2/b3, b1/b3 and b3/b4), inversion, and duplications resulting from non-allelic homologous recombination have been reported [4]–[11]. Among them, the most clinically relevant type is gr/gr deletion including two copies of the DAZ gene and one copy of CDY1 gene. DAZ and CDY1 have been known to be the most important candidates related to spermatogenesis in the AZFc region [12]–[14]. Recently, the gr/gr deletion has been newly defined as ‘gr/gr deletion rearrangements’ and was divided into five rearrangement types, simple gr/gr deletion, gr/gr deletion-b2/b4 duplication, gr/gr deletion-b2/b4 multiple duplication, gr/gr deletion-CDY1 and DAZ amplification [6], [15]. In addition, several other studies showed the relationship between gr/gr deletion subtypes and spermatogenetic impairment but the results were different among populations. In some populations, the deletion showed a significant risk factor for spermatogenetic failure but not in others [4], [7], [14], [16]–[22].
Therefore, this study was designed to characterize the partial AZFc deletion patterns and their clinical consequences in Korean population. So, we carried out AZFc-STS analysis, gene copy analysis and gene dosage analysis and Y- haplogroup analysis as well.
Materials and Methods
Ethics Statement
This study was approved by the Institutional Review Board of CHA Gangnam Medical Center (IRB number: 09–06), and written informed consent was obtained from all participants.
Study Population
A total of 619 men who were born to ethnic Korean parents, were analyzed for classical AZF deletions and partial AZFc region deletions in the Fertility Center of CHA Gangnam Medical Center between January 2009 and December 2010. Of these, 210 patients were excluded because they had either numerical or structural chromosome abnormalities, known causes of spermatogenic failure (such as obstruction of the vas deferens, history of orchitis and active orchitis, or history of unilateral, bilateral cryptochidism and varicocele) or insufficient clinical data. In addition, 32 patients with classical AZF deletions were also excluded. Thus, the subjects were composed of 377 men with azoospermia or oligozoospermia (sperm concentration of <20×106/ml, in all three semen analyses). The normozoospermic control group comprised 217 men who consulted the same hospital for a routine fertility work-up. All of the control subjects were clinically healthy and possessed sperm concentrations of >20×106/ml, normal sperm motility and morphology, and hormonal parameters. Semen analysis was performed according to the World Health Organization criteria [23].
Characterization of the Partial AZFc Deletions
Genomic DNAs were extracted from peripheral blood samples using the QIAamp® DNA Blood Mini Kit (QIAGEN, Hilden, Germany). Multiplex PCR reactions were performed using three STSs for the AZFc region (sY1191, sY1291, sY1206) and organized into two multiplex PCRs including a PCR control marker (SOHLH2). The amplification conditions were 94°C for 5 min, and then 30 cycles of 95°C for 30 sec, 61°C for 30 sec, and 72°C for 30 sec and a final elongation at 72°C for 10 min. The PCR reaction was always performed with a male control sample, a female sample, and a blank sample. The reaction products were analyzed by electrophoresis on 2% agarose gel. We identified partial AZFc deletions by the following STS results: the absence of sY1191, sY1291, sY1191/sY1291, and sY1206 represents the b2/b3, gr/gr, b1/b3, and b3/b4 deletions, respectively [7], [19].
YAP Haplotype Analysis
Two haplogroups, YAP+ lineage (hgr.DE) and YAP− lineage [hgr. Y*(xDE)], were analyzed in samples with gr/gr deletion. Insertion/deletion polymorphisms of the YAP element was detected by PCR amplification using flanking primers as described in Hammer and Horai [24].
gr/gr Subtypes Analysis
Deletion copy types of CDY1 and DAZ gene were analyzed by the previous described method [14]. For DAZ, a sequence family variant (SFV), STS-sY587 placed in intron 10, which discriminates DAZ1/2 from DAZ3/4, was used. And for CDY1, we used a C/A SFV located in 7750 bp upstream from the CDY1 translation start codon (CDY1-7750), which distinguishes CDY1a from CDY1b. SFVs were analyzed by PCR followed by restriction enzyme digestion: DAZ-sY587/DraI (DAZ1/2 cut); CDY1-7750/PvuII (CDY1b cut). For quantitative analysis of CDY1 and DAZ, quantitative fluorescent-PCR (QF-PCR) was performed according to the previously described method [14], [25]. Briefly, CDY1 and DAZ were co-amplified with CDY2 and DAZL, respectively as a control with known gene copies. One primer (forward primer) in each set was labeled with 6-FAM (fluorescein amidite) fluorescent dye. The amplified products were loaded on the ABI 3130xl genetic analyzer and analyzed with the GeneMapper ID®version 3.2 (Applied Biosystems, Foster City, CA). The copy numbers of the genes were estimated based on the relative DAZ/DAZL and CDY1/CDY2 ratios.
Statistical Analysis
Statistical analysis was performed using the statistical package SPSS for Windows (version 20, Chicago, IL, USA) software. The frequency of azoo-/oligozoospermia compared with normozoospermia was analyzed using both the chi-square test and Fisher’s exact test (two-tailed). Continuous variables were analyzed by the t- test for independent samples. Probability (p) values <0.05 were considered statistically significant.
Results
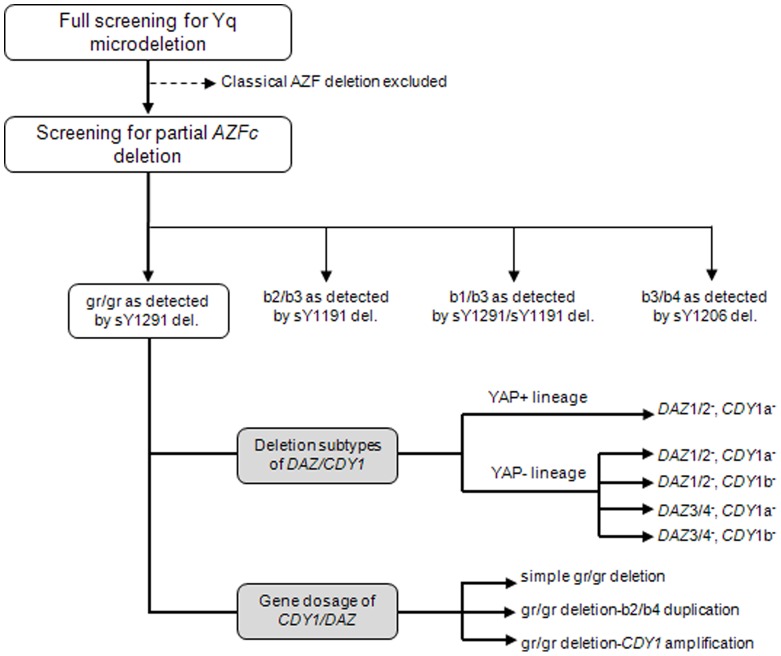
A total of 377 azoo-/oligozoospermic patients and 217normozoospermic men were analyzed by this combined method described in Figure 1. Four types of partial AZFc deletions, the gr/gr, b2/b3, b1/b3, and b3/b4 deletions, were identified in this study (Table 1). Partial AZFc deletions were more frequently found in men with spermatogenic impairment than in the control group [59/377, 15.6% vs. 14/217, 6.5%, p = 0.001; odds ratio (OR) = 2.690; 95% confidence interval(CI) = 1.388–4.239]. Among them, the frequencies of gr/gr deletions were significantly higher in men with azoo-/oligozoospermia than normozoospermic men (32/377, 8.5% vs. 5/217, 2.3%, p = 0.003; OR = 3.933; 95% CI = 1.509–10.250), while the frequency of the b2/b3 deletion did not differ between men with azoo-/oligozoospermia and normozoospermia (22/377, 5.8% vs. 9/217, 4.1%). The b1/b3 (4/377, 1.1%) and b3/b4 (1/377, 0.3%) deletions were observed in the azoo-/oligozoospermic groups, but not in the normozoospermic controls.
Figure 1. Analysis scheme.
Table 1. The distribution of partial AZFc deletion in groups with different spermatogenic status.
| Partial deletion type | Azoo-/oligozoospermia (n = 377) | Normozoospermia (n = 217) | P-values | OR | 95% CI |
| gr/gr deletion | 32 (8.5%) | 5 (2.3%) | 0.003 | 3.933 | 1.509–10.250 |
| b2/b3 deletion | 22 (5.8%) | 9 (4.1%) | 0.373 | 1.432 | 0.647–3.169 |
| b1/b3 deletion | 4 (1.1%) | 0 | 0.302 | 0.989 | 0.979–1.000 |
| b3/b4 deletion | 1 (0.3%) | 0 | 1.000 | 0.997 | 0.992–1.003 |
| Total | 59 (15.6%) | 14 (6.5%) | 0.001 | 2.690 | 1.388–4.239 |
Compared between the groups with azoo-/oligozoospermia and normozoospermia, significant of P<0.05 are marked in bold.
As shown in Figure 2, men with gr/gr deletion were divided into two subgroups based on Y-haplogroups, YAP+ (14cases) and YAP− (23 cases). The frequency of men with gr/gr deletion/YAP+ haplogroup was the similar distributions in both groups; azoo-/oligozoospermia (11/377, 2.9%) and normozoospermic group (3/217, 1.4%). While the frequency of men with gr/gr deletion/YAP− haplogroup was significantly higher in the azoo-/oligozoospermic men than in the normozoospermic men (21/377, 5.6% vs. 2/217, 0.9%, p = 0.004; OR = 6.341; 95% CI = 1.472–27.312).
Figure 2. Comparison of gr/gr deletion frequencies between azoo-/oligozoospermic and normozoospermic groups in YAP+ and YAP− haplogroups.
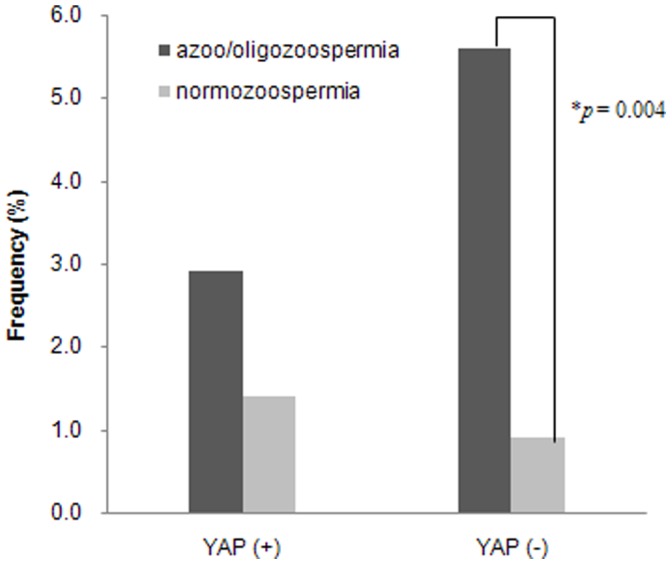
* Fisher’s exact test (two-tailed), OR = 6.341 (95% CI = 1.472–27.312), significant at P<0.05.
For further characterization of gr/gr deletions, we classified gr/gr deletions into four additional subtypes based on the deletion types of CDY1 and DAZ gene copies (Table 2). The YAP+ lineage carried only one deletion subtype, DAZ1/2−, CDY1a−, with similar frequency in both azoo-/oligozoospermic and normozoospermic groups. On the other hand, in YAP− lineage, there were four deletion subtypes, DAZ1/2−, CDY1a−; DAZ1/2−, CDY1b −; DAZ3/4−, CDY1a− and DAZ3/4−, CDY1b−. Two types, DAZ1/2−, CDY1a− and DAZ3/4−, CDY1a−, were found in only spermatogenetic impairment group.
Table 2. The frequency of gr/gr deletion subtypes by gene copy types of DAZ-CDY1, divided on the basis of their Y-haplogroup.
| Group | YAP+ (n = 14) | YAP− (n = 580) | ||
| Azoo-/oligozoospermia(n = 11) | Normozoospermia(n = 3) | Azoo-/oligozoospermia(n = 366) | Normozoospermia(n = 214) | |
| DAZ1/DAZ2−, CDY1a− | 11(100%) | 3 (100%) | 5 (1.4%) | 0 |
| DAZ1/DAZ2−,CDY1b− | 0 | 0 | 5 (1.4%) | 1 (0.5%) |
| DAZ3/DAZ4−,CDY1a− | 0 | 0 | 3 (0.8%) | 0 |
| DAZ3/DAZ4−, CDY1b− | 0 | 0 | 8 (2.2%) | 1 (0.5%) |
Compared between the groups with azoo-/oligozoospermia and normozoospermia, no significant difference by fisher’s exact test (two-tailed), P>0.05.
Quantitative analysis of CDY1 and DAZ showed three gr/gr rearrangements, simple gr/gr deletion, gr/gr deletion-b2/b4 duplication, and gr/gr deletion-CDY1 amplifications (Fig. 3). The frequency of the simple gr/gr deletion, which is the presence of one CDY1 and two DAZ copies, were significantly different between azoo-/oligozoospermic (26/377, 6.9%) and normozoospermic (5/217, 2.3%) (p = 0.015, OR = 3.141; 95% CI = 1.188–8.303). Whereas, the gr/gr deletion-b2/b4 duplication (gr/gr deletion followed by b2/b4 duplication with the presence of two CDY1 and four DAZ copies) and gr/gr deletion-CDY1 amplication (gr/gr deletion with the presence of two CDY1 and two DAZ copies) were found in only azoo-/oligozoospermic group with frequencies of 1.3% (5/377) and 0.3% (1/377), respectively (Table 3). We also compared the mean total sperm concentration in azoo-/oligozoospermic groups. There was no significant difference between the subjects with simple gr/gr deletion and gr/gr deletion-b2/b4 duplication (5.6±15.8×106/ml vs. 5.0±11.2×106/ml, respectively).
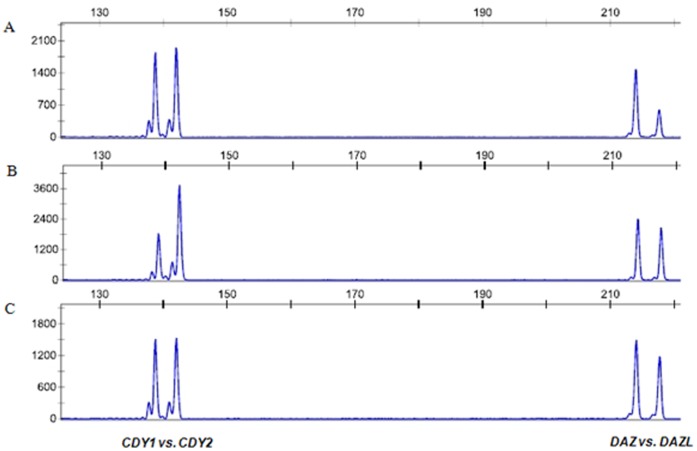
Figure 3. Examples of electrophoretograms showing different gene dosages of CDY1/CDY2 and DAZ/DAZL genes.
The x-axis shows length of PCR products in base pairs and y-axis shows fluorescent intensity. The gene dosage of CDY1 and DAZ can be calculated by the comparison of peak area with CDY2 and DAZL, respectively, as internal standard with known number of copies. (A) Gene dosage of CDY1 and DAZ showed the 1∶1 of CDY1/CDY2 and 2∶1 of DAZ/DAZL patterns (in cases of no deletion according to the reference sequence or gr/gr deletion-b2/b4 duplication) (B) Gene dosage of CDY1 and DAZ identified by the 0.5∶1 of CDY1/CDY2 and 1∶1 of DAZ/DAZL patterns (in case of gr/gr deletion) (C) Gene dosage of CDY1 and DAZ identified by the 1∶1 of CDY1/CDY2 and 1∶1 of DAZ/DAZL patterns (in case of gr/gr deletion-CDY1 amplification).
Table 3. The frequency of gr/gr rearrangements according to analyses of DAZ-CDY1 gene copy number.
| Rearrangement type | Azoo-/oligozoospermia (n = 377) | Normozoospermia (n = 217) | P-values | OR | 95% CI |
| gr/gr del | 26 (6.9%) | 5 (2.3%) | 0.015 | 3.141 | 1.188–8.303 |
| gr/gr del-b2/b4 dupl | 5 (1.3%) | 0 | 0.164 | 0.987 | 0.975–0.998 |
| gr/gr del-CDY1 ampl | 1 (0.3%) | 0 | 1.000 | 0.997 | 0.992–1.003 |
Compared between the groups with azoo-/oligozoospermia and normozoospermia, significant of P<0.05 are marked in bold.
Discussion
In this study, we investigated the types of partial AZFc deletions and their clinical implications in Korean population. Firstly, we screened Yq microdeletions using STS markers for AZFa, AZFb and AZFc region. And then, gr/gr deletions were classified according to the Y-haplogroup, deletion copy types and gene copy number of CDY1 and DAZ genes.
Regardless of deletion type, as we expected, the overall frequency of partial AZFc deletions in azoo-/oligozoospermic men was higher than in normozoospermic men (15.6% vs. 6.5%, p = 0.001). This result suggested that such mutations could be a risk factor for impaired spermatogenesis in the Korean population. Four types of partial AZFc deletions were identified in our population and only gr/gr deletions were statistically associated with impaired spermatogenesis (Table 1). Our result is consistent with two recent meta-analyses [20], [26] and with the largest study on Caucasians [21]. However, several other studies showed no association between gr/gr deletions and azoo-/oligozoospermia in Caucasian [10], [27], Asian [4], [28]–[31] and admixed ethnic populations [32]. The phenotype of gr/gr deletion carriers is reported in Table S1. For the other three types, the frequency of b2/b3deletion was not significant difference in between azoo-/oligozoospermic males and normozoospermic controls, which suggested that b2/b3 deletion is not associated with spermatogenetic impairment in our study population. Similar observations have been reported in various studies [6], [19], [27], [33]. Data from very large study populations from China and, very recently, a North African population suggest that b2/b3 deletion is a risk factor for impaired sperm production [28], [29], [34]. However, the limited number of subjects with this deletion in our study does not allow to define its role in the Korean population. The b1/b3 and b3/b4 deletions were identified only in patients with azoo-/oligozoospermia indicating that deletions might affect on spermatogenesis with mechanism yet to be revealed. The results, however, were limited due to the small number of cases. We compared the clinical features including total sperm count, combined testicular volume, FSH and testosterone levels between subjects with gr/gr deletion and others (Table S2). No statistically significant differences in sperm count were found when comparing sperm count of gr/gr deletion carriers versus non carriers. This may derive from the peculiar composition of our study population which shows a high prevalence of azoospermic men. Gr/gr deletion carriers are mainly oligozoospermic and in fact this deletion is more likely to be associated with oligozoospermia than with azoospermia [21], [26].
The overall frequency of gr/gr deletions in Korean patient with spermatogenetic failure (8.5%) was higher than Europeans (∼4.5%) and similar to Han Chinese populations (10.0 and 10.6%) [11], [26]. This might be resulting from different origins of the study populations. It has been reported that some Asian populations, including Korean, Japanese, and Tibetan, showed the higher frequency of YAP+ haplogroup compared to other populations [35], [36]. And the haplogroup D2b derived from YAP+ lineage always possessed gr/gr deletion and showed normal phenotype [31], [37]. So, we reclassified gr/gr deletions based on YAP haplogroups, YAP+ (hgr.DE) and YAP− [hgr.Y*(xDE)] lineages. Our data showed that the frequency of gr/gr deletion with YAP+ lineage was not significantly different between azoo-/oligozoospermic males and normozoospermic controls. However, the frequency of gr/gr deletion with YAP− lineage was much higher in azoo-/oligozoospermic males than in controls (Fig. 2). So, we concluded that only gr/gr deletion with YAP− lineage was associated with spermatogenetic impairment in Korean population. Hereby, we demonstrated that the gr/gr deletion might effect on spermatogenetic impairment inY haplogroup-dependent manner.
We also investigated gr/gr deletion subtypes according to deletion patterns of DAZ and CDY1 gene copies. Normally, four copies of DAZ gene and two copies of CDY1 gene are assigned in the AZFc region. Several studies related to gr/gr deletion subtypes and spermatogenetic impairments have presented different conclusions. Some studies showed DAZ1/2 deletions were associated with spermatogenetic impairment [17], [19], [21], [38], [39], and Machev et al. [14] presented DAZ3/4-CDY1a deletions were linked to the phenotype. More recently, no association between subtypes of gr/gr deletion and phenotypic abnormalities was also reported [6]. In our study, subjects with YAP+ lineage showed a homogeneous pattern of gr/gr deletion, DAZ1/2−, CDY1a− and no significant difference between azoo-/oligozoospermic and normozoospermic controls. This result was the same as a previous report [37]. In YAP− lineage, deletion of the CDY1a was found only in spermatogenetic impairment group (Table 2). Although our result might not be sufficient to verify the association between these deletion subtypes and clinical consequences, this phenomenon is similar to previous studies in a correlation between the absence of CDY1a and male infertility [14], [39]. So, further studies will be required.
Finally, we carried out CDY1 and DAZ gene copy number analysis to identify gr/gr rearrangement types (Table 3). Krausz et al. [6] reported that gr/gr deletions could be classified into five rearrangement types based on the copy number of CDY1 and DAZ gene. In our study, three out of five rearrangement types were identified and the majority of gr/gr deletions (83.8%, 31/37) were simple gr/gr deletion type, one copy of CDY1 and two copies of DAZ. This result was similar to European population (80%, 128/160) [6]. The other two types, gr/gr deletion-b2/b4 duplications (four copies of DAZ and two copies of CDY1) and gr/gr deletion-CDY1 amplification (two copies of DAZ and two copies of CDY1), were found in patients with spermatogenetic impairment with frequencies of 13.5% (5/37) and 2.7% (1/37), respectively, but none in normozoospermic controls. These results suggested that regardless of having an additional duplication of AZFc region, the simple gr/gr deletion might be associated with spermatogenetic impairment in Korean population. Recently, Lu et al. [22] reported that b2/b3 deletion with secondary duplication was also a risk factor for spermatogenetic impairment in Han Chinese population. Meanwhile, two possible mechanisms to generate the gr/gr deletion-CDY1 amplification are proposed and the recombinant products resulting from both ways were not distinguishable as shown in Figure S1. So, we could not verify what happened first with current technology.
In conclusion, we analyzed 377patients with spermatogenetic impairments and 217normozoospermic controls. As far as we know, this is the first report that only gr/gr deletion with YAP− lineage, among several types of partial AZFc deletions, was associated with spermatogenetic impairment in Korean population. Although the role of CDY1 and DAZ on spermatogenesis is still not clear, further studies on other genes related to spermatogenesis and larger scale population study will be assured to understand the spermatogenesis pathology.
Supporting Information
Two possible mechanisms of gr/gr del- CDY1 amplification; one is that the g1/g2 recombination resulting in gr/gr deletion arises first and then the CDY1 amplification occurs and the other is vice versa. The recombinant products from both ways are not distinguishable.
(TIF)
Clinical features of patients bearing gr/gr deletions and their deletion patterns based on the type and number of DAZ - CDY 1copies deleted.
(DOCX)
Means±SD of the total sperm count, testicular volume and hormonal levels in patients and in controls.
(DOCX)
Funding Statement
This study was supported by a grant (A084923) of the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare and Family Affairs, Republic of Korea. The funders had no role in study design, data collection and analysis, decision to publish,or preparation of the manuscript.
References
- 1. Vogt PH, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, et al. (1996) Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet 5: 933–943. [DOI] [PubMed] [Google Scholar]
- 2. Repping S, Skaletsky H, Lange J, Silber S, Van Der Veen F, et al. (2002) Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am J Hum Genet 71: 906–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Noordam MJ, Repping S (2006) The human Y chromosome: a masculine chromosome. Curr Opin Genet Dev 16: 225–232. [DOI] [PubMed] [Google Scholar]
- 4. Lin YW, Hsu LC, Kuo PL, Huang WJ, Chiang HS, et al. (2007) Partial duplication at AZFc on the Y chromosome is a risk factor for impaired spermatogenesis in Han Chinese in Taiwan. Hum Mutat 28: 486–494. [DOI] [PubMed] [Google Scholar]
- 5. Yen P (2001) The fragility of fertility. Nat Genet 29: 243–244. [DOI] [PubMed] [Google Scholar]
- 6. Krausz C, Giachini C, Xue Y, O’Bryan MK, Gromoll J, et al. (2009) Phenotypic variation within European carriers of the Y-chromosomal gr/gr deletion is independent of Y-chromosomal background. J Med Genet 46: 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Repping S, Skaletsky H, Brown L, van Daalen SK, Korver CM, et al. (2003) Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet 35: 247–251. [DOI] [PubMed] [Google Scholar]
- 8. Repping S, van Daalen SK, Korver CM, Brown LG, Marszalek JD, et al. (2004) A family of human Y chromosomes has dispersed throughout northern Eurasia despite a 1.8-Mb deletion in the azoospermia factor c region. Genomics 83: 1046–1052. [DOI] [PubMed] [Google Scholar]
- 9. Lynch M, Cram DS, Reilly A, O’Bryan MK, Baker HW, et al. (2005) The Y chromosome gr/gr subdeletion is associated with male infertility. Mol Hum Reprod 11: 507–512. [DOI] [PubMed] [Google Scholar]
- 10. Stouffs K, Tournaye H, Van der Elst J, Haentjens P, Liebaers I, et al. (2008) Do we need to search for gr/gr deletions in infertile men in a clinical setting? Hum Reprod 23: 1193–1199. [DOI] [PubMed] [Google Scholar]
- 12. Burd CG, Dreyfuss G (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science 265: 615–621. [DOI] [PubMed] [Google Scholar]
- 13. Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, et al. (2001) The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet 29: 279–286. [DOI] [PubMed] [Google Scholar]
- 14. Machev N, Saut N, Longepied G, Terriou P, Navarro A, et al. (2004) Sequence family variant loss from the AZFc interval of the human Y chromosome, but not gene copy loss, is strongly associated with male infertility. J Med Genet 41: 814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shahid M, Dhillon VS, Khalil HS, Sexana A, Husain SA (2011) Associations of Y-chromosome subdeletion gr/gr with the prevalence of Y-chromosome haplogroups in infertile patients. Eur J Hum Genet 19: 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferras C, Fernandes S, Marques CJ, Carvalho F, Alves C, et al. (2004) AZF and DAZ gene copy-specific deletion analysis in maturation arrest and Sertoli cell-only syndrome. Mol Hum Reprod 10: 755–761. [DOI] [PubMed] [Google Scholar]
- 17. Fernandes S, Huellen K, Goncalves J, Dukal H, Zeisler J, et al. (2002) High frequency of DAZ1/DAZ2 gene deletions in patients with severe oligozoospermia. Mol Hum Reprod 8: 286–298. [DOI] [PubMed] [Google Scholar]
- 18. Yang Y, Ma M, Li L, Su D, Chen P, et al. (2010) Differential effect of specific gr/gr deletion subtypes on spermatogenesis in the Chinese Han population. Int J Androl 33: 745–754. [DOI] [PubMed] [Google Scholar]
- 19. Ferlin A, Tessari A, Ganz F, Marchina E, Barlati S, et al. (2005) Association of partial AZFc region deletions with spermatogenic impairment and male infertility. J Med Genet 42: 209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Visser L, Westerveld GH, Korver CM, van Daalen SK, Hovingh SE, et al. (2009) Y chromosome gr/gr deletions are a risk factor for low semen quality. Hum Reprod 24: 2667–2673. [DOI] [PubMed] [Google Scholar]
- 21. Giachini C, Laface I, Guarducci E, Balercia G, Forti G, et al. (2008) Partial AZFc deletions and duplications: clinical correlates in the Italian population. Hum Genet 124: 399–410. [DOI] [PubMed] [Google Scholar]
- 22. Lu C, Zhang F, Yang H, Xu M, Du G, et al. (2011) Additional genomic duplications in AZFc underlie the b2/b3 deletion-associated risk of spermatogenic impairment in Han Chinese population. Hum Mol Genet 20: 4411–4421. [DOI] [PubMed] [Google Scholar]
- 23.WHO (1999) Labortory manual for the examination of human semen and sperm-cervical mucus interaction, 4th edn. Cambridge, UK: Cambridge University Press.
- 24. Hammer MF, Horai S (1995) Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet 56: 951–962. [PMC free article] [PubMed] [Google Scholar]
- 25. Plaseski T, Noveski P, Trivodalieva S, Efremov GD, Plaseska-Karanfilska D (2008) Quantitative fluorescent-PCR detection of sex chromosome aneuploidies and AZF deletions/duplications. Genet Test 12: 595–605. [DOI] [PubMed] [Google Scholar]
- 26. Stouffs K, Lissens W, Tournaye H, Haentjens P (2010) What about gr/gr deletions and male infertility? Systematic review and meta-analysis. Hum Reprod Update 17: 197–209. [DOI] [PubMed] [Google Scholar]
- 27. Hucklenbroich K, Gromoll J, Heinrich M, Hohoff C, Nieschlag E, et al. (2005) Partial deletions in the AZFc region of the Y chromosome occur in men with impaired as well as normal spermatogenesis. Hum Reprod 20: 191–197. [DOI] [PubMed] [Google Scholar]
- 28. Lu C, Zhang J, Li Y, Xia Y, Zhang F, et al. (2009) The b2/b3 subdeletion shows higher risk of spermatogenic failure and higher frequency of complete AZFc deletion than the gr/gr subdeletion in a Chinese population. Hum Mol Genet 18: 1122–1130. [DOI] [PubMed] [Google Scholar]
- 29. Wu B, Lu NX, Xia YK, Gu AH, Lu CC, et al. (2007) A frequent Y chromosome b2/b3 subdeletion shows strong association with male infertility in Han-Chinese population. Hum Reprod 22: 1107–1113. [DOI] [PubMed] [Google Scholar]
- 30. Zhang F, Li Z, Wen B, Jiang J, Shao M, et al. (2006) A frequent partial AZFc deletion does not render an increased risk of spermatogenic impairment in East Asians. Ann Hum Genet 70: 304–313. [DOI] [PubMed] [Google Scholar]
- 31. de Carvalho CM, Zuccherato LW, Fujisawa M, Shirakawa T, Ribeiro-dos-Santos AK, et al. (2006) Study of AZFc partial deletion gr/gr in fertile and infertile Japanese males. J Hum Genet 51: 794–799. [DOI] [PubMed] [Google Scholar]
- 32. Ravel C, Chantot-Bastaraud S, El Houate B, Mandelbaum J, Siffroi JP, et al. (2006) GR/GR deletions within the azoospermia factor c region on the Y chromosome might not be associated with spermatogenic failure. Fertil Steril 85: 229–231. [DOI] [PubMed] [Google Scholar]
- 33. Fernando L, Gromoll J, Weerasooriya TR, Nieschlag E, Simoni M (2006) Y-chromosomal microdeletions and partial deletions of the Azoospermia Factor c (AZFc) region in normozoospermic, severe oligozoospermic and azoospermic men in Sri Lanka. Asian J Androl 8: 39–44. [DOI] [PubMed] [Google Scholar]
- 34. Eloualid A, Rhaissi H, Reguig A, Bounaceur S, El Houate B, et al. (2012) Association of spermatogenic failure with the b2/b3 partial AZFc deletion. PLoS One 7: e34902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jin HJ, Kwak KD, Hammer MF, Nakahori Y, Shinka T, et al. (2003) Y-chromosomal DNA haplogroups and their implications for the dual origins of the Koreans. Hum Genet 114: 27–35. [DOI] [PubMed] [Google Scholar]
- 36. Kim W, Shin DJ, Harihara S, Kim YJ (2000) Y chromosomal DNA variation in east Asian populations and its potential for inferring the peopling of Korea. J Hum Genet 45: 76–83. [DOI] [PubMed] [Google Scholar]
- 37. Sin HS, Koh E, Shigehara K, Sugimoto K, Maeda Y, et al. (2010) Features of constitutive gr/gr deletion in a Japanese population. Hum Reprod 25: 2396–2403. [DOI] [PubMed] [Google Scholar]
- 38. Yang Y, Xiao C, Zhang S, Zhoucun A, Li X (2006) Preliminary study of the relationship between DAZ gene copy deletions and spermatogenic impairment in Chinese men. Fertil Steril 85: 1061–1063. [DOI] [PubMed] [Google Scholar]
- 39. Giachini C, Guarducci E, Longepied G, Degl’Innocenti S, Becherini L, et al. (2005) The gr/gr deletion(s): a new genetic test in male infertility? J Med Genet 42: 497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Two possible mechanisms of gr/gr del- CDY1 amplification; one is that the g1/g2 recombination resulting in gr/gr deletion arises first and then the CDY1 amplification occurs and the other is vice versa. The recombinant products from both ways are not distinguishable.
(TIF)
Clinical features of patients bearing gr/gr deletions and their deletion patterns based on the type and number of DAZ - CDY 1copies deleted.
(DOCX)
Means±SD of the total sperm count, testicular volume and hormonal levels in patients and in controls.
(DOCX)