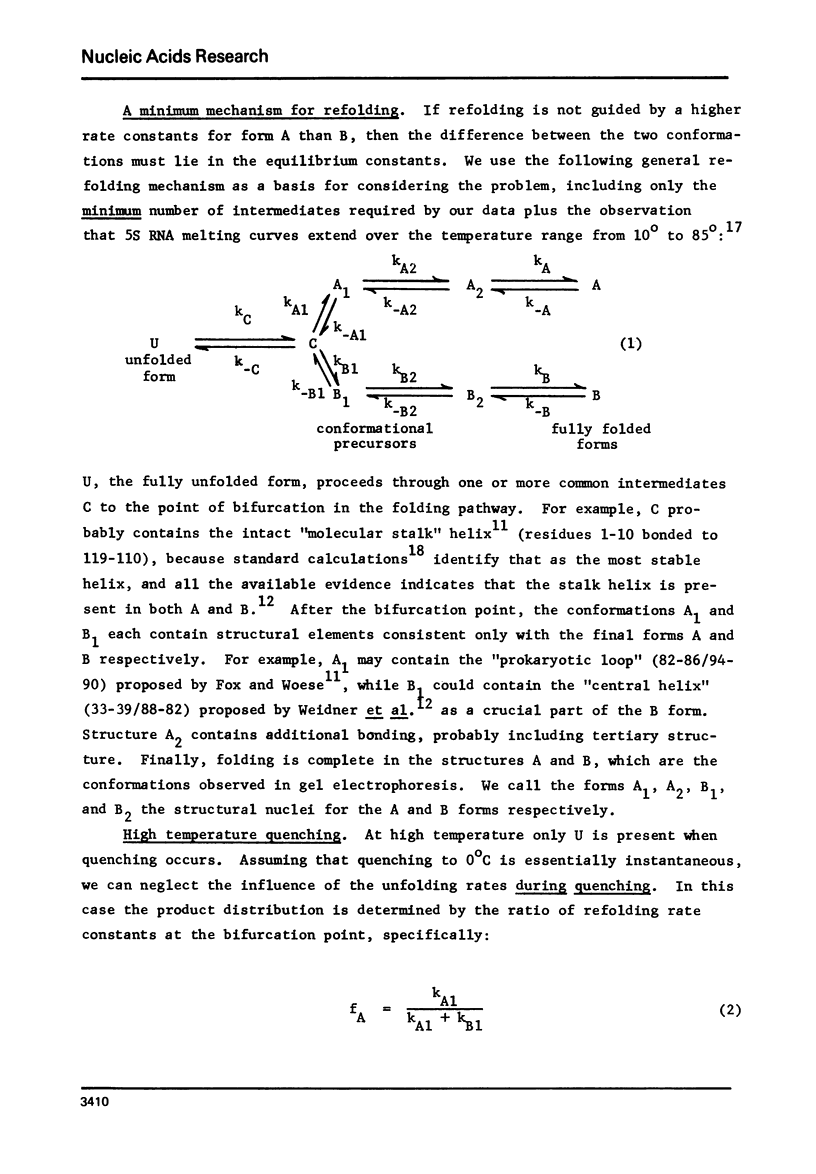
Abstract
The refolding of 5S RNA into its two conformational states has been examined as a function of solvent composition and annealing conditions. The results show that the product distribution depends on the folding pathway. Quick cooling from high temperature produces roughly equal amounts of the two forms, even in the presence of 1 mm Mg++. However annealing by slow cooling to intermediate temperatures (50 degrees--60 degrees C) in Mg++-containing buffers, followed by quick cooling, allows formation of a structure which guides the refolding path to the "native" conformation. The stability of this structural nucleus for the "native" conformation depends strongly on Mg++ concentration. We conclude that the A ("native") conformation differs from the B conformation not in rate of refolding, but rather in having a lower enthalpy and a also a smaller rate of unfolding for the critical structural nucleus. The order of folding during biosynthesis may be crucial for forming the "native" conformation.
Full text
PDF



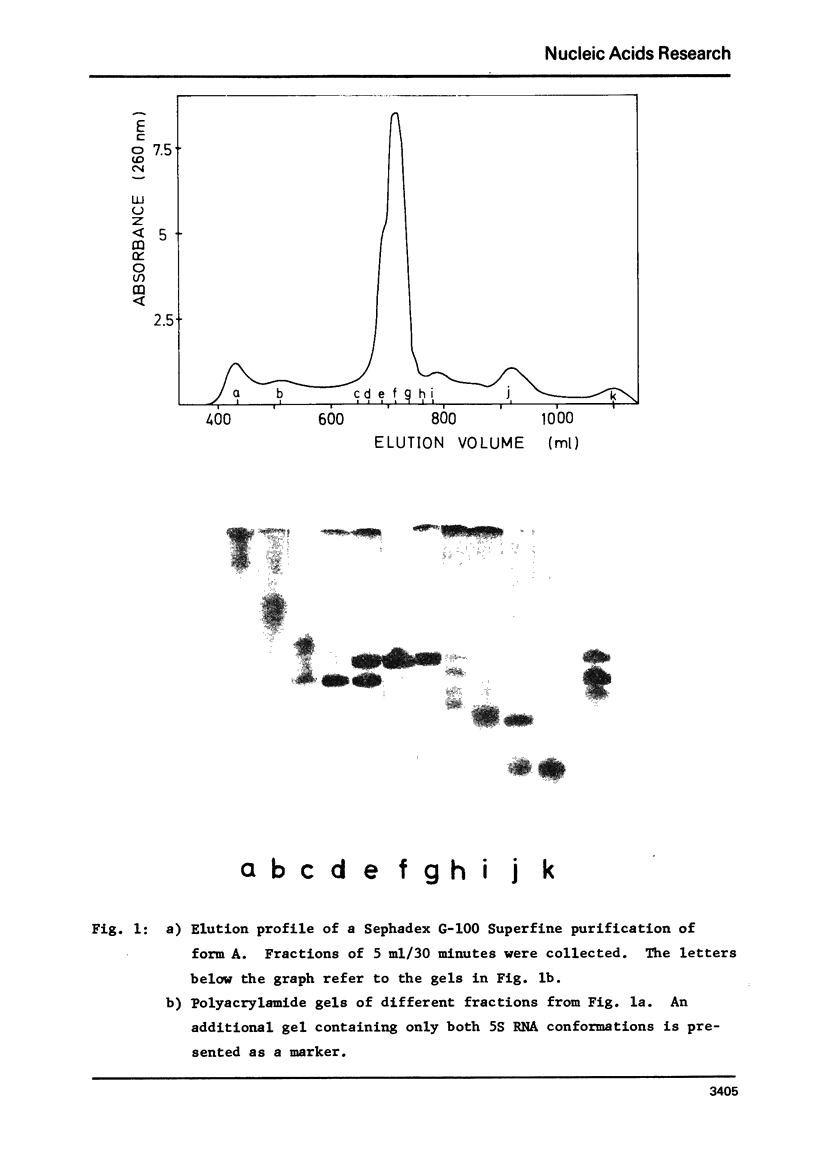

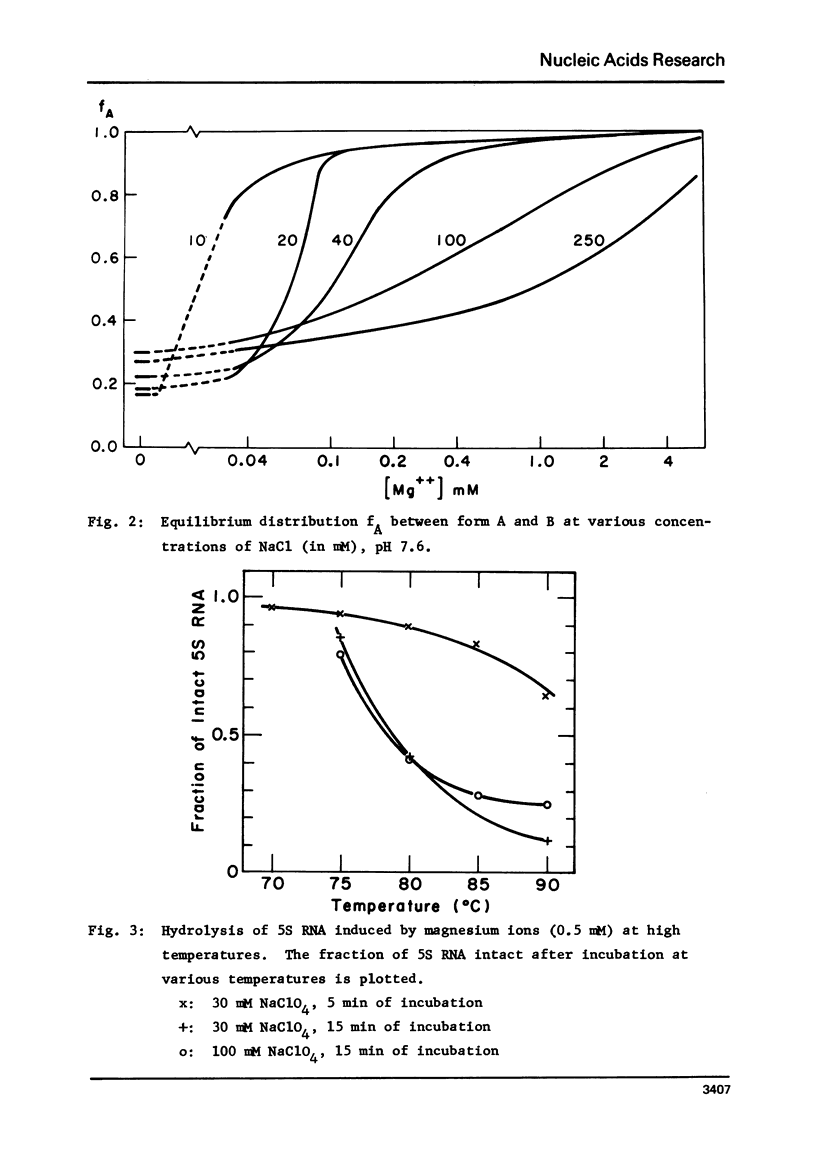

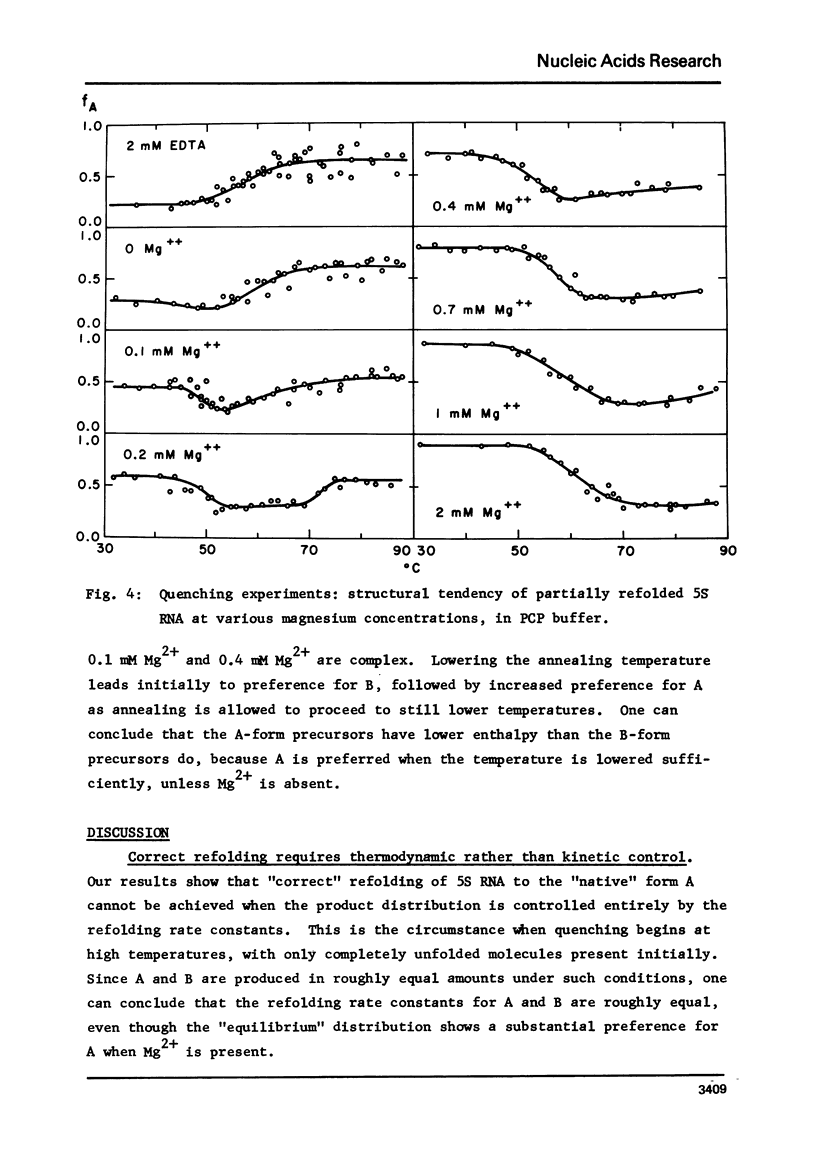




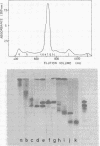
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aubert M., Scott J. F., Reynier M., Monier R. Rearrangement of the conformation of Escherichia coli 5S RNA. Proc Natl Acad Sci U S A. 1968 Sep;61(1):292–299. doi: 10.1073/pnas.61.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crothers D. M., Cole P. E., Hilbers C. W., Shulman R. G. The molecular mechanism of thermal unfolding of Escherichia coli formylmethionine transfer RNA. J Mol Biol. 1974 Jul 25;87(1):63–88. doi: 10.1016/0022-2836(74)90560-9. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A., Sprinzl M., Pongs O. The involvement of 5S RNA in the binding of tRNA to ribosomes. Biochem Biophys Res Commun. 1973 Oct 1;54(3):942–948. doi: 10.1016/0006-291x(73)90785-7. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A. Structure and function of 5S and 5.8 S RNA. Prog Nucleic Acid Res Mol Biol. 1976;18:45–90. [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Gralla J., Crothers D. M. Free energy of imperfect nucleic acid helices. II. Small hairpin loops. J Mol Biol. 1973 Feb 5;73(4):497–511. doi: 10.1016/0022-2836(73)90096-x. [DOI] [PubMed] [Google Scholar]
- Gray P. N., Monier R. Partial localization of the 5S RNA binding site on 23S RNA. Biochimie. 1972;54(1):41–45. doi: 10.1016/s0300-9084(72)80036-1. [DOI] [PubMed] [Google Scholar]
- Herr W., Noller H. F. A fragment of 23S RNA containing a nucleotide sequence complementary to a region of 5S RNA. FEBS Lett. 1975 May 1;53(2):248–252. doi: 10.1016/0014-5793(75)80030-5. [DOI] [PubMed] [Google Scholar]
- Horne J. R., Erdmann V. A. Isolation and characterization of 5S RNA-protein complexes from Bacillus stearothermophilus and Escherichia coli ribosomes. Mol Gen Genet. 1972;119(4):337–344. doi: 10.1007/BF00272091. [DOI] [PubMed] [Google Scholar]
- Kearns D. R., Wong Y. P. Investigation of the secondary structure of Escherichia coli 5 S RNA by high-resolution nuclear magnetic resonance. J Mol Biol. 1974 Aug 25;87(4):755–774. doi: 10.1016/0022-2836(74)90083-7. [DOI] [PubMed] [Google Scholar]
- Lecanidou R., Richards E. G. The thermodynamics and kinetics of conformational changes in 5-S RNA from Escherichia coli. Eur J Biochem. 1975 Sep 1;57(1):127–133. doi: 10.1111/j.1432-1033.1975.tb02283.x. [DOI] [PubMed] [Google Scholar]
- Richards E. G., Lecanidou R., Geroch M. E. The kinetics of renaturation of 5-S RNA from Escherichia coli in the presence of Mg 2+ ions. Eur J Biochem. 1973 Apr;34(2):262–267. doi: 10.1111/j.1432-1033.1973.tb02755.x. [DOI] [PubMed] [Google Scholar]
- Römer R., Hach R. tRNA conformation and magnesium binding. A study of a yeast phenylalanine-specific tRNA by a fluorescent indicator and differential melting curves. Eur J Biochem. 1975 Jun 16;55(1):271–284. doi: 10.1111/j.1432-1033.1975.tb02160.x. [DOI] [PubMed] [Google Scholar]
- Scott J. F., Monier R., Aubert M., Reynier M. Some optical properties of 5S-RNA from E. coli. Biochem Biophys Res Commun. 1968 Dec 9;33(5):794–800. doi: 10.1016/0006-291x(68)90230-1. [DOI] [PubMed] [Google Scholar]
- Staehelin T., Maglott D. M., Monro R. E. On the catalytic center of peptidyl transfer: a part of the 50 S ribosome structure. Cold Spring Harb Symp Quant Biol. 1969;34:39–48. doi: 10.1101/sqb.1969.034.01.008. [DOI] [PubMed] [Google Scholar]
- Stein A., Crothers D. M. Conformational changes of transfer RNA. The role of magnesium(II). Biochemistry. 1976 Jan 13;15(1):160–168. doi: 10.1021/bi00646a025. [DOI] [PubMed] [Google Scholar]
- Weidner H., Yuan R., Crothers D. M. Does 5S RNA function by a switch between two secondary structures? Nature. 1977 Mar 10;266(5598):193–194. doi: 10.1038/266193a0. [DOI] [PubMed] [Google Scholar]
- Wintermeyer W., Zachau H. G. Mg 2+ -katalysierte, spezifische Spaltung von tRN. Biochim Biophys Acta. 1973 Feb 23;299(1):82–90. [PubMed] [Google Scholar]