Abstract
During DNA synthesis base stacking and Watson-Crick (WC) hydrogen bonding increase the stability of nascent base-pairs when they are in a ternary complex. To evaluate the contribution of base stacking to the incorporation efficiency of dNTPs when a DNA polymerase encounters an abasic site, we varied the Penultimate Base-pairs (PBs) adjacent to the abasic site using all 16 possible combinations. We then determined pre-steady state kinetic parameters with an RB69 DNA polymerase variant and solved nine structures of the corresponding ternary complexes. The incorporation efficiency for incoming dNTPs opposite an abasic site varied between 2 and 210 fold depending on the identity of the PB. We propose that the A rule can be extended to encompass the fact that DNA polymerase can bypass dA/abasic sites more efficiently than other dN/abasic sites. Crystal structures of the ternary complexes show that the surface of the incoming base was stacked against the PB’s interface and that the kinetic parameters for dNMP incorporations were consistent with specific features of base-stacking, such as surface area and partial charge-charge interactions between the incoming base and the PB. Without a templating nucleotide residue, an incoming dNTP has no base with which it can hydrogen bond and cannot be desolvated so that these surrounding water molecules become ordered and remain on the PB’s surface in the ternary complex. When these water molecules are on top of a hydrophobic patch on the PB, they destabilize the ternary complex and the incorporation efficiency of incoming dNTPs is reduced.
Abasic sites are the most common lesions found in DNA. They occur under physiological conditions, with up to 10,000 abasic sites formed in each cell per day due to spontaneous hydrolysis of the N-glycosidic bond between the base and the deoxy- ribosyl moiety.(1–4) In most cases, these lesions are rapidly removed via Base Excision Repair (BER) pathways.(5) Left unrepaired, abasic sites provide strong blocks to DNA synthesis due to lack of a templating base to instruct correct nucleotide incorporation.(6–9) In the absence of coding information, most replicative polymerases preferentially incorporate dAMP opposite an abasic site. This strong preference for dATP is known as the “A-rule”.(8, 10–20) However, the mechanistic basis for the A-rule still remains elusive despite the large number of kinetic and structural studies that have been reported on non-instructive lesion bypass or translesion synthesis by DNA polymerases.(21–23) It has been speculated that the preference for A is due to the fact that purines stack better than pyrimidines,(24–25) however this explanation does not account for why A is inserted more efficiently than G. If the extent of base stacking alone was the only determining factor for dNTP insertion opposite an abasic site, then dGTP should be incorporated as efficiently as dATP. Since this does not happen with most DNA pols, it implies that there are other interactions between the incoming dNTP and the PB (for the location of the PB in the primer-template sequence, see Fig. 1A) that influence the incorporation efficiency of a dNMP opposite an abasic site.
Figure 1.
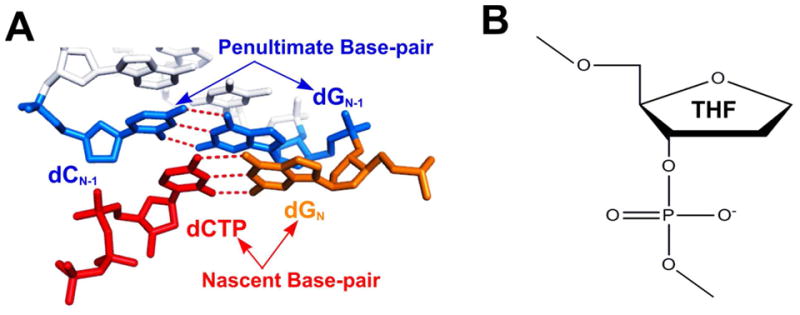
Nascent/penultimate base pairs and an abasic analogue. (A) Illustration of the positions of Nascent Base-pair (NB) and Penultimate Base-pair (PB) in the NBP. (B) Structure of Tetrahydrofuran (THF) in DNA.
Reineks and Berdis(26) have identified a non-natural dNTP analogue, 5-nitro-1-indolyl-2′-deoxyriboside-5′-triphosphate (5-NITP), that was incorporated opposite an abasic site 1000 fold more efficiently than dAMP by T4 DNA polymerase (T4pol), a close relative of RB69pol. Because the size and shape of 5-NITP and dATP are similar, the large increase in incorporation efficiency was attributed to the nitro group at the 5 position on the indole ring of 5-NITP. Presumably it provides greater π–π interactions between the incoming nucleotide analogue and the aromatic amino acid side chains present in the NPB compared to the case where the incoming dNTP was dATP. This explanation was deemed insufficient and required modification when the ternary complex, that had 5-NITP opposite an abasic site, was determined.(27) In addition to enhanced π–π stacking interaction, there appeared to be a positive contribution from dipole-dipole interactions between the nitro group of 5-NITP and N4 of the cytosine residue situated 3′ to the abasic site in the template strand.(27) This result demonstrated that base-stacking and π–π interactions were only part of the reason that 5-NITP was inserted more efficiently than dATP when it was opposite an abasic site. It should be noted that the extent of this preference appears to be dependent on the particular DNA polymerase. For example, when 5-NITP was opposite an abasic site in the presence of Klenow Fragment there was only a small difference in the insertion efficiency of 5-NITP compared to dATP.(26) Whereas, in our attempts to understand more about how this selectivity is achieved with a B family DNA polymerase, we used RB69pol and primer-templates with tetrahydrofuran (THF, Fig 1B) as a stable abasic site mimic, an analogue has almost always been used in studies of translesion synthesis by DNA polymerases.(16, 22, 28–31)
After placing a THF in the templating position we determined pre-steady-state kinetic parameters for all four incoming dNTPs with wild type (wt) RB69pol, in an exo− background, and with an exo− RB69pol triple mutant (tm) that had the Y567A/L561A/S565G substitutions. We designed this mutant so that it had same pre-steady-state kinetic parameters as wt RB69pol for insertion of correct dNTPs although it had reduced base selectivity.(32) Most importantly, this RB69pol mutant allowed us to capture relevant ternary complexes with THF in the templating position for crystallographic studies, and made it possible to correlate and rationalize the kinetic behavior with structures of the corresponding ternary complexes. In addition, for each incoming dNTP, we varied the identity of the PB employing all 16 base-pair combinations. Although the kinetics for incorporation of dNMPs opposite an abasic site by T4 pol have been studied previously without varying the PB’s identity,(26, 29, 31, 33–35) the results could not be fully explained solely by differences in base-stacking or desolvation when insertion efficiencies of the incoming dNTPs were compared.(31, 33–35) Our kinetic data and structural results show that, while base-stacking is important, partial charge interactions between the incoming dNTP and the interface of the PB contribute significantly to the pre-steady-state kinetic paramaters for dNMP insertion opposite an abasic site in the context of the tm RB69pol. We also propose that the A rule can be extended to include the observation that B family DNA polymerases, as exemplified by RB69pol and Phi29pol, can bypass dA/abasic sites more efficiently than any other dN/abasic lesion.
EXPERIMENTAL PROCEDURES
Chemicals
All chemicals were the highest quality commercially available; dNTPs were purchased from Roche (Roche Applied Science, Indianapolis, IN). [γ-32P]ATP was purchased from MP Biomedicals (Irvine, CA).
Enzymes
wt RB69pol and tm RB69pol, in an exonuclease-deficient background (D222A and D327A), were over-expressed in E. coli strain BL21(DE), purified, and stored as previously described.(36) The Phi29pol (exo−) was over-expressed in E. coli strain BL21(DE), purified as previously described.(37)
DNA Substrates
Oligonucleotides were synthesized at the Keck facilities (Yale University). The sequence of the primer-template (P/T) used in this study was as follows: 5′-GCGGACTGCTTAY (primer) and 5′-TCAXZTAAGCAGTCCGCG, with X = THF, Y = C, G, A, or T, and Z = G, C, T, or A for Y–Z Watson-Crick base pairs. For kinetic studies, the primer was labeled at the 5′-end with 32P using T4 polynucleotide kinase and [γ-32P]-ATP and annealed to unlabeled templates by heating to 95 °C, followed by slowly cooling down to 25 °C to give fully duplexed P/Ts. For crystallization, Y = ddC, ddG, ddA, or ddT to prevent phosphoryl transfer.
Chemical Quench Experiments
Rapid chemical quench experiments were performed at 23°C with a buffer solution containing 66 mM Tris-HCl (pH 7.4) and 10mM MgSO4 using a KinTek RFQ-3 instrument (KinTek Corp., University Park, PA). Experiments were performed under single-turnover conditions, with a 10-fold excess RB69pol over the P/T. The final concentrations after mixing were: 1 μM enzyme, 83 nM 32P-labeled P/T, and 10 mM Mg2+. Reaction mixtures were quenched with 0.5 M EDTA (pH 8.0). Substrates and products were separated by 19:1% (w/v) PAGE containing 8M urea, visualized using a MD Strom 860 imager (Molecular Imaging), and quantified using ImageQuaNT software. For each Kd and kpol determination, seven different [dNTP] were used. The amount of product formed versus time was plotted for each [dNTP] and fitted by nonlinear regression to the following equation: , where A is the observed amplitude of product formation, and kobs is the observed rate constant. The kinetic parameters kpol (the maximum rate of dNMP incorporation) and Kd,app (defined as the [dNTP] at which the rate of dNMP incorporation reaches 1/2 kpol), were obtained by plotting kobs versus [dNTP] to a hyperbolic equation: , where kobs represents the observed rate at a given [dNTP].
Crystallization of dNTP/THF-containing tm RB69pol ternary complexes
Dideoxy-terminated oligos were used for crystallization. The tm RB69pol (110 μM final concentration) was mixed with an equimolar ratio of freshly annealed duplex DNA. A solution of dNTP was then added to give a final concentration of 2 mM. The complex containing dGTP was prepared using the soaking replacement method from the preformed dCTP-containing crystals. Crystals was grown under oil using a microbatch procedure.(38) A solution of 100 mM CaCl2, 15% (w/v) PEG 350 monomethyl ether (MME), and 100 mM sodium cacodylate (pH 6.5) was mixed with an equal volume of the protein complex. The rectangular-shaped crystals typically grew in 2 days at 20°C and had dimensions of about 0.1 mm × 0.1 mm × 0.2 mm. Crystals were transferred from the mother liquor to a cryoprotectant solution containing 30% (w/v) PEG 350 MME, 100 mM CaCl2, and 100 mM Na cacodylate (pH 6.5) prior to freezing in liquid nitrogen.
Data collection, structure determination, and refinement
X-ray diffraction data were collected at 110K, using synchrotron radiation sources at beam line 24ID-E, Northeast Collaborative Access Team (NECAT), Advanced Photon Source, Argonne National Laboratory (APS, ANL, Chicago, IL). The data were processed using HKl2000 program suite (Table 1 and 2). The structures were solved by molecular replacement using Phaser,(39) starting with the pol structure from the ternary complex of wt RB69pol with PDB accession number 3NCI,35 and refined using REFMAC5.(40) The P/T duplex and dNTP were built into electron density maps using the program COOT.(41) Structure refinement statistics are summarized in Table 1 and 2. Figures were made using the program Pymol.(42)
Table 1.
Crystallographic Statistics for Data Collection and Structure Refinement of dNTP/THF-Containing Ternary Complexes Formed by tm RB69pol with ddC/dG as the PBP.
| dATP vs THF (Triple mutant) | dCTP vs THF (Triple mutant) | dGTP vs THF (Triple mutant) | dTTP vs THF (Triple mutant) | |
|---|---|---|---|---|
| Space group | P212121 | P212121 | P212121 | P212121 |
| Unit cell (a,b,c (Å)) | 75.0,120.1,130.5 | 75.0,120.1,130.7 | 74.9,120.0,130.6 | 75.4,120.2,131.6 |
| Resolution (Å) | 50.0-2.04 | 50.0-1.96 | 50.0-1.87 | 50.0-1.84 |
| No of unique reflections | 75,390 | 107,757 | 97,840 | 100,249 |
| Redundancy | 4.7 (3.4) | 4.7 (4.0) | 4.7 (3.1) | 4.4 (2.9) |
| Completeness (%) | 99.3 (96.2) | 98.3 (90.4) | 99.3 (92.4) | 97.1 (80.2) |
| Rmerge (%) | 10.1 (98.6) | 4.5 (14) | 7.8 (80.7) | 6.4 (65.5) |
| I/σ | 11.8 (1.0) | 27.6 (10.9) | 16.0 (1.2) | 19.4 (1.5) |
| Final model | ||||
| Amino acid residues | 903 | 903 | 903 | 903 |
| Water molecules | 610 | 883 | 798 | 795 |
| Ca 2+ ions | 4 | 4 | 6 | 4 |
| Template nucleotides | 18 | 18 | 18 | 18 |
| Primer nucleotides | 13 | 13 | 13 | 13 |
| dNTP | 1 | 1 | 1 | 1 |
| Refinement Statistics | ||||
| Reflections | 71,342 | 79,816 | 92,567 | 94,921 |
| R (%) | 18.1 (27.3) | 16.7 (16.8) | 17.1 (27.3) | 17.2 (26.6) |
| Rfree (%) r.m.s.d |
22.8 (33.7) | 20.8 (22.4) | 20.2 (29.4) | 20.5 (30.6) |
| Bond length (Å) | 0.009 | 0.008 | 0.008 | 0.008 |
| Bond angles (°) | 1.187 | 1.112 | 1.089 | 1.102 |
| PDB code | 4DTR | 4DTS | 4DTU | 4DTX |
Statistics for the highest resolution shell are in parenthesis.
Rmerge = ΣhklΣj|Ij(hkl)−<Ij(hkl)>|/Σ hkl Σj <I(hkl)>, statistics for merging all observations for given reflections.
R = Σhkl|Fobs(hkl)−Fcalc(hkl)|/Σhkl Fobs(hkl), statistics for crystallographic agreement between the measured and model-calculated amplitudes. Rfree is the agreement for cross-validation data set.
Root mean square deviations (rmsd) to ideal values.
Table 2.
Crystallographic Statistics for Data Collection and Structure Refinement of dNTP/THF-Containing Ternary Complexes Formed by tm RB69pol with Different PBs.
| ddT/dA/dTTP | ddG/dC/dCTP | ddA/dT/dATP | ddA/dT/dCTP | ddA/dT/dGTP | |
|---|---|---|---|---|---|
| PB | ddT/dA | ddG/dC | ddA/dT | ddA/dT | ddA/dT |
| dNTP | dTTP | dCTP | dATP | dCTP | dGTP |
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 |
| Unit cell (a,b,c (Å)) | 75.1, 119.8, 130.5 | 75.2, 120.5, 130.8 | 78.3, 118.1, 130.2 | 78.2, 119.3, 130.9 | 78.5, 118.3, 130.7 |
| Resolution (Å) | 50.0 - 1.90 | 50.0 - 1.95 | 50.0 - 1.96 | 50.0 - 2.05 | 50.0 - 2.04 |
| No of unique reflections | 92,548 | 86,912 | 86,518 | 78,651 | 75,856 |
| Redundancy | 3.6 (3.4) | 3.6 (3.7) | 3.9 (3.9) | 3.6 (3.7) | 3.6 (3.6) |
| Completeness (%) | 99.1 (99.0) | 99.6 (99.9) | 99.7 (100.0) | 99.5 (100.0) | 99.2 (99.9) |
| Rmerge (%) | 6.0 (24.5) | 9.2 (74.4) | 9.8 (83.2) | 8.7 (70.6) | 11.4 (71.5) |
| I/σ | 18.7 (4.0) | 12.8 (1.3) | 12.5 (1.3) | 12.9 (1.3) | 10.7 (1.6) |
| Final model | |||||
| Amino acid residues | 903 | 903 | 903 | 903 | 903 |
| Water molecules | 906 | 700 | 645 | 562 | 597 |
| Ca 2+ ions | 5 | 5 | 5 | 5 | 5 |
| Template nucleotides | 18 | 18 | 18 | 18 | 18 |
| Primer nucleotides | 13 | 13 | 13 | 13 | 13 |
| dNTP | 1 | 1 | 1 | 1 | 1 |
| Refinement Statistics | |||||
| Reflections | 87,843 | 82,442 | 82,125 | 74,651 | 71,956 |
| R (%) | 17.1 (21.4) | 18.4 (30.5) | 18.4 (27.6) | 18.4 (27.1) | 18.9 (25.4) |
| Rfree (%) r.m.s.d |
20.2 (25.4) | 22.0 (36.2) | 21.6 (32.4) | 22.5 (32.6) | 22.9 (30.3) |
| Bond length (Å) | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 |
| Bond angles (°) | 1.106 | 1.085 | 1.050 | 1.078 | 1.112 |
| PDB code | 4DTJ | 4DTM | 4DTN | 4DTO | 4DTP |
Statistics for the highest resolution shell are in parenthesis.
Rmerge = ΣhklΣj|Ij(hkl)−<Ij(hkl)>|/Σ hklΣj <I(hkl)>, statistics for merging all observations for given reflections.
R = Σhkl|Fobs(hkl)−Fcalc(hkl)|/Σ hkl Fobs(hkl), statistics for crystallographic agreement between the measured and model-calculated amplitudes. Rfree is the agreement for cross-validation data set.
Root mean square deviations (rmsd) to ideal values.
PDB accession numbers
Coordinates and structure factors for the THF-containing tm RB69pol ternary complex structure have been deposited in the Protein Data Bank under accession code 4DTR, 4DTS, 4DTU, 4DTX, 4DTJ, 4DTM, 4DTN, 4DTO, and 4DTP.
RESULTS AND DISCUSSION
Kinetic Parameters for Incorporation of dNMPs Opposite THF with dC/dG at the PBP by the wt and tm RB69pols
We have determined pre-steady-state kinetic parameters for incorporation of each of the four dNMPs opposite an abasic (THF) site with dC/dG at the PBP catalyzed by the wt and tm RB69pols. The order of incorporation efficiencies (kpol/Kd) catalyzed by wt RB69pols, shown in Table 3, is consistent with the generally accepted A-rule namely that DNA polymerases preferentially insert dAMP opposite an abasic site. Similarly, the order of kpol/Kd values exhibited by the tm RB69pol follows the same pattern as seen with wt RB69pol but the differences in incorporation among the incoming dNTPs is greatly decreased (Table 3). There is a 280-fold difference in incorporation efficiency between incoming dATP compared to dCTP when opposite THF with wt RB69pol. In contrast, only an 18-fold difference in incorporation efficiency was observed between dAMP and dCMP opposite THF with the tm RB69pol (Table 3). Clearly, the tm RB69pol reduced the A-rule selection gap by fifteen fold relative to wt RB69pol. This change appears to be due to elimination of the rigid hydrogen bonding network present in the NBP of wt RB69pol so that the tm RB69pol has increased flexibility in the NBP.(38, 43) In addition, it is interesting that the tm RB69pol incorporates dAMP and dGMP opposite an abasic site with almost equal efficiency, deviating from the classic A rule.
Table 3.
Pre-Steady-State Kinetic Parameters for Incorporation of dNTPs Opposite THF by wt and tm RB69pol when the dC/dG is in the PBP.a
| pol | dNTP/THF | kpol (s−1) | Kd (μM) | kpol/Kd (μM−1 s−1) | Ratiob |
|---|---|---|---|---|---|
| wt | dATP/THF | 0.30 ± 0.01 | 1400 ± 100 | 2.1 × 10−4 | 313 |
| wt | dGTP/THF | 0.17 ± 0.01 | 1800 ± 200 | 9.4 × 10−5 | 140 |
| wt | dTTP/THF | N.A.D | N.A.D | 2.7 × 10−6 | 4 |
| wt | dCTP/THF | N.A.D | N.A.D | 6.7 × 10−7 | 1 |
|
| |||||
| tm | dATP/THF | 360 ± 9 | 250 ± 24 | 1.44 | 18 |
| tm | dGTP/THF | 240 ± 12 | 170 ± 30 | 1.41 | 17 |
| tm | dTTP/THF | 210 ± 14 | 830 ± 140 | 0.25 | 3 |
| tm | dCTP/THF | 150 ± 10 | 1900 ± 250 | 0.08 | 1 |
The corresponding progress curves and plots of kobs vs [dNTP] are shown in Fig S1 and S2.
Incorporation efficiency ratio relative to the lowest value for each enzyme.
N.A.D stands for Not Accurately Determined. The resulting kpol/Kd value is either from an extrapolation of data fitting or the slope of the plot of kobs vs [dNTP].
Kinetic Parameters for Incorporation of dNMPs Opposite THF by the tm and wt RB69pol with various PBs
To determine the effect of different PBs on the incorporation efficiency of an incoming dNTP, we determined the pre-steady-state kinetic parameters for incorporation of each of the four dNTPs opposite an abasic site by tm RB69pol with all 16 possible PB combinations (Table 4). We were surprised to find that there was nearly a 210 fold difference in incorporation efficiency depending on the identities of the incoming dNTP and the PB. The highest incorporation efficiency (1.47 μM−1s−1) was observed when the PB was dC/dG and the incoming dNTP was dATP. The lowest incorporation efficiency (0.007 μM−1s−1) occured when the PB was dA/dT and the incoming dNTP was dCTP. These results suggest that there are specific interactions between the base of incoming dNTPs and the PB. The influence of PB’s identity on incorporation efficiency was not known or obvious from previous kinetic and structural data when incoming dNTPs were opposite complementary templating nucleotide residues as no significant differences were observed with either the tm RB69pol or the wt enzyme.(32)
Table 4.
Pre-Steady-State Kinetic Parameters for Incorporation of dNTPs Opposite THF by the tm RB69pol with Different Penultimate Base-pairs.a
| Penultimate BPb | Incoming dNTP | kpol (s−1) | Kd (μM) | kpol/Kd (μM−1s−1) | Ratioc |
|---|---|---|---|---|---|
| dG/dC | dCTP | 170 ± 5 | 590 ± 50 | 0.29 | 41 |
| dC/dG | 150 ± 10 | 1900 ± 250 | 0.08 | 11 | |
| dT/dA | N.A.D | N.A.D | 0.015 | 2 | |
| dA/dT | N.A.D | N.A.D | 0.007 | 1 | |
|
| |||||
| dC/dG | dTTP | 210 ± 14 | 830 ± 140 | 0.25 | 31 |
| dG/dC | N.A.D | N.A.D | 0.04 | 5 | |
| dA/dT | N.A.D | N.A.D | 0.01 | 1 | |
| dT/dA | N.A.D | N.A.D | 0.008 | 1 | |
|
| |||||
| dC/dG | dATP | 360 ± 9 | 250 ± 24 | 1.44 | 11 |
| dG/dC | 120 ± 2 | 150 ± 13 | 0.80 | 6 | |
| dT/dA | 130 ± 7 | 380 ± 80 | 0.34 | 3 | |
| dA/dT | 150 ± 6 | 1100 ± 130 | 0.13 | 1 | |
|
| |||||
| dC/dG | dGTP | 240 ± 12 | 170 ± 30 | 1.41 | 35 |
| dG/dC | 230 ± 6 | 660 ± 60 | 0.35 | 9 | |
| dT/dA | 120 ± 7 | 1100 ± 210 | 0.10 | 3 | |
| dA/dT | N.A.D | N.A.D | 0.04 | 1 | |
N.A.D stands for Not Accurately Determined. The resulting kpol/Kd value is either from an extrapolation of data fitting or the slope of the plot of kobs vs [dNTP]. The corresponding progress curves and plots of kobs vs [dNTP] are shown in Fig S3 and S4.
With the penultimate BP, the base in the primer strand is shown on the left, and the base in the template strand is shown on the right.
Incorporation efficiency ratio relative to the lowest value for each dNTP.
For a given incoming dNTP, the difference in incorporation efficiency varies from 11 fold (when the incoming dNTP is dATP) to 41 fold (when the incoming dNTP is dCTP) (Table 4). From these results it is clear that interactions other than base stacking between the incoming dNTP and the PB must be involved. Analysis of these kinetic variations revealed the following pattern: (i) for a given PB, dAMP is always incorporated more efficiently than the other three dNMPs (the A-rule); (ii) for a given incoming dNTP, the incorporation efficiency when the PB is dC/dG or dG/dC is higher than when the PB is dA/dT or dT/dA. This is consistent with the fact that there is more “breathing” in duplex DNA that has A/T rich sequences at the P/T junction; (iii) for purine dNTPs (dPuNTPs), the incorporation efficiency is the highest when the PB is dC/dG, and is lowest when the PB is dA/dT (Table 4).
To see if the same pattern would hold for wt RB69pol, we determined corresponding kinetic parameters for incorporation of dCTP and dTTP opposite an abasic site with 8 possible PB combinations. When dTTP is the incoming dNTP, the incorporation efficiency is the highest when the PB is dC/dG, and is lowest when the PB is dT/dA (Table 5). This trend is consistent with that found with the tm RB69pol. However when dCTP is the incoming dNTP, the order of incorporation efficiency with 4 possible PB combinations follows a different pattern compared to that observed with the tm RB69pol (Table 4 and Table 5). It is important to note that the ratio of differences between the highest and lowest incorporation efficiency for each dNTP by wt enzyme is 3 to 6 fold less than that found with the mutant. Therefore, certain interactions between the incoming dNTP and the PB must be amplified in the presence of the tm RB69pol.
Table 5.
Pre-Steady-State Kinetic Parameters for Incorporation of dNTPs Opposite THF by the wt RB69pol with Different Penultimate Base-pairs.
| Penultimate BPb | Incoming dNTP | kpol/Kd (μM−1 s−1)a | Ratiob |
|---|---|---|---|
| dC/dG | dCTP | 6.7 × 10−7 | 7 |
| dA/dT | 3.6 × 10−7 | 4 | |
| dG/dC | 3.3 × 10−7 | 3 | |
| dT/dA | 1.0 × 10−7 | 1 | |
|
| |||
| dC/dG | dTTP | 2.7 × 10−6 | 9 |
| dG/dC | 1.2 × 10−6 | 4 | |
| dA/dT | 1.2 × 10−6 | 4 | |
| dT/dA | 3.3 × 10−7 | 1 | |
The kpol/Kd value was calculated by using kpol/[dNTP]sub, with [dNTP] = 5mM. The kpol/Kd values for dC/dG with incoming dCTP and dTTP were taken from Table 1.
Incorporation efficiency ratio relative to the lowest value for each dNTP.
Kinetic Parameters for Incorporation of dAMP Opposite dT when Bypassing a dN/THF Lesion
Unlike other DNA pols, wt RB69pol inserts dGMP opposite an abasic site (THF) almost as efficiently as dAMP with only a 2 fold difference which is insignificant with respect to the A-rule selection. With the tm RB69pol, dAMP and dGMP are incorporated opposite an abasic site with equal efficiency (violating the classic A rule, Table 3). To further investigate the extent to which the tm RB69pol was more permissive than wt RB69pol, we determined the pre-steady-state kinetics parameters for incorporation of dAMP opposite dT when bypassing a dN/THF lesion catalyzed by the tm RB69pol (Table 6). Interestingly, the tm RB69pol was able to bypass a dA/THF lesion 14 fold more efficiently than a dG/THF site, 49 fold more efficiently than a dT/THF site, and almost 100 fold more efficiently than a dC/THF lesion. As shown in Table 6, the same trend also obtained for wt RB69pol and Phi29pol, another B family pol. This trend is more important for burying the abasic site to allow for uninterrupted DNA replication. Based on these results we propose that the A-rule can be extended, namely that B family polymerases can bypass dA/THF sites much more efficiently than other dN/THF lesions.
Table 6.
Pre-Steady-State Kinetic Parameters for Incorporation of dATP Opposite dT bypassing a dN/THF Lesion by the wt and tm RB69pol.
| pol | Bypass dN/THF | kpol/Kd (μM−1 s−1)a | Ratiob |
|---|---|---|---|
| RB69pol tm | dA/THF | 7.9 × 10−5 | 98 |
| RB69pol tm | dG/THF | 6.4 × 10−6 | 8 |
| RB69pol tm | dT/THF | 1.8 × 10−6 | 2 |
| RB69pol tm | dC/THF | 8.0 × 10−7 | 1 |
|
| |||
| RB69pol wt | dA/THF | 1.0 × 10−6 | 24 |
| RB69pol wt | dG/THF | 5.6 × 10−8 | 1 |
| RB69pol wt | dC/THF | 4.8 × 10−8 | 1 |
| RB69pol wt | dT/THF | 4.2 × 10−8 | 1 |
|
| |||
| Phi29pol | dA/THF | 9.0 × 10−7 | 45 |
| Phi29pol | dG/THF | 4.4 × 10−8 | 2 |
| Phi29pol | dC/THF | 2.7 × 10−8 | 1 |
| Phi29pol | dT/THF | 2.0 × 10−8 | 1 |
The kpol/Kd values for RB69pol wt and Phi29pol were calculated by using kpol/[dNTP]sub, with [dNTP] = 5mM. The corresponding progress curves and plots of kobs vs [dNTP] by RB69pol tm are shown in Fig S5.
Incorporation efficiency ratio relative to the lowest value for each enzyme.
Overview of Crystal Structures of tm RB69pol Ternary Complexes Containing Each of the Four dNTPs Opposite an Abasic Site
In our initial structural studies, a standard P/T was used where the PB was ddC/dG.(44) With this P/T, we have obtained four crystal structures of tm RB69pol ternary complexes containing an abasic site opposite each of the four incoming dNTPs at resolutions varying from 1.84 to 2.04 Å (Table 1). These structures have been refined with cross-validation R-factors varying from 20.2 to 22.8%. Following kinetic studies with different PBs, we have determined five additional structures of tm RB69pol complexes containing an abasic site in the DNA substrate, again opposite each of the four incoming dNTPs. These five new ternary complexes were chosen to represent ones displaying the highest and lowest incorporation efficiencies for each incoming dNTP and were determined at resolutions varying from 1.9 to 2.0 Å with free R factors varying from 20.2 to 22.9% (Table 2). The overall structures of all nine ternary complexes containing an abasic site are identical to the dCTP/dG-containing wt RB69pol ternary complex. The root-mean-squares Cα deviation between the highest resolution ternary complex structure of the tm RB69pol (containing dTTP/THF, 1.8 Å) and the wt RB69pol (containing dCTP/dG, 1.8 Å) is 0.12 Å, suggested that the three amino acid substitutions in the NBP of tm RB69pol did not alter the overall structure. In all cases, the electron densities for P/Ts containing a templating THF, the incoming dNTP and the surrounding network of ordered water molecules were well-defined (Fig. 2).
Figure 2.
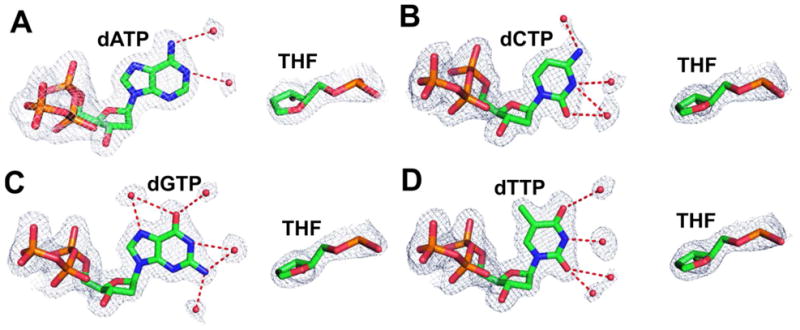
Structures of the dNTP/THF nascent base-pairs in the ternary complexes of tm RB69pol with ddC/dG at the PBP superimposed onto the final 2Fo-Fc electron density maps contoured at 1.2 σ. (A) The dATP/THF structure at 2.04-Å resolution. (B) The dCTP/THF structure at 1.96-Å resolution. (C) The dGTP/THF structure at 1.87-Å resolution. (D) Final The dTTP/THF structure at 1.84-Å resolution.
Structural Basis for the Differences in Incorporation Efficiences of Purine Versus Pyrimidine dNMPs Opposite an Abasic Site
Our kinetic data have shown that the catalytic efficiency for incorporating purine dNTPs (dPuNTP) opposite an abasic site by tm RB69pol are seven to eighteen fold higher than that of pyrimidine dNTPs (dPyNTP). In addition, the tm RB69pol incorporates dGMP as efficiently as dAMP opposite an abasic site. These kinetic observations are consistent with the corresponding structures. Thus, in all four dNTP/THF ternary complexes, with ddC/dG at the PBP, the base of the incoming dNTP is completely stacked onto the PB. Superposition of the Cα coordinates of all four dNTP/THF structures shows that the triphosphate tail and the ribosyl moiety of the dNTPs interact with the pol in an identical manner (Fig. 3A). Because dPuTPs are able to stack better against PBs in ternary complexes than dPyTPs, this could be one reason why dPuTPs are incorporated more efficiently than dPyTPs opposite an abasic site. Among the two dPuTPs, the base of dATP overlaps almost completely with that of dGTP when the two complexes are superimposed (Fig. 3A). Both dPuTPs stack equally well against the PB and could account for the fact that the tm RB69pol incorporates both dPuTPs with equal efficiency. Among the two dPyTPs, the base of dTTP is tilted more toward its triphosphate tail compared to dCTP (Fig. 3B). The interatomic distance between the C6 and O5′ atoms of the incoming dTTP is 3.27 Å, and the corresponding distance for the incoming dCTP is 3.54 Å. As a result, the base of dTTP stacks right on top of ddC which is the 3′ terminal base in the primer strand of the PB, and the partial negatively charged O4 of dTTP lies exactly over the partial positively charged N4 of ddC (Fig. 3C). In contrast, the base of dCTP stacks on top of the surface of the ddC/dG pair near its base-pairing interface at the primer-template junction, and N4 of dCTP is equidistant from N4 of ddC and O6 of dG (Fig. 3D). Therefore, better π-π stacking between the dTTP and ddC, together with interactions of the complementary partial charges between O4 of dTTP and N4 of ddC could explain why the tm RB69pol incorporates dTTP three fold more efficiently than dCTP when opposite an abasic site.
Figure 3.
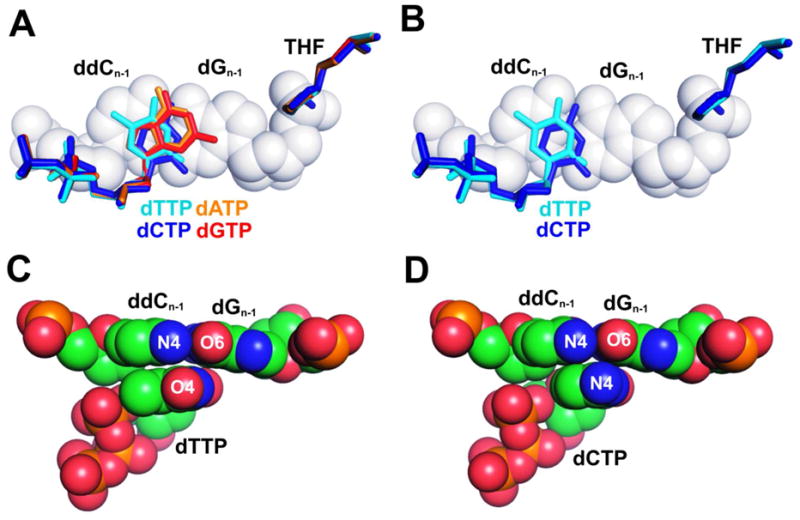
Geometry of base stacking and partial charge interactions with fixed PB’s dCTP/dG. (A) Superposition of tm RB69pol Cα coordinates of the four complexes (dATP/THF, orange; dCTP/THF, blue; dGTP/THF, red; and dTTP/THF, cyan). The PB’s ddC/dG was shown in space filling model. (B) Superposition of tm RB69pol Cα coordinates of the two pyrimidine/THF-containing complexes (dCTP/THF, blue and dTTP/THF, cyan). (C) dTTP and the PB’s ddC/dG in the dTTP/THF-containing tm RB69pol. (D) dCTP and the PB’s ddC/dG in the dCTP/THF-containing tm RB69pol.
Due to the smaller stacking surface of pyrimidines compared to purines, we suspect that the partial charge interactions between base of the incoming dNTP and the base in the primer strand at the PBP would have a more significant effect on the insertion efficiency of dPyTPs than dPuTPs. This speculation is consistent with what we have observed in structures with different PBs. The incorporation efficiency of dCMP opposite an abasic site is highest when the PB is dG/dC (0.29μM−1s−1), and lowest when the PB is dA/dT (0.007μM−1s−1). As shown in Fig 4A, the partial positively charged N4 of dCTP lies on top of the partial negatively charged O6 of ddG, and the partial negatively charged O2 of dCTP is situated right over the partial positively charged N2 of ddG. The positive and negative partial charge interactions between these four atoms help to stabilize the ternary complex. In contrast, the partial positively charged N4 of dCTP is positioned just adjacent to the partial positively charged N6 of ddA in the dCTP-containing structure with ddA/dT at the PBP (Fig. 4B). The resulting repulsive interactions would likely destabilize the ternary complex. Therefore, the differences in partial charge interactions between dCTP and the PBs are likely to account for the 41 fold difference in the catalytic efficiencies for dCMP incorporation in these two contexts. The same phenomena were observed when the incoming dNTP was dTTP. The incorporation efficiency of dTMP opposite an abasic site is highest when the PB is dC/dG (0.26μM−1s−1), and is lowest when the PB is dT/dA (0.013μM−1s−1) (Table 4). In the ternary structure with ddC/dG at the PBP, the partial negatively charged O4 and O2 of dTTP stack neatly on the surface of the partial positively charged N4 of ddC and N2 of dG respectively (Fig. 4C). In contrast, the partial negatively charged O4 of dTTP stacks right over the partial negatively charged O4 of ddT in the structure where ddT/dA is at the PBP (Fig. 4D).
Figure 4.
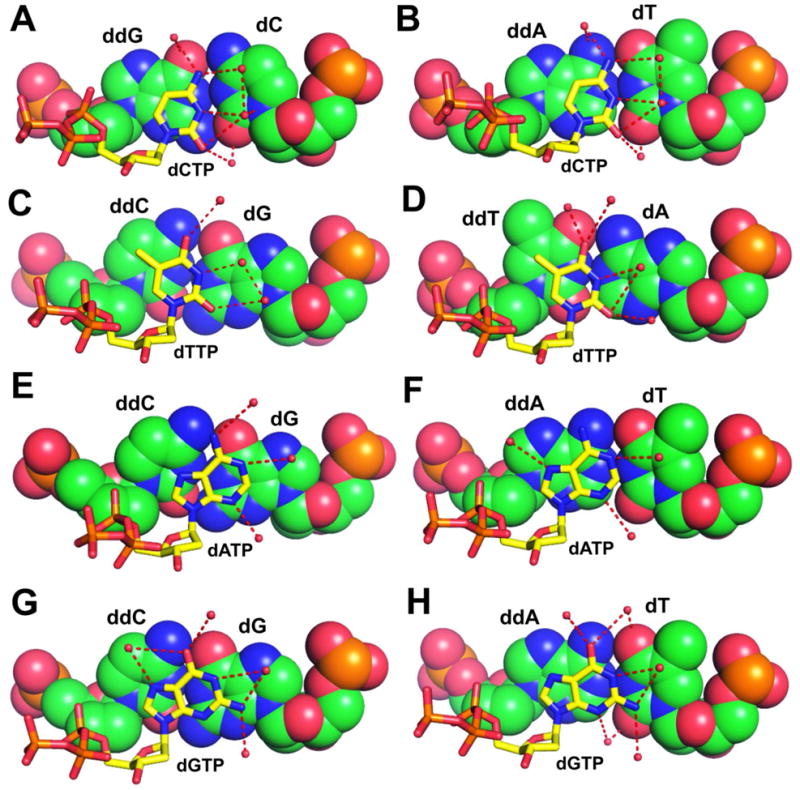
Base-stacking of the incoming dNTP with different PBs. The incoming dNTP is shown in thick stick, and the PB is shown with a space filling model. (A) dCTP with ddG/dC. (B) dCTP with ddA/dT. (C) dTTP with ddA/dT. (D) dTTP with ddT/dA. (E) dATP with ddC/dG. (F) dATP with ddA/dT. (G) dGTP with ddC/dG. (H) dGTP with ddA/dT.
When the incoming dNTP is a dPuTP, its incorporation efficiency opposite an abasic site is highest when the PB is ddC/dG, and lowest when the PB is dA/dT. As shown in Fig 4E and Fig 4G, both the N6 of dATP and the O6 of dGTP are positioned just over the interface of the ddC/dG pair at the primer-template junction. When the PB is ddA/dT, the partial positively charged N6 of dATP stacks on the surface of the partial positively charged N6 of ddA (Fig. 4F), and the partial negative charged O6 of dGTP stacks on top of the partial positively charged N6 of ddA (Fig. 4H). Therefore, partial charge interactions between the dNTPs and the PBs are not likely to be the major factors influencing the incoporation efficiencies of dPuTPs opposite an abasic site. Upon examination of all the THF-containing structures, a common feature is that the bases of the incoming dNTPs remain partially hydrated in the NBP (Figs. 2 and 4). Nearly all HB donors and acceptors of dNTPs have at least one ordered water molecule within HB distance. These ordered water molecules are located on the PB surface as part of an ordered hydration network. Thus, without a templating base, the incoming dNTP failed to dehydrate its W-C base-pairing face when forming a ternary complex relative to the ternary complex with a W-C templating base. When the PB is ddA/dT and the incoming dNTP is a dPuTP, the ordered water molecules are located just over the C atoms of the PB. Such hydration of incoming dNTPs would be expected to destabilize the ternary complex. In contrast, when the PB is ddC/dG, the ordered water molecules are located very close to the N atoms of the dG base. Given this proximity, these ordered water molecules would be expected to help stabilize the ternary complex. Therefore, the extent of hydration around the incoming base and PBP is likely to be a key determinant for the observed kinetic variations with respect to incoming dPuTPs (Table 3).
We haven’t been able to obtain crystals of the ternary complex of wt RB69pol with THF at the n position of the template strand due to the high apparent dissociation constant. Our previous structural studies with the dGTP/dC-containing wt RB69pol have shown that there is a rigid HB network at the minor groove of the nascent P/T duplex. This network includes five ordered water molecules and the side chain hydroxyl groups of Y567, Y416, and T622. Also, Y567 forms a HB with the O2 or N3 atom of PB at the template strand via a water molecule. In contrast, the Y567 to Ala substitution in the tm RB69pol disrupted this rigid HB network and the space generated by removing the hydroxyphenyl group of Y567 is occupied by additional water molecules which are no longer within HB distance to the O2 or N3 atom of PB. Therefore, the charge distribution around the exocyclic groups of the PB might be slightly different between the wt pol and tm pol. Together with the flexibility of NBP in the tm RB69pol, the partial charge interactions between the PB and the incoming dNTP might somehow be amplified in the context of the tm pol. That might be the reason that the difference in incorporation efficiency with different PBs for each dNTP by wt enzyme is 3 to 6 fold less than that of the mutant, although we cannot rule out other possibilities without the structure of dNTP/THF-containing ternary complex of wt RB69pol.
Structural Comparison of THF-Containing Ternary Complexes with DNA Polymerases from Other Families
Studies of replication past abasic lesions in DNA have been conducted extensively with DNA polymerases from several families and it is clear that different mechanisms are used by the various pols to bypass abasic sites.(16, 22–23, 26, 45–46) In addition to the structures reported in this study, structures of DNA polymerases from A, B, X and Y families bound to THF-containing duplex DNA are available.(22)
As a representative of A family polymerases, KlenTaq, the large fragment of T. aquaticus DNA polymerase I, follows the A-rule by utilizing an amino acid templating mechanism. As shown in Fig 5A, Y671, a highly conserved residue in the A family DNA pols, forms a hydrogen bond with N3 of the incoming dATP. This interaction facilitates nucleotide incorporation by mimicking a pyrimidine nucleobase for directing dAMP incorporation opposite an abasic site.(23) R283 of pol β, a family X polymerase, is another example of where an amino acid residue directs the incorporation of dAMP opposite THF.(8) In this case, R283 was observed to make a hydrogen bond with the THF phosphate backbone (Fig. 5B). It was proposed that misinsertion may occur through a similar “abasic site intermediate”. In contrast to KlenTaq and pol β, Dpo4, a family Y polymerase, can efficiently replicate DNA past an abasic lesion by looping out the abasic site.(47) As shown in Fig 5C, the abasic site remains extrahelical and the incoming dCTP pairs with a base 5′ to the lesion. Primer extension then generates a minus 1 frameshift.(47) E. coli DNA Pol II, a family B polymerase, is another DNA pol that employs a looping-out mechanism. A small cavity outside the NBP of Pol II can accommodate a looped-out template 2 bp upstream. As a result, the incoming dATP base-pairs with dT at the N+3 position of the template strand (Fig. 5D).(46) By contrast, in all nine structures of THF-containing ternary complexes of tm RB69pol reported here, neither insertion of a protein side chain nor looping-out of the abasic site was observed. Rather, the abasic lesion in all nine ternary complexes of tm RB69pol was locked in place by two hydrogen bonds: one between N572 and a phosphate oxygen on the 3′ side of the lesion, and the other between the backbone amide hydrogen of I362 and the phosphate oxygen of the lesion (Fig. 5E). These two hydrogen bond interactions are probably why the abasic lesion does not loop out in RB69pol although the overall structure of RB69pol is quite similar to that of Pol II.
Figure 5.
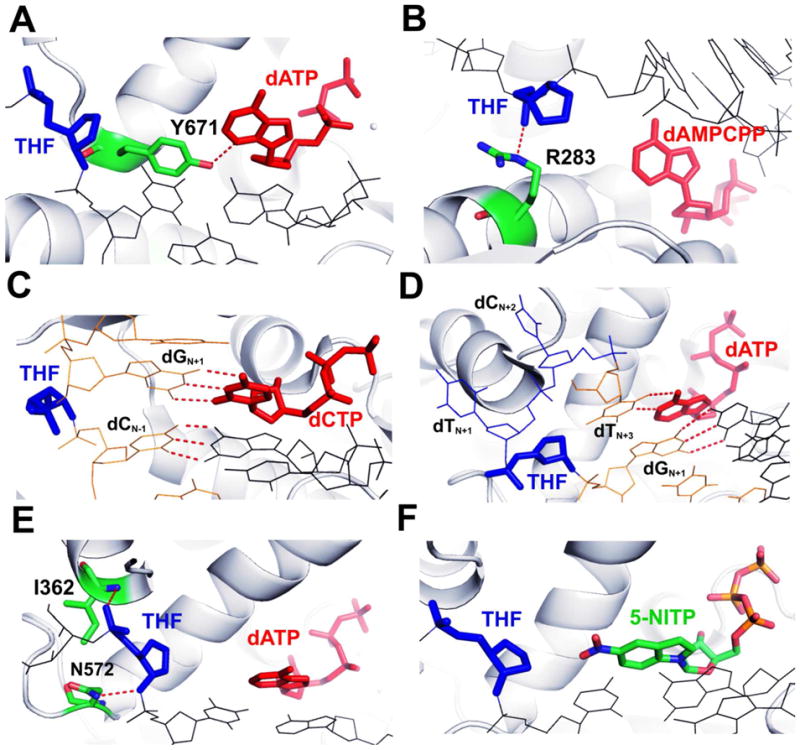
Structural comparison with other known THF-containing ternary complexes. (A) The NBP of the dATP/THF-containing ternary complex of KlenTaq. (B) The NBP of the dAMPCPP/THF-containing ternary complex of pol β. (C) The NBP of the dCTP/THF-containing ternary complex of Dpo4. (D) The NBP of the dATP/THF-containing ternary complex of PolII. (E) The NBP of the dATP/THF-containing ternary complex of tm RB69pol. (F) The NBP of the 5-NITP/THF-containing ternary complex of wt RB69pol. The incoming dNTP is shown in red, the THF moiety is shown in blue, and the protein residues are shown in green. The polymerase is shown in grey cartoon.
Zahn, et al,(27) have reported a structure of wt RB69pol ternary complex containing an abasic site paired with an unnatural purine triphosphate analogue, 5-NITP (Fig. 5F). A rigid hydrogen bonding network involving the minor groove of the P/T duplex and Y567 was also observed in the 5-NITP-containing structure (Fig. 6A). Least squares superimposition of the 5-NITP-containing ternary complex of wt RB69pol with the dATP/THF-containing ternary complex of tm RB69pol shows that: (i) the bases of 5-NITP and dATP are in the same plane, but the indole ring of 5-NITP tilts more toward the minor groove to maximize the stacking interaction with the ddG/dC at the PBP (Fig. 6B); (ii) the A567 and G568 main-chain amide linkage shifts 0.7 Å laterally toward the Y416 side chain (Fig. 6B); (iii) the W1 water molecule was absent in tm RB69pol complex (Fig. 6A), and the rigid hydrogen bonding network was disrupted. This is consistent with the increased incorporation efficiency for all the dNMPs opposite an abasic site, 104 to 105 fold exhibited by tm RB69pol relative to wt. Interestingly, a partial charge interaction between the negatively charged O5 of 5-NTIP and the positively charged N4 of dC was observed (Fig. 6C). According to the explanation of Zahn et al,27 this partial charge interaction was probably why 5-NITP is incorporated more efficiently than dAMP opposite an abasic site, and is consistent with our observation of tm RB69pol that partial charge interactions between the incoming dNTPs and the PB play a dominant role in determining the value of the corresponding kinetic parameters.
Figure 6.
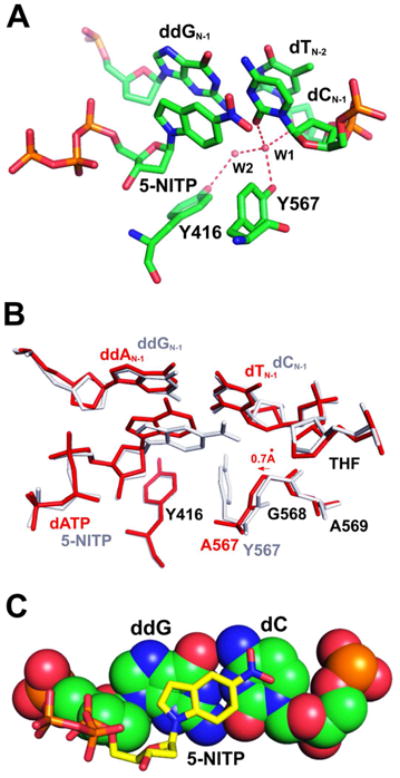
Structural comparison of the THF-containing ternary complexes of wt and tm RB69pols. (A) The HB network in the NBP of the 5-NITP/THF-containing ternary complex of wt RB69pol. (B) Superposition of Cα coordinates of the dATP/THF-containing ternary complex of tm RB69pol (shown in red stick) with the 5-NITP/THF-containing ternary complex of wt RB69pol (shown in white stick). (C) Base-stacking of the incoming 5-NITP (shown in yellow) with the PB (shown with a space filling model).
To summarize, we have shown that, in addition to base stacking, partial charge interactions between the incoming dNTP and the penultimate base-pair make significant contributions to the incorporation efficiencies of dNTPs opposite abasic sites in the context of the tm RB69pol. Although this trend might be specific to tm RB69pol, it provides the first example that partial charge interactions between the PB and incoming dNTP can modulate insertion efficiency. In addition, we also propose an extension of the A rule namely that B family polymerases can bypass a dA/THF site much more efficiently than any other dN/THF pair. Taken together the results presented here add another dimension to the extensive literature on the structural basis for bypassing abasic sites by DNA pols.
Supplementary Material
Acknowledgments
This work was supported by NIH RO1-GM063276-09 (to W.H.K.) and by SCSB-GIST (to S.H.E & J.W.).
We thank the staff of the NE-CAT beamline 24-ID-E at the Advanced Photon Source of Argonne National Laboratory.
ABBREVIATIONS
- pol
DNA polymerase
- RB69pol
bacteriophage RB69 DNA polymerase
- THF
tetrahydrofuran, an abasic nucleotide residue
- NBP
Nascent basepair-Binding Pocket
- P/T
primer/template DNA duplex
- HB
hydrogen bond
- wt
wild type
- tm
triple mutant harboring L561A/S565G/Y567A substitutions
- dNTPs
2′-deoxynucleoside 5′-triphosphates
- dNMPs
2′-deoxynucleoside 5′-mono-phosphate
- dPuTPs
purine dNTPs
- dPyTPs
pyrimidine dNTPs
- PEG 350 MME
polyethylene glycol 350 monomethyl ether
- 5-NITP
5-nitro-1-indolyl; 2′-deoxyriboside-5′-triphosphate
- PB
Penultimate Base-pair
- PBP
Penultimate Base-pair Position
Footnotes
Additional five figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura J, Walker VE, Upton PB, Chiang SY, Kow YW, Swenberg JA. Highly sensitive apurinic/apyrimidinic site assay can detect spontaneous and chemically induced depurination under physiological conditions. Cancer Res. 1998;58:222–225. [PubMed] [Google Scholar]
- 3.Nakamura J, Swenberg JA. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999;59:2522–2526. [PubMed] [Google Scholar]
- 4.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 5.Seeberg E, Eide L, Bjoras M. The base excision repair pathway. Trends Biochem Sci. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 6.Hatahet Z, Zhou M, Reha-Krantz LJ, Ide H, Morrical SW, Wallace SS. In vitro selection of sequence contexts which enhance bypass of abasic sites and tetrahydrofuran by T4 DNA polymerase holoenzyme. J Mol Biol. 1999;286:1045–1057. doi: 10.1006/jmbi.1998.2520. [DOI] [PubMed] [Google Scholar]
- 7.Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 8.Beard WA, Shock DD, Batra VK, Pedersen LC, Wilson SH. DNA polymerase beta substrate specificity: side chain modulation of the “A-rule”. J Biol Chem. 2009;284:31680–31689. doi: 10.1074/jbc.M109.029843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 10.Boiteux S, Laval J. Coding properties of poly(deoxycytidylic acid) templates containing uracil or apyrimidinic sites: in vitro modulation of mutagenesis by deoxyribonucleic acid repair enzymes. Biochemistry. 1982;21:6746–6751. doi: 10.1021/bi00269a020. [DOI] [PubMed] [Google Scholar]
- 11.Sagher D, Strauss B. Insertion of nucleotides opposite apurinic/apyrimidinic sites in deoxyribonucleic acid during in vitro synthesis: uniqueness of adenine nucleotides. Biochemistry. 1983;22:4518–4526. doi: 10.1021/bi00288a026. [DOI] [PubMed] [Google Scholar]
- 12.Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 13.Randall SK, Eritja R, Kaplan BE, Petruska J, Goodman MF. Nucleotide insertion kinetics opposite abasic lesions in DNA. J Biol Chem. 1987;262:6864–6870. [PubMed] [Google Scholar]
- 14.Lawrence CW, Borden A, Banerjee SK, LeClerc JE. Mutation frequency and spectrum resulting from a single abasic site in a single-stranded vector. Nucleic Acids Res. 1990;18:2153–2157. doi: 10.1093/nar/18.8.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mozzherin DJ, Shibutani S, Tan CK, Downey KM, Fisher PA. Proliferating cell nuclear antigen promotes DNA synthesis past template lesions by mammalian DNA polymerase delta. Proc Natl Acad Sci U S A. 1997;94:6126–6131. doi: 10.1073/pnas.94.12.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shibutani S, Takeshita M, Grollman AP. Translesional synthesis on DNA templates containing a single abasic site. A mechanistic study of the “A rule”. J Biol Chem. 1997;272:13916–13922. doi: 10.1074/jbc.272.21.13916. [DOI] [PubMed] [Google Scholar]
- 17.Avkin S, Adar S, Blander G, Livneh Z. Quantitative measurement of translesion replication in human cells: evidence for bypass of abasic sites by a replicative DNA polymerase. Proc Natl Acad Sci U S A. 2002;99:3764–3769. doi: 10.1073/pnas.062038699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strauss BS. The “A” rule revisited: polymerases as determinants of mutational specificity. DNA Repair (Amst) 2002;1:125–135. doi: 10.1016/s1568-7864(01)00014-3. [DOI] [PubMed] [Google Scholar]
- 19.Pages V, Johnson RE, Prakash L, Prakash S. Mutational specificity and genetic control of replicative bypass of an abasic site in yeast. Proc Natl Acad Sci U S A. 2008;105:1170–1175. doi: 10.1073/pnas.0711227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaaper RM, Kunkel TA, Loeb LA. Infidelity of DNA synthesis associated with bypass of apurinic sites. Proc Natl Acad Sci U S A. 1983;80:487–491. doi: 10.1073/pnas.80.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor JS. New structural and mechanistic insight into the A-rule and the instructional and non-instructional behavior of DNA photoproducts and other lesions. Mutat Res. 2002;510:55–70. doi: 10.1016/s0027-5107(02)00252-x. [DOI] [PubMed] [Google Scholar]
- 22.Zahn KE, Wallace SS, Doublie S. DNA polymerases provide a canon of strategies for translesion synthesis past oxidatively generated lesions. Curr Opin Struct Biol. 2011;21:358–369. doi: 10.1016/j.sbi.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fiala KA, Hypes CD, Suo Z. Mechanism of abasic lesion bypass catalyzed by a Y-family DNA polymerase. J Biol Chem. 2007;282:8188–8198. doi: 10.1074/jbc.M610718200. [DOI] [PubMed] [Google Scholar]
- 25.Obeid S, Blatter N, Kranaster R, Schnur A, Diederichs K, Welte W, Marx A. Replication through an abasic DNA lesion: structural basis for adenine selectivity. EMBO J. 2010;29:1738–1747. doi: 10.1038/emboj.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reineks EZ, Berdis AJ. Evaluating the contribution of base stacking during translesion DNA replication. Biochemistry. 2004;43:393–404. doi: 10.1021/bi034948s. [DOI] [PubMed] [Google Scholar]
- 27.Zahn KE, Belrhali H, Wallace SS, Doublie S. Caught bending the A-rule: crystal structures of translesion DNA synthesis with a non-natural nucleotide. Biochemistry. 2007;46:10551–10561. doi: 10.1021/bi7008807. [DOI] [PubMed] [Google Scholar]
- 28.Greenberg MM, Weledji YN, Kroeger KM, Kim J. In vitro replication and repair of DNA containing a C2′-oxidized abasic site. Biochemistry. 2004;43:15217–15222. doi: 10.1021/bi048360c. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, Rhee C, Bebenek A, Drake JW, Wang J, Konigsberg W. The L561A substitution in the nascent base-pair binding pocket of RB69 DNA polymerase reduces base discrimination. Biochemistry. 2006;45:2211–2220. doi: 10.1021/bi052099y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheriff A, Motea E, Lee I, Berdis AJ. Mechanism and dynamics of translesion DNA synthesis catalyzed by the Escherichia coli Klenow fragment. Biochemistry. 2008;47:8527–8537. doi: 10.1021/bi800324r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabouri N, Johansson E. Translesion synthesis of abasic sites by yeast DNA polymerase epsilon. J Biol Chem. 2009;284:31555–31563. doi: 10.1074/jbc.M109.043927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, Beckman J, Wang J, Konigsberg W. RB69 DNA polymerase mutants with expanded nascent base-pair-binding pockets are highly efficient but have reduced base selectivity. Biochemistry. 2009;48:6940–6950. doi: 10.1021/bi900422b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berdis AJ. Dynamics of translesion DNA synthesis catalyzed by the bacteriophage T4 exonuclease-deficient DNA polymerase. Biochemistry. 2001;40:7180–7191. doi: 10.1021/bi0101594. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X, Lee I, Berdis AJ. The use of nonnatural nucleotides to probe the contributions of shape complementarity and pi-electron surface area during DNA polymerization. Biochemistry. 2005;44:13101–13110. doi: 10.1021/bi050585f. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, Lee I, Zhou X, Berdis AJ. Hydrophobicity, shape, and pi-electron contributions during translesion DNA synthesis. J Am Chem Soc. 2006;128:143–149. doi: 10.1021/ja0546830. [DOI] [PubMed] [Google Scholar]
- 36.Wang M, Xia S, Blaha G, Steitz TA, Konigsberg WH, Wang J. Insights into base selectivity from the 1.8 A resolution structure of an RB69 DNA polymerase ternary complex. Biochemistry. 2011;50:581–590. doi: 10.1021/bi101192f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berman AJ, Kamtekar S, Goodman JL, Lazaro JM, de Vega M, Blanco L, Salas M, Steitz TA. Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases. EMBO J. 2007;26:3494–3505. doi: 10.1038/sj.emboj.7601780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia S, Eom SH, Konigsberg WH, Wang J. Structural Basis for Differential Insertion Kinetics of dNMPs Opposite a Difluorotoluene Nucleotide Residue. Biochemistry. 2012;51:1476–1485. doi: 10.1021/bi2016487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.The PyMOL Molecular Graphics System, Version 1.2r3pre. Schrodinger, LLC; [Google Scholar]
- 43.Xia S, Konigsberg WH, Wang J. Hydrogen-bonding capability of a templating difluorotoluene nucleotide residue in an RB69 DNA polymerase ternary complex. J Am Chem Soc. 2011;133:10003–10005. doi: 10.1021/ja2021735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang M, Xia S, Blaha G, Steitz TA, Konigsberg WH, Wang J. Insight into base selectivity from the 1.8Å resolution structure of an RB69 DNA polymerase ternary complex. Biochemistry. 2010 doi: 10.1021/bi101192f. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. DNA synthesis across an abasic lesion by human DNA polymerase iota. Structure. 2009;17:530–537. doi: 10.1016/j.str.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang F, Yang W. Structural insight into translesion synthesis by DNA Pol II. Cell. 2009;139:1279–1289. doi: 10.1016/j.cell.2009.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ling H, Boudsocq F, Woodgate R, Yang W. Snapshots of replication through an abasic lesion; structural basis for base substitutions and frameshifts. Mol Cell. 2004;13:751–762. doi: 10.1016/s1097-2765(04)00101-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


