Abstract
Gram-negative bacteria are impervious to many drugs and environmental stresses because they possess an outer membrane (OM) containing lipopolysaccharide (LPS). LPS is biosynthesized at the cytoplasmic (inner) membrane (IM) and is transported to the OM by an unknown mechanism involving the LPS transport proteins, LptA-G. These proteins have been proposed to form a bridge between the two membranes; however, it is not known how this bridge is assembled to prevent mistargeting of LPS. We use in vivo photocrosslinking to reveal the specific protein-protein interaction sites that give rise to the Lpt bridge. We also show that the formation of this transenvelope bridge cannot proceed before the correct assembly of the LPS translocon in the OM. This ordered sequence of events may ensure that LPS is never transported to the OM if it cannot be translocated across it to the cell surface.
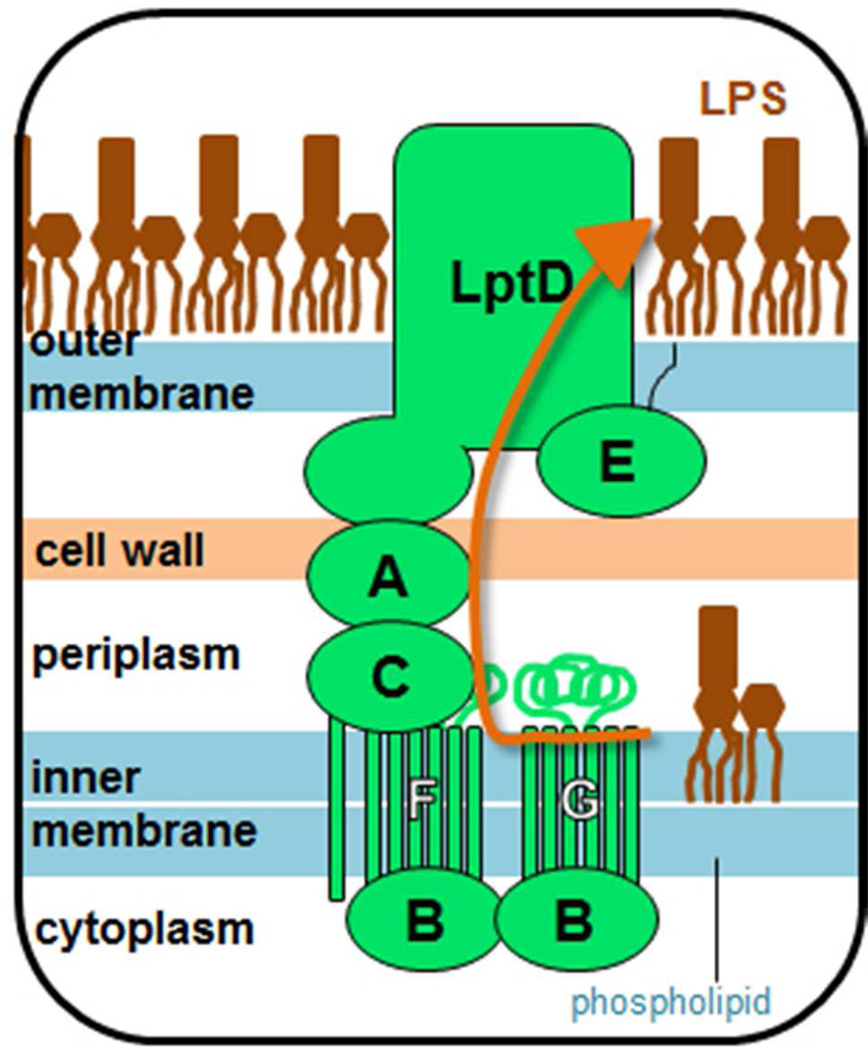
The cell envelope of Gram-negative bacteria is characterized by the presence of an outer membrane (OM) surrounding the cytoplasmic (inner) membrane (IM) and the peptidoglycan cell wall (Fig. 1). The OM is a unique asymmetric bilayer with an inner leaflet consisting of phospholipid and an outer leaflet consisting of lipopolysaccharide (LPS).1–3 Anionic LPS molecules are bridged by divalent cations in vivo to create an impervious layer that blocks the influx of hydrophobic molecules, such as antibiotics, bile salts and detergents.4 LPS is biosynthesized at the cytoplasmic face of the IM5 and then flipped to the periplasmic side by MsbA.6 Subsequently, the molecule is transported across the periplasm and the OM and positioned exclusively at the cell surface.
Fig. 1.
The lipopolysaccharide exporter consists of seven essential proteins, LptA-G, that form a transenvelope complex spanning the inner membrane (IM), periplasm, and outer membrane (OM).
In Escherichia coli, this LPS transport process is known to require the seven essential Lpt proteins,7 but how these proteins move large, amphipathic LPS molecules across both membranes and the periplasmic space is not understood. The Lpt proteins are localized in three distinct compartments of the cell envelope (Fig. 1). In the IM, LptBFG comprise an ATP-binding cassette (ABC) transporter that is proposed to facilitate release of LPS from the IM. LptBFG associate with LptC, an integral IM protein whose function is not well understood.8–11 In the OM, LptD12,13 consists of a large N-terminal periplasmic domain of unknown function and a C-terminal transmembrane β-barrel domain that binds the lipoprotein LptE.14–16 In its functional form, LptD contains two interdomain disulfide bonds whose proper formation requires LptE, but which are not required for the stability of the LptD/E complex.16–18 The LptD/E complex is proposed to serve as the translocon that facilitates the passage of LPS across the OM bilayer.
LPS has been suggested to transit the aqueous periplasmic space via a transenvelope bridge. In support of this hypothesis, LPS transport has been observed in spheroplasts, bacterial cells in which the soluble periplasmic contents have been lost.19 Moreover, all seven Lpt proteins co-fractionate and co-purify, suggesting that they are found in a single complex.20 Depletion of any Lpt protein causes LPS accumulation at the periplasmic face of the IM,9 consistent with a model in which all seven proteins function in a highly cooperative manner to prevent mistargeting of LPS by defective transport machines. It is not known how the Lpt proteins are assembled into a transenvelope structure that ensures proper LPS export.
In this work, we establish the architecture of the transenvelope Lpt bridge in vivo. We define the interaction sites of the periplasmic protein LptA with LptC in the IM and with the N-terminal domain of LptD in the OM. These structurally homologous domains interact in a conserved manner involving the edges of their β-sheets. We also establish how the assembly of this bridge is regulated. We demonstrate that formation of the Lpt bridge requires the presence of correct disulfide bonds in LptD, which itself depends on the proper assembly of the LptD/E translocon in the OM. This regulatory mechanism may prevent mistargeting of LPS by Lpt complexes that are not fully functional.
Experimental Procedures
In vivo photocrosslinking and whole-cell lysate analysis
To construct E. coli strains for in vivo photocrosslinking, specific codons within the lptA, lptC, and lptD sequences were replaced with the TAG codon, and amber suppression was used to incorporate the UV-photocrosslinkable unnatural amino acid para-benzoylphenylalanine (pBPA) at these positions21,22 (see Supporting Information for details of strain construction). Diploid strains were used for all experiments except where otherwise noted. Overnight cultures grown in LB supplemented with pBPA and antibiotics, as appropriate, were diluted 1:100 into 100 mL of the same media and grown to mid-log phase at 37°C. Each culture was split in half, and each sample was pelleted and either used directly or resuspended in 10 mL ice-cold TBS (20 mM Tris pH 8.0, 150 mM NaCl) and irradiated with UV light at 365 nm for 10 min in an LM-26 benchtop transilluminator (UVP). All cells were resuspended in 2.5 mL ice-cold TBS-B (20 mM Tris pH 8.0, 300 mM NaCl, 20 mM imidazole) containing 1% Anzergent 3–14 (Anatrace), 100 µg/mL lysozyme, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 50 µg/mL DNase I, lysed by sonication, and centrifuged at 18,500 × g in a table-top centrifuge for 30 min. Nickel affinity purification was performed as described.15 Eluates were concentrated to a final volume of approximately 40 µL using 10,000 Da cut-off Amicon centrifugal concentrators (Millipore). SDS-PAGE buffer was added to a final volume of 80 µL for SDS-PAGE and immunoblotting. Sample loading was normalized by cell density except where stated otherwise.
In vivo photocrosslinking and total membrane analysis
1.5 L cell cultures were grown, harvested, and UV-irradiated (or not) as described in the previous section. All samples were resuspended in 10 mL ice-cold TBS supplemented with 100 µg/mL lysozyme, 1 mM PMSF, and 50 µg/mL DNase I. Cells were lysed and membranes were collected, extracted, and subjected to TALON affinity chromatography as described.20 The final eluates were concentrated to ~150 µL using a 3,000 Da cutoff filter (Millipore), and samples were mixed with an equal volume of SDS-PAGE buffer for SDS-PAGE and immunoblotting.
Additional methods
Bacterial strains, constructs, antibodies, antibiotics, SDS-PAGE and other experimental methods are described in the Supporting Information.
Results
The Lpt bridge forms a head-to-tail oligomer of structurally homologous domains in vivo
We have previously suggested that the Lpt proteins form a single complex that spans the IM, periplasm and OM.20 Here, we have used in vivo photocrosslinking to define which proteins form this transenvelope bridge and how they are connected. To do this, we used E. coli strains in which proteins substituted with the UV-photocrosslinkable residue para-benzoylphenylalanine (pBPA)21,22 were expressed at low levels. Each of these modified proteins could support cell growth in the absence of the relevant wild-type chromosomal allele, establishing that they are functional. By testing a variety of positions within each protein of interest, we could map the sites at which it interacts with other members of the Lpt assembly in vivo.
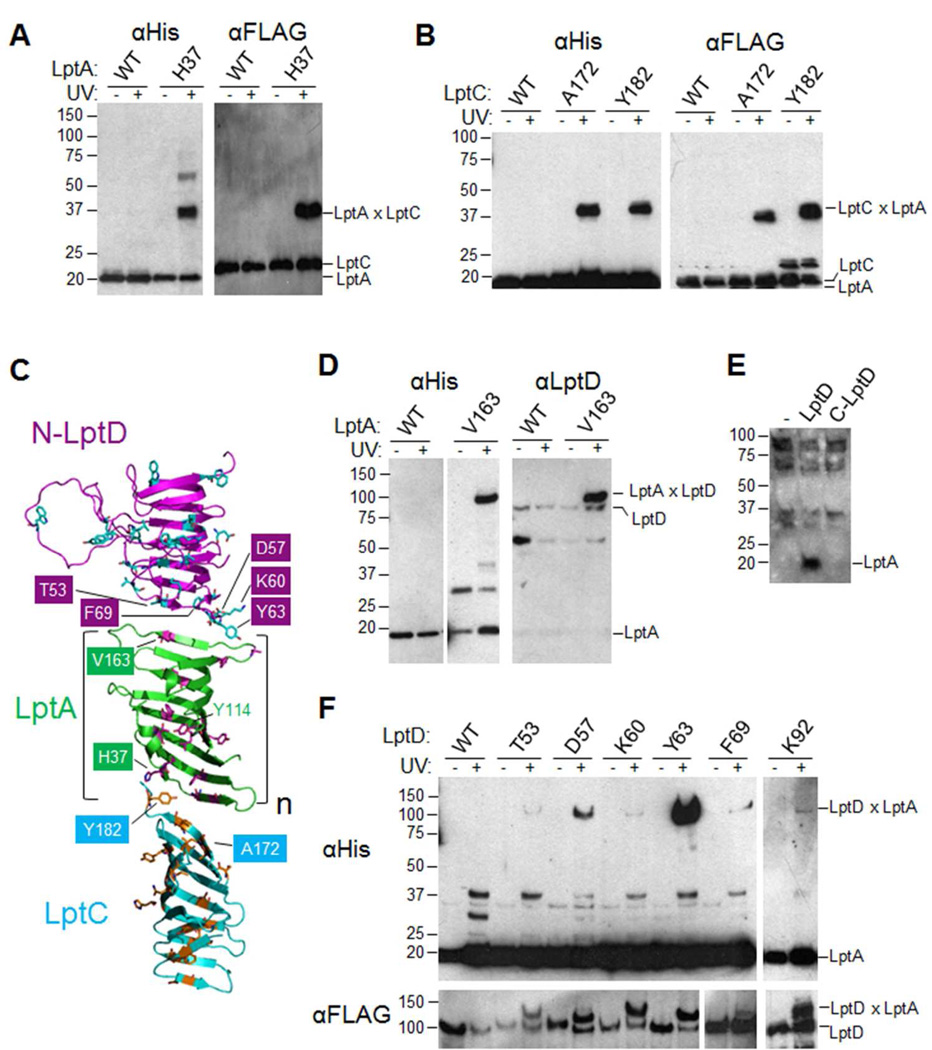
We initially focused on LptA. This protein has been proposed to physically link the IM and the OM because it is the only periplasmic Lpt component without a membrane tether and because it fractionates with both membranes.20 Also, one intermolecular interaction of LptA has already been established: LptA co-purifies with overexpressed LptC,23 and, when purified separately, the two proteins stably associate.24 To determine if our in vivo photocrosslinking system could identify specific sites of interaction between LptA and LptC, we generated variants of His-tagged LptA (LptA-His) substituted with pBPA at 14 sites distributed throughout the three-dimensional structure of LptA.25 Upon co-expressing these variants with FLAG-tagged LptC (FLAG-LptC), we observed robust in vivo photocrosslinking between FLAG-LptC and LptA-His substituted with pBPA at position H37 (Fig. 2A and Fig. S1 in the Supporting Information). This result shows that the N-terminal end of LptA (Fig. 2C) physically interacts with LptC in the Lpt complex in vivo.
Fig. 2.
The Lpt bridge is an oligomer of structurally homologous domains: LptC, LptA and the N-terminal domain of LptD.
(A) E. coli expressing low levels of N-terminally FLAG-tagged LptC (FLAG-LptC) and either wild-type C-terminally His-tagged LptA (LptA-His) or LptA-His containing pBPA at position H37 (numbering includes the LptA signal sequence, a.a. 1–27) were either left untreated or irradiated with UV light. After cell lysis, nickel affinity chromatography, and SDS-PAGE, samples were blotted with either αHis (left) or αFLAG (right) antibodies. See Fig. S1.
(B) Cells expressing LptA-His and the indicated pBPA-substituted variant of FLAG-LptC were treated as in Fig. 2A. See Fig. S2.
(C) To model the interactions within the Lpt bridge, one monomer in the tetrameric crystal form of LptA25 was manually replaced with the crystal structure of LptC (cyan),26 while another monomer was replaced with a model structure of N-LptD (magenta), generated by HHPred and MODELLER41 on the basis of the LptA structure. The LptC, LptA and LptD residues analyzed in Fig. 2 and Fig. S1–3 are shown in contrasting colors, and residues where crosslinking was observed are labeled. The number (n) of LptA molecules present in the bridge remains unclear (see main text).
(D) Cells expressing the indicated pBPA-substituted LptA-His variant were either left untreated or irradiated with UV light. Membrane extracts from each cell sample were subjected to TALON affinity chromatography, followed by immunoblotting with indicated antibodies.
(E) Membrane extracts from cells expressing full-length LptD-His, C-LptD-His (where C-LptD denotes a.a. 203–784, equipped with the native LptD N-terminal signal sequence, a.a. 1–24), or neither (−) were subjected to TALON affinity chromatography, followed by immunoblotting with αLptA antibody.
(F) Cells expressing LptA-His and the indicated pBPA-substituted variants of full-length LptD-3×FLAG (numbering includes the LptD signal sequence, a.a. 1–24) were treated and analyzed as in Fig. 2A. Sample loading for the αFLAG blot was adjusted to compensate for the unequal translation efficiency of various pBPA-containing protein constructs. See Fig. S3.
Having shown that LptA-LptC interactions can be detected by in vivo photocrosslinking, we sought to define the residues of LptC responsible for the interaction with LptA. We tested 23 positions distributed throughout the three-dimensional structure of LptC (Fig. S2)26 and found that LptC substituted with pBPA at positions A172 or Y182 could be crosslinked to LptA in vivo (Fig. 2B and S2). These LptA-interacting residues are found at the C-terminal edge of the LptC β-jellyroll (Fig. 2C), consistent with the previous observation that mutants in the C-terminus of LptC disrupt the association with LptA.23 Thus, LptC and LptA interact via their C- and N-terminal edges, respectively (Fig. 2C), similarly to the interactions within LptA oligomers in the crystal structure of LptA.25 These observations explain how LptA is anchored at the IM.
Whereas interactions between LptA and LptC at the IM had been observed previously, nothing was known about how LptA is anchored at the OM. During our survey of potential interaction sites throughout LptA, we observed that LptA-His-V163pBPA could form a crosslinked product of over 100 kDa (Fig. S1). This molecular weight is consistent with a covalent adduct of LptA-His (18 kDa) and LptD (87 kDa). Indeed, immunoblotting with αLptD antiserum confirmed strong crosslinking between LptA-His and LptD (Fig. 2D). Thus, while the N-terminus of LptA interacts with LptC at the IM, the C-terminus of LptA interacts with LptD at the OM (Fig. 2C). However, these results do not clarify whether a single LptA molecule can engage in both interactions simultaneously (vide infra).
LptD consists of a periplasmic N-terminal domain (N-LptD) and a C-terminal transmembrane β-barrel domain (C-LptD).15 We asked which of these domains serves as the OM anchor for LptA. We previously showed that isolated C-LptD is folded because it can associate with LptE as efficiently as can full-length LptD.15 By contrast, we found that, unlike full-length LptD, isolated C-LptD did not associate with LptA (Fig. 2E). Thus, elements of N-LptD are required for the interaction between LptD and LptA.
N-LptD belongs to the same OstA structural superfamily as LptA and LptC,25,27,28 but its three-dimensional structure is unknown. We generated a homology-based structural model for N-LptD (Fig. 2C) and probed whether the interactions between N-LptD and LptA resemble those between LptC and LptA. To do this, we incorporated pBPA at 22 positions throughout the N-terminal domain of a full-length, functional LptD. Indeed, LptD and LptA could be crosslinked when pBPA was incorporated at several positions at the N-terminal edge of the predicted β-jellyroll of N-LptD (Fig. 2C, 2F and S3). These results show that the N-terminal edge of N-LptD interacts with the C-terminal edge of LptA. Therefore, all three OstA superfamily domains interact in a similar, ordered fashion, with the N- and C-termini of each domain oriented toward the IM and OM, respectively.
It has been proposed that LptA may function as an oligomer on the basis of its crystal structure,25 and concentration-dependent head-to-tail oligomerization of LptA in vitro has been reported.29 However, it is not known whether LptA oligomers exist in vivo. When we co-expressed FLAG-tagged LptA (LptA-FLAG) with pBPA-substituted LptA-His, we observed UV-dependent covalent LptA dimers in vivo. The residues involved in this interaction were found at the N- and C-terminal edges of the LptA β-jellyroll (Fig. S4A-B), consistent with the LptA crystal structure25 and in vitro chemical crosslinking data.29 In particular, residue H37 at the N-terminal edge of LptA can participate in both homodimerization and heterodimerization with LptC (Fig. 2A), whereas residue V163 at the C-terminal edge of LptA can participate in both homodimerization and heterodimerization with LptD (Fig. 2D). Therefore, LptA can form head-to-tail homodimers in vivo. Nevertheless, it is also possible that a single LptA molecule can bridge the periplasm by contacting both LptC and N-LptD.
Assembly of the Lpt bridge requires the presence of native disulfide bonds in LptD
The OM translocon is composed of the transmembrane β-barrel protein LptD and the lipoprotein LptE.14,15 In the functional translocon, LptE resides within the lumen of the LptD β-barrel.16 In the absence of LptE, LptD cannot form the two non-consecutive, interdomain disulfide bonds, C31–C724 and C173–C725, at least one of which must be present for LptD to function.17 Defects in any Lpt component lead to buildup of LPS in the IM; therefore, transport across the periplasm and the OM is somehow coupled to prevent LPS mistargeting. We reasoned that the cell may accomplish this by coordinating the assembly of the Lpt bridge with that of the LptD/E translocon. One possibility is that assembly of the transenvelope bridge is required for the formation of correct disulfide bonds in LptD. In this case, assembly of the active OM translocon would require two distinct signals: the presence of LptE and that of an intact transenvelope bridge. Disrupting the bridge should then compromise the formation of correct LptD disulfide bonds.
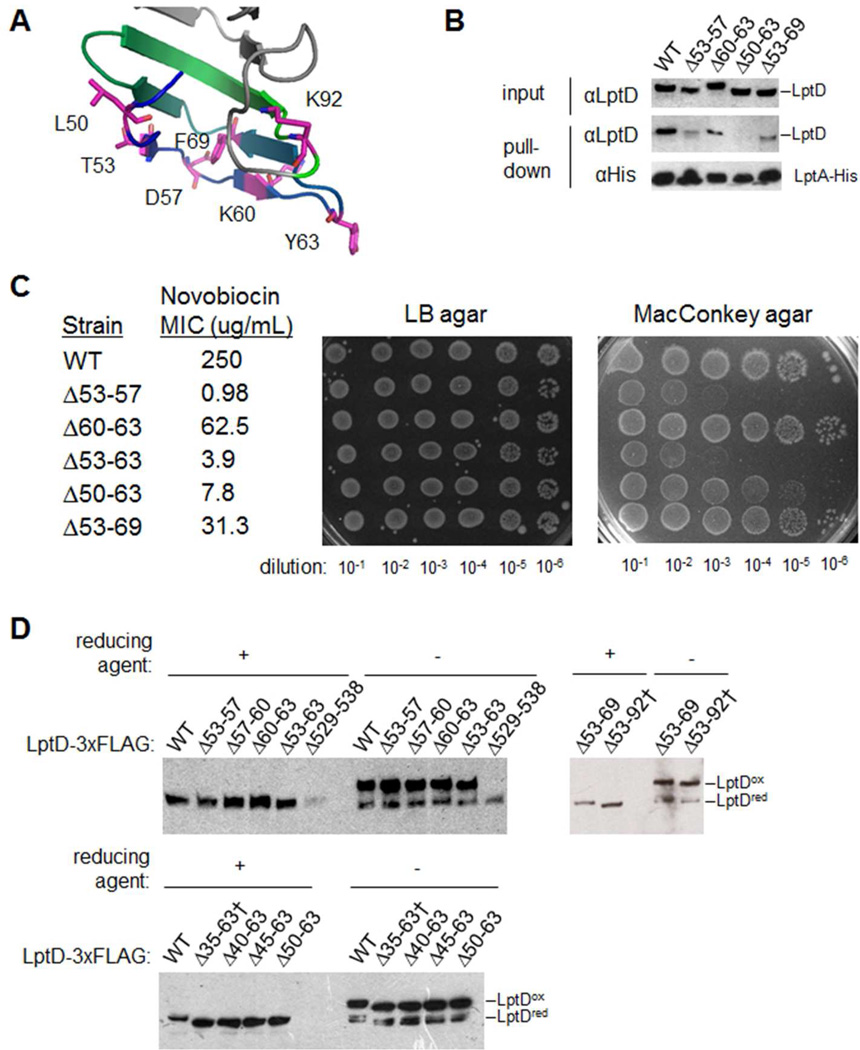
To test this possibility, we constructed a series of ten mutant LptD proteins lacking specific portions of the LptA-binding site in N-LptD (Fig. 2C,F and 3A). Progressively larger deletions in this region decreased the affinity of LptD for LptA-His, confirming the importance of these residues in LptD-LptA interactions (Fig. 3B). In cells, small- to moderate-sized deletions (e.g., Δ53–57 and Δ53–63) compromised OM integrity, as judged by sensitivity to MacConkey agar and novobiocin (Fig. 3C),4 whereas large deletions (Δ35–63 and Δ53–92) led to a loss of cell viability (not shown), consistent with the role of these residues in forming the transenvelope bridge. However, all the deletion mutant proteins, even those with a complete loss of function, still adopted a wild-type-like disulfide bond configuration (Fig. 3D). Therefore, the ability to form a transenvelope bridge is not necessary for LptD to form the correct disulfide bonds.
Fig. 3.
The OM translocon can form independently of the transenvelope bridge.
(A) Model structure of the N-terminal domain of LptD (see Fig. 2C), magnified to highlight the LptA-interacting residues (see Fig. 2F and S3).
(B) Membrane extracts from chromosomal ΔlptD::kan cells expressing LptA-His and the indicated variant of LptD were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies, either before (input) or after TALON affinity chromatography.
(C) The OM integrity of chromosomal ΔlptD::kan cells expressing the indicated lptD variants was tested by determination of the minimal inhibitory concentration (MIC) of novobiocin in LB media and by serial dilution onto either LB agar or MacConkey agar.
(D) Total lysates from cells expressing the indicated variant of LptD-3×FLAG were analyzed by SDS-PAGE in the presence or absence of the reducing agent β-mercaptoethanol, followed by immunoblotting with αFLAG antibody. The correctly oxidized form of LptD, LptDox, migrates at a higher apparent molecular weight than the reduced form, LptDred (ref. 17). The previously characterized Δ529–538 variant is an example of an incorrectly oxidized mutant protein.16 Dagger (†) indicates non-functional LptD variants.
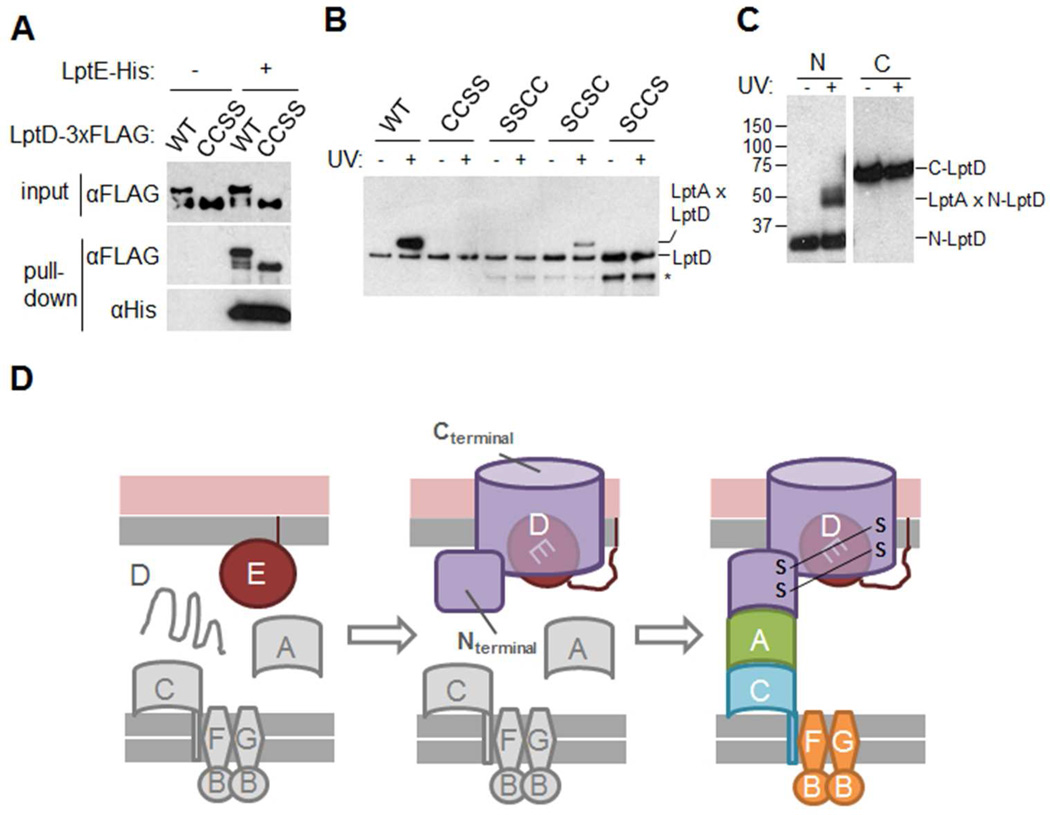
An alternative possibility is that proper assembly of LptD disulfides is a prerequisite for formation of the Lpt bridge. To test this hypothesis, we used several different LptD mutant proteins that each contained only one disulfide bond. Wild-type LptD contains four cysteine residues at positions 31, 173, 724, and 725 (denoted CCCC); in each mutant, two of these were replaced by serine (S). Like wild-type LptD, each of these mutant variants could still associate with LptE (Fig. 4A and S5A), showing that these proteins are stably expressed and correctly folded and inserted into the OM. The SCSC protein can support cell viability because it contains a C173–C725 disulfide bond, one of the two disulfides found in wild-type LptD; by contrast, the other three variants (CCSS, SSCC, and SCCS) cannot form either of the two native disulfide bonds and are non-functional.17
Fig. 4.
Formation of the transenvelope bridge requires a fully assembled OM translocon.
(A) Lysates from cells expressing the indicated variant of LptD-3×FLAG with or without LptE-His were analyzed by non-reducing SDS-PAGE and immunoblotting with the indicated antibodies, either before (input) or after (pulldown) nickel affinity chromatography. C (cysteine) and S (serine) represent the residues present at positions 31, 173, 724, and 725 of LptD, respectively. See Fig. S5A.
(B) Cells expressing LptA-His-V163pBPA and the indicated variant of LptD-3×FLAG (labeled as in Fig. 4A) were analyzed as in Fig. 2A and immunoblotted with αFLAG antibody. To facilitate visualization of both crosslinked and free LptD, this experiment was performed under conditions in which LptD-3×FLAG is retained non-specifically on nickel resin. The asterisk (*) represents degradation products of LptD-3×FLAG. See Fig. S5B-C.
(C) Cells expressing LptA-His-V163pBPA and either N-LptD-3×FLAG (a.a. 1–203) or C-LptD-3×FLAG (see Fig. 2E) were analyzed as in Fig. 2A and immunoblotted with αFLAG antibody. See Fig. S5D-E.
(D) Ordered assembly of the Lpt machinery. Proper association of LptD with LptE is required for folding and insertion of the LptD β-barrel into the OM (first step) and for formation of native disulfide bonds in LptD (second step).15–17 The presence of native disulfides in turn allows the N-terminal domain of LptD to engage LptA, enabling the formation of the transenvelope bridge. How the LptC-LptA interaction may be regulated is unknown.
To determine which of these mutant LptD proteins could associate with LptA, we used LptA-HisV163pBPA, the LptA variant that photocrosslinks to wild-type LptD (Fig. 2D). We found that only the SCSC variant could be crosslinked by LptA, whereas the CCSS, SSCC, and SCCS variants could not (Fig. 4B). Notably, the SCCS mutant could not interact with LptA despite the presence of a C173–C724 disulfide bond (Fig. S5B), which differs by only one residue from the C173–C725 configuration found in the functional SCSC mutant. These results show that native disulfide bonds are required in order for LptD to interact with LptA, explaining why mutant LptD proteins that lack these bonds are non-functional. We also tested LptDΔ529–538, which contains all four cysteine residues, but fails to form native disulfide bonds due to a defect in interaction with LptE;16 this mutant also could not be crosslinked by LptA-His-V163pBPA (Fig. S5C). Taken together, these results indicate that LptD must adopt a functional disulfide bond configuration before the transenvelope bridge can be assembled.
Based on our study of the LptD-LptA interaction (Fig. 2D-F), we had expected that N-LptD would contain all the structural determinants necessary for LptA binding. However, our finding that LptA cannot crosslink to incorrectly disulfide-bonded variants of full-length LptD (Fig. 4B) indicates that N-LptD must also be properly connected to C-LptD to form the bridge. We imagined that correct disulfide bonds might confer an N-LptD conformation that enables LptA binding. If so, N-LptD should not be capable of interacting with LptA if expressed separately from C-LptD (i.e., as “isolated N-LptD”). Surprisingly, however, we found that isolated N-LptD could be crosslinked by LptA-His-V163pBPA (Fig. 4C). This interaction was specific, since isolated C-LptD could not be crosslinked despite being expressed at a far higher level (Fig. 4C and S5D); also, an LptA-His variant substituted with pBPA at position Y114, away from the C-terminal edge of LptA (Fig. 2C), could not be crosslinked to isolated N-LptD (Fig. S5E). These results show that isolated N-LptD can fold into a conformation that contains the key structural features for LptA binding, despite the absence of native disulfide bonds. The fact that LptA cannot interact with the N-terminal domain of a full-length LptD protein lacking correct disulfide bonds (Fig. 4B and S5C) suggests that, in full-length LptD, C-LptD prevents N-LptD from binding to LptA until the correct disulfide bonds are established.
Discussion
We have defined the architecture of the transenvelope bridge responsible for LPS transport across the periplasm, as well as a regulatory mechanism to control bridge assembly in vivo. We have shown how periplasmic LptA contacts both LptC at the IM and the N-terminal domain of LptD at the OM, creating a continuous bridge of anti-parallel β-strands between the membranes (Fig. 2C). Our results establish the essential role of N-LptD in orchestrating the biogenesis of the transenvelope bridge, which can only occur after proper assembly of the OM LPS translocon. In this way, the cell synchronizes the assembly of components in the OM and the periplasm, ensuring proper coordination of their function.
Several lines of evidence indicate that the intermolecular contacts we have identified occur in the functional Lpt bridge in vivo. We tested 59 positions throughout LptC, LptA, and N-LptD, each of which belongs to the OstA structural superfamily; without exception, the residues involved in UV-photocrosslinking in vivo were found only at the exposed edges of these β-sheet structures (Fig. 2C), in close agreement with the crystallographic LptA oligomer25 and in vitro studies.29 We (Fig. 3C) and others23,25 have also shown that genetic disruption of these edge regions compromises the assembly and function of the Lpt bridge. The observed N- to C-terminal orientation of the domains within the bridge is consistent with genomic data: many Gram-negative organisms encode predicted fusion proteins containing as many as four OstA-superfamily domains.28 Our data do not resolve the number of LptA molecules present in each transenvelope bridge. We can observe LptA dimers (Fig. S4A), but their physiological relevance is difficult to establish because the regions of the protein involved in dimerization also interact with LptC and LptD. Structural arguments have been used to suggest that four OstA-superfamily domains are necessary to span the width of the periplasm.25 However, the periplasm may be constricted in the vicinity of Lpt bridges; we have previously shown that the entire transenvelope complex is associated with an unusual membrane fraction containing both IM and OM components,20 suggesting that Lpt bridges might exist within zones of membrane adhesion.30,31 It is also possible that the transenvelope bridge is not a static structure, but can vary in length to accommodate varying intermembrane distances under different cellular conditions or in different organisms.
We have shown that N-LptD performs the essential function of linking the OM translocon to the transenvelope bridge in a regulated manner. The N-terminal domain of a full-length LptD protein can interact with LptA (Fig. 2F), but only if at least one native disulfide bond is present within LptD (Fig. 4B and S5C). The formation of these native disulfides depends on proper assembly of LptD with LptE.15–18 Thus, LptD disulfide bonds report on the presence of a correctly assembled LptD/E translocon in the OM, which then enables the formation of the transenvelope bridge (Fig. 4D). This regulatory mechanism ensures that the cell does not couple Lpt bridges to non-functional OM translocons, consistent with the observation that depletion of LptD or LptE results in LptA degradation23 and causes LPS to accumulate at the IM.9 The presence of active transenvelope bridges coupled to defective OM translocons might be harmful to the cell because LPS would be mistargeted to the periplasm.
It is intriguing that, in full-length, non-functional LptD molecules, the C-terminal β-barrel prevents the N-terminal domain from interacting with LptA: N-LptD can interact with LptA on its own (Fig. 4C), but not in the context of a full-length LptD molecule lacking correct disulfide bonds (Fig. 4B and S5C). LptD is one of the largest OM β-barrels, predicted to contain as many as 24 β-strands.32,33 Every structurally characterized β-barrel protein of this size is occluded by a plug domain encoded on the same polypeptide.34 Previously, we established that mature LptD is plugged by a separate protein, LptE.16 However, these two proteins are assembled at the OM by distinct pathways – the β-barrel assembly pathway35 and the localization of lipoproteins system,36 respectively. Details of how LptD and LptE form into a complex remain to be elucidated. Here, we have found that N-LptD is not accessible until LptE properly associates with the C-LptD β-barrel and enables the formation of correct disulfide bonds (Fig. 4D).17 A regulatory mechanism of this type has been demonstrated for the pilus usher FimD: in the presence of substrate, a plug domain found near the N-terminus of FimD rotates out of the lumen of the FimD β-barrel, opening the channel for translocation of pilus subunits.37
Whereas Gram-negative pathogenesis and virulence can require numerous transenvelope complexes such as efflux pumps and protein secretion machines,38,39 the Lpt proteins constitute the only such complex known to be essential for Gram-negative cell viability. A recently discovered Pseudomonas-specific peptidomimetic antibiotic binds to LptD, and resistance is conferred by a mutation in the N-terminal domain of P. aeruginosa LptD.40 It is tempting to speculate that this drug disrupts the function or regulation of the transenvelope Lpt bridge in Pseudomonas. Understanding the assembly of the Lpt machinery can shed light on how its components function in transporting LPS and how these functions can be disrupted by antibiotics.
Supplementary Material
Acknowledgments
The plasmid pSup-BpaRS-6TRN was a generous gift from Prof. Peter G. Schultz (The Scripps Research Institute, La Jolla, CA). The authors also thank Z. Yao for MIC measurements, R.M. Davis for technical assistance in generating the ΔlptA::kan allele, and Dr. S.-S. Chng, Dr. L.S. Gronenberg, M. Xue, and C. Liu for reagents.
This work was supported by NIAID Grant AI081059 (D.K.). E.F. was a graduate fellow of the Fannie & John Hertz Foundation.
Footnotes
Supporting Information Available. Additional experimental procedures and supporting data are included in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kamio Y, Nikaido H. Outer membrane of Salmonella typhimurium: Accessibility of phospholipid headgroups to phospholipase C and cyanogen bromide activated dextran in the external medium. Biochemistry. 1976;15:2561–2570. doi: 10.1021/bi00657a012. [DOI] [PubMed] [Google Scholar]
- 2.Funahara Y, Nikaido H. Asymmetric localization of lipopolysaccharides on the outer membrane of Salmonella typhimurium. J. Bacteriol. 1980;141:1463–1465. doi: 10.1128/jb.141.3.1463-1465.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mühlradt PF, Golecki JR. Asymmetrical distribution and artefactual reorientation of lipopolysaccharide in the outer membrane bilayer of Salmonella typhimurium. Eur. J. Biochem. 1975;51:343–352. doi: 10.1111/j.1432-1033.1975.tb03934.x. [DOI] [PubMed] [Google Scholar]
- 4.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doerrler WT. Lipid trafficking to the outer membrane of Gram-negative bacteria. Mol. Microbiol. 2006;60:542–552. doi: 10.1111/j.1365-2958.2006.05130.x. [DOI] [PubMed] [Google Scholar]
- 7.Sperandeo P, Dehò G, Polissi A. The lipopolysaccharide transport system of Gram-negative bacteria. Biochim. Biophys. Acta. 2009;1791:594–602. doi: 10.1016/j.bbalip.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 8.Sperandeo P, et al. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J. Bacteriol. 2007;189:244–253. doi: 10.1128/JB.01126-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sperandeo P, et al. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J. Bacteriol. 2008;190:460–4469. doi: 10.1128/JB.00270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narita S, Tokuda H. Biochemical characterization of an ABC transporter LptBFGC complex required for the outer membrane sorting of lipopolysaccharides. FEBS Lett. 2009;583:2160–2164. doi: 10.1016/j.febslet.2009.05.051. [DOI] [PubMed] [Google Scholar]
- 12.Braun M, Silhavy TJ. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol. Microbiol. 2002;45:1289–1302. doi: 10.1046/j.1365-2958.2002.03091.x. [DOI] [PubMed] [Google Scholar]
- 13.Bos MP, Tefsen B, Geurtsen J, Tommassen J. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9417–9422. doi: 10.1073/pnas.0402340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu T, et al. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chng SS, Ruiz N, Chimalakonda G, Silhavy TJ, Kahne D. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc. Natl. Acad. Sci. U.S.A. 2010;107:5363–5368. doi: 10.1073/pnas.0912872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freinkman E, Chng SS, Kahne D. The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2486–2491. doi: 10.1073/pnas.1015617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruiz N, Chng SS, Hiniker A, Kahne D, Silhavy TJ. Nonconsecutive disulfide bond formation in an essential integral outer membrane protein. Proc. Natl. Acad. Sci. U.S.A. 2010;107:12245–12250. doi: 10.1073/pnas.1007319107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chimalakonda G, et al. Lipoprotein LptE is required for the assembly of LptD by the beta-barrel assembly machine in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2492–2497. doi: 10.1073/pnas.1019089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tefsen B, Geurtsen J, Beckers F, Tommassen J, de Cock H. Lipopolysaccharide transport to the bacterial outer membrane in spheroplasts. J. Biol. Chem. 2005;280:4504–4509. doi: 10.1074/jbc.M409259200. [DOI] [PubMed] [Google Scholar]
- 20.Chng SS, Gronenberg LS, Kahne D. Proteins required for lipopolysaccharide assembly in Escherichia coli form a transenvelope complex. Biochemistry. 2010;49:4565–4567. doi: 10.1021/bi100493e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chin JW, Schultz PG. In vivo photocrosslinking with unnatural amino acid mutagenesis. Chem Bio Chem. 2002;11:1135–1137. doi: 10.1002/1439-7633(20021104)3:11<1135::AID-CBIC1135>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 22.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 23.Sperandeo P, et al. New insights into the Lpt machinery for lipopolysaccharide transport to the cell surface: LptA-LptC interaction and LptA stability as sensors of a properly assembled transenvelope complex. J. Bacteriol. 2011;193:1042–1053. doi: 10.1128/JB.01037-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowyer A, Baardsnes J, Ajamian E, Zhang L, Cygler M. Characterization of interactions between LPS transport proteins of the Lpt system. Biochem. Biophys. Res. Commun. 2011;40:1093–1098. doi: 10.1016/j.bbrc.2010.12.121. [DOI] [PubMed] [Google Scholar]
- 25.Suits MD, Sperandeo P, Dehò G, Polissi A, Jia Z. Novel structure of the conserved Gram-negative lipopolysaccharide transport protein A and mutagenesis analysis. J. Mol. Biol. 2008;380:476–488. doi: 10.1016/j.jmb.2008.04.045. [DOI] [PubMed] [Google Scholar]
- 26.Tran AX, Dong C, Whitfield C. Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J. Biol. Chem. 2010;285:33529–33539. doi: 10.1074/jbc.M110.144709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bos MP, Robert V, Tommassen J. Biogenesis of the Gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- 28.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merten JA, Schultz KM, Klug CS. Concentration-dependent oligomerization and oligomeric arrangement of LptA. Protein Sci. 2012;21:211–218. doi: 10.1002/pro.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bayer ME. Areas of adhesion between wall and membrane of Escherichia coli. J. Gen. Microbiol. 1968;53:395–404. doi: 10.1099/00221287-53-3-395. [DOI] [PubMed] [Google Scholar]
- 31.Bayer ME. Zones of membrane adhesion in the cryofixed envelope of Escherichia coli. J. Struct. Biol. 1991;107:268–280. doi: 10.1016/1047-8477(91)90052-x. [DOI] [PubMed] [Google Scholar]
- 32.Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ. PRED-TMBB: a web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic Acids Res. 2004;32:W400–W404. doi: 10.1093/nar/gkh417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayat S, Elofsson A. BOCTOPUS: Improved topology prediction of transmembrane β barrel proteins. Bioinformatics. 2012;28:516–522. doi: 10.1093/bioinformatics/btr710. [DOI] [PubMed] [Google Scholar]
- 34.Fairman JW, Noinaj N, Buchanan SK. The structural biology of β-barrel membrane proteins: a summary of recent reports. Curr. Opin. Struct. Biol. 2011;21:523–531. doi: 10.1016/j.sbi.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hagan CL, Silhavy TJ, Kahne D. β-Barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem. 2011;80:189–210. doi: 10.1146/annurev-biochem-061408-144611. [DOI] [PubMed] [Google Scholar]
- 36.Okuda S, Tokuda H. Lipoprotein sorting in bacteria. Annu. Rev. Microbiol. 2011;65:239–259. doi: 10.1146/annurev-micro-090110-102859. [DOI] [PubMed] [Google Scholar]
- 37.Phan G, et al. Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate. Nature. 2011;474:49–53. doi: 10.1038/nature10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Misra R, Bavro VN. Assembly and transport mechanism of tripartite drug efflux systems. Biochim. Biophys. Acta. 2009;1794:817–825. doi: 10.1016/j.bbapap.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holland IB. The extraordinary diversity of bacterial protein secretion mechanisms, in Protein Secretion. In: Economou A, editor. Methods in Molecular Biology. Vol. 619. New York: Springer; 2010. pp. 1–20. [DOI] [PubMed] [Google Scholar]
- 40.Srinivas N, et al. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science. 2010;327:1010–1013. doi: 10.1126/science.1182749. [DOI] [PubMed] [Google Scholar]
- 41.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.