Abstract
The complexity and rapid growth of genetic data demand investment in information technology to support effective use of this information. Creating infrastructure to communicate genetic information to health care providers and enable them to manage that data can positively affect a patient’s care in many ways. However, genetic data are complex and present many challenges. We report on the usability of a novel application designed to assist providers in receiving and managing a patient’s genetic profile, including ongoing updated interpretations of the genetic variants in those patients. Because these interpretations are constantly evolving, managing them represents a challenge. We conducted usability tests with potential users of this application and reported findings to the application development team, many of which were addressed in subsequent versions. Clinicians were excited about the value this tool provides in pushing out variant updates to providers and overall gave the application high usability ratings, but had some difficulty interpreting elements of the interface. Many issues identified required relatively little development effort to fix suggesting that consistently incorporating this type of analysis in the development process can be highly beneficial. For genetic decision support applications, our findings suggest the importance of designing a system that can deliver the most current knowledge and highlight the significance of new genetic information for clinical care. Our results demonstrate that using a development and design process that is user focused helped optimize the value of this application for personalized medicine.
Keywords: clinical decision support, electronic health records, genomics, personalized medicine
1. Introduction
Personalized medicine--the incorporation of an individual’s genetic makeup into the management of their care--has the potential to play a major role in the future of healthcare. While there are few successful examples of the routine use of genetic information in guiding patient care to date, the prospective benefits are great. As evidenced-based use of genetic data becomes more common for a variety of purposes, there is a growing need for information technology (IT) to support the management of the rapidly expanding knowledge [1, 2].
One specific challenge that genetic counselors, geneticists, and clinicians regularly face today is how to track the changing state of knowledge about genetic variants, including those of unknown significance. Our understanding of the clinical implications of variants evolves as new scientific evidence becomes available, so obtaining new information on previously reported variants is important for clinical care [3].
For example, with hypertrophic cardiomyopathy, new knowledge about variants may critically influence the treatment of patients and their families and, in some circumstances, help prevent sudden cardiac death [4]. However, typical processes in place for learning about and managing variant updates (changes in knowledge relating to the clinical significance of variants) have many challenges. Currently, it is not standard practice for labs to continually re-evaluate variants and contact treating clinicians [5]. However, for laboratories that do, there are a large number of variants to track as scientific knowledge changes over time, and the cost to the lab to amend old reports with every variant update is high, particularly since this is not currently reimbursed by payers. In addition, while treating clinicians are ultimately responsible for managing the care of patients, in most cases they do not have the capacity, given their case load, to remain up-to-date on all variants affecting their patients [6, 7]. Laboratories may be in the best position to monitor the current state of knowledge on the variants they have identified [8]. For these reasons, the development and design of IT infrastructure and applications to support clinicians and laboratories in this process are necessary. Designing systems that satisfy the needs of the users and are flexible enough to handle the continued evolution of this complex and emerging area is an important step in realizing the benefits of personalized medicine.
Involving treating clinicians early in the design process and generating requirements based on user research can reduce development and support costs, increase user satisfaction, and ensure the user’s long term commitment to the application [9, 10]. In addition, using principles of human factors and usability to evaluate systems can help prevent errors, delays and frustrations that, left unaddressed, may result in underuse or even abandonment of systems [11–14]. Designing interfaces that support the user’s natural process for decision making, problem solving and information processing will allow the user to conduct their work with a minimum amount of unnecessary cognitive effort [15].
While user-centered design has rather rapidly diffused into many industries in the last decade, challenges specific to healthcare applications have resulted in developers and vendors of these applications being slower to implement this philosophy [15–17]. A number of reports have been published recently by the Agency for Healthcare Research and Quality and National Institute of Standards and Technology in hopes of creating guidelines around incorporating human factors and usability in the design of health IT [16, 18]. More limited access to users and the complexity and sensitivity of patient data are just some of the challenges faced by those developing such applications [19, 20].
The volume and continual evolution of variant knowledge requires a system and interface that is designed to support complex decision-making and knowledge that changes frequently. For example, in the last six years, the Laboratory for Molecular Medicine (LMM) at the Partners Center for Personalized Genetic Medicine (PCPGM) has made approximately 214 category changes to previously reported variants associated with just one condition, hypertrophic cardiomyopathy, impacting nearly 756 patients [21, 22]. In order to effectively manage the growing number of genetic test results and changing knowledge of variants, clinicians will need interfaces that will allow them to efficiently review genetic test results, provide the most up-to-date information to their patients with confidence and easily identify patients and families that may require changes to their treatment and testing plans.
Understanding how clinicians use these tools and evaluating them early is critical to successful development and use of these applications. As part of a broader study evaluating the usability and utility of an application for tracking variants and the value of more timely genetic variant information, we conducted a usability assessment of the first version of a novel application prior to its implementation in a healthcare clinic. Results of these analyses were used to further enhance the application prior to additional development and broader distribution.
2. Methods
2.1 GeneInsight Clinic Application Description
The Partners HealthCare Center for Personalized Genetic Medicine developed the GeneInsight suite of applications to provide infrastructure around managing genetic data [23]. The GeneInsight Lab component provides IT infrastructure for the laboratory, assisting in genetic knowledge management and report generation. To support the management and communication of genetic test results and variant updates to clinicians, a new innovative web-based application was developed. GeneInsight Clinic (GIC), as part of the GeneInsight suite, was created with the goal of providing health care providers the ability to fully manage their patient genetic profiles [6]. GeneInsight delivers structured electronic genetic reports and generates physician alerts when new knowledge is identified on variants in their patients. One important element in accomplishing the goals of this new tool and process is to ensure that the design of GIC satisfies the needs of the users and is flexible to handle continued evolution of this emerging area.
GIC utilizes a web-based interface that allows the clinician to access a complete electronic summary and history of a patient’s genetic profile. In addition to the application, a new process was developed to deliver patient-specific alerts notifying clinicians about the availability of genetic reports in the system or updates to their patients’ variants. The clinicians receive these alerts through email with a link to the information in GIC. Currently, each GIC contains information on all patients within the clinic receiving genetic testing through the LMM. Email notifications of final reports and variant updates regarding their patients are sent to all providers within the clinic. To assist in determining what updates should be emailed to the clinician and at what frequency, variant updates are classified based on the significance of the change in knowledge into one of three alert categories: high, medium, or low (see appendix for types of variant changes and their alert levels). The lab updates the category in the GeneInsight Lab component of the system. The approval of the change in variant category triggers the logic that generates the update and sends the email to the appropriate clinic. High variant change alerts are emailed to the clinician as soon as the updates are approved by the lab, and medium and low alerts are emailed to the clinician in a weekly summary email, along with a list of any unreviewed high alerts or new report alerts
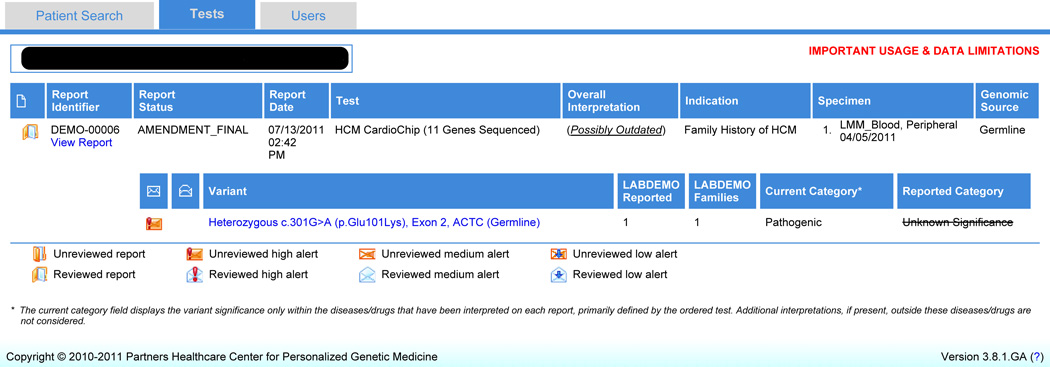
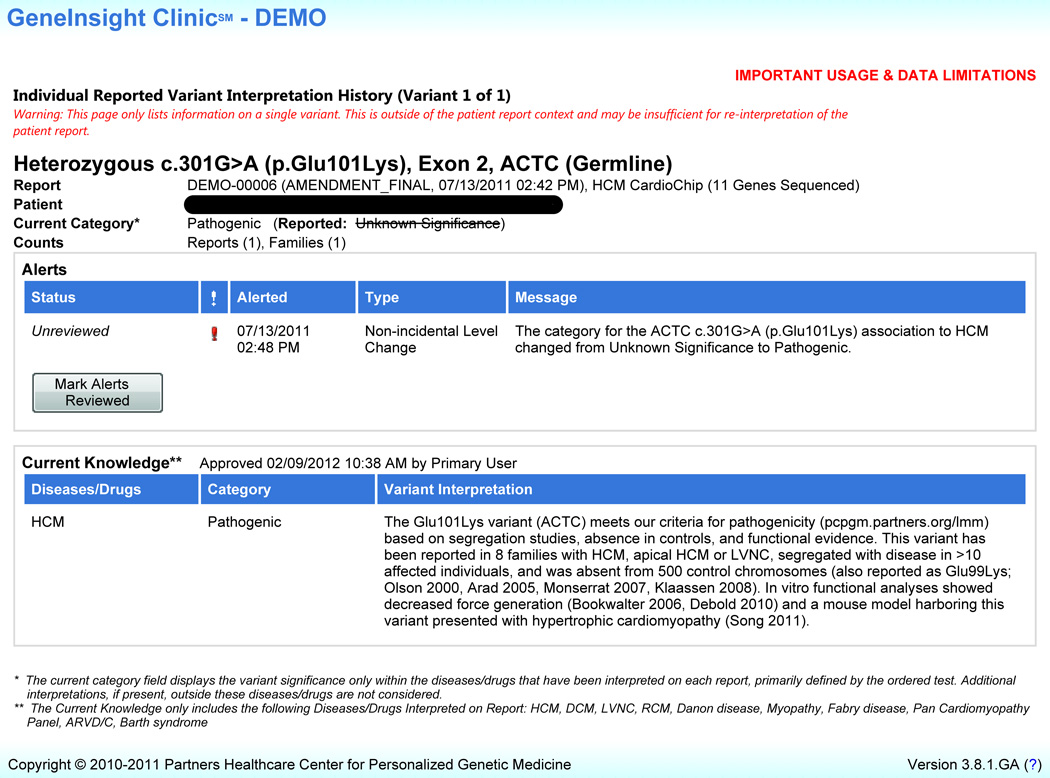
The GIC interface includes four main pages. The Search page allows clinicians to conduct: 1. a patient search, or 2. a variant search resulting in a list of patients in that clinic presenting with that variant. Once a single patient is selected, the Tests page (Figure 1) has detailed information on the patient’s genetic test results including reported variants and any updated variant information. The Case Report Details page includes some structured elements of the patient’s report as well as a portable document format (.pdf) copy of the full final report issued by the lab. The Individual Reported Variant Interpretation History page (Figure 2) includes more specific information on each variant for the patient including supporting evidence for any category changes to the variant, as well as the current state of knowledge on the variant. Users have the ability to mark that reports have been reviewed by clicking a “mark reviewed” button on the Tests page. Similarly, users can mark that variant updates have been reviewed by using a “mark reviewed” button on the Individual Reported Variant Interpretation History page [21].
Figure 1.
GeneInsight Clinic patient reports view (Tests page) (Version 3.8). – This page displays a summary of genetic test report results and variant knowledge updates on an individual patient. Users can view the report and obtain specific information about the variants listed by clicking on the variant links. Users can also mark a report reviewed by using the button provided.
Figure 2.
GeneInsight Clinic Individual Reported Variant Interpretation History Page (Version 3.8) - Users navigate to this page by clicking on the variant name on the patient reports view for an individual patient. Information regarding the individual variant’s history including alerts on variant knowledge changes and current interpretation is provided on this page. The user can mark alerts reviewed using the button provided.
Some of the initial designs for the GIC interface were evaluated by the usability specialist (PN) prior to participant recruitment and usability testing. The usability specialist on the research team conducted an expert review of some of the early designs and provided a short report of recommendations to the development team based on standard usability heuristics [24]. In addition, the usability specialist provided informal reviews of subsequent prototypes. The design evaluated during the usability tests was the first version tested with users.
2.2 Participants and Recruitment
The evaluation was approved by the Partners HealthCare System Human Research Committee and was conducted at two academic medical centers.
We requested participation in the broader study from physicians, genetic counselors, nurse practitioners and nurses at two locations where GIC was being implemented. Both clinics order a large number of genetic cardiomyopathy tests from the Laboratory for Molecular Medicine. Participants had the opportunity to opt out of the broader study and subsequently, if they did not choose to opt out, an email was sent to request participation in the usability study component. Seven clinicians from a clinic in the Partners HealthCare System and two clinicians from an academic medical center in a Midwestern state consented to participate in the first round of usability testing. No remuneration was provided for participation.
2.3 Task Scenario Development
Before the testing sessions, usability testing scenarios were developed from screen shots and use cases with the assistance of subject-matter experts (MV, HR). The scenarios were identical for all participants, regardless of their clinical role. The scenarios were reviewed by all research team members to ensure that the content, format, and presentation were representative of real expected use and addressed the major functional components of the application. Most of the common and critical tasks were placed at the beginning of the testing session.
A GIC tool development environment was set up with test patients that were appropriate for each clinical scenario. The test patients and clinical scenarios were reviewed by members of the tool’s development team and subject matter expert, and the usability test was piloted by a genetic counselor on the research team (SB). The pilot participant was not involved in the development of the tasks and did not see the tasks before doing the usability test. After the test, the pilot participant offered feedback on the usability testing process, as well as the content and wording of task scenarios. As a result of the feedback, some task scenarios and test patients’ content were revised.
2.4 Testing Environment & Equipment
All usability tests were conducted on-site in a conference room at the participant’s offices at both clinic sites. Two members of the research team were present, the usability specialist (PN) served as the moderator and an observer (SP) transcribed the session and annotated task markers and other metrics using the Morae Observer software [25]. The participant’s interaction with the interface was observed and moderated by the usability specialist who sat next to and slightly behind the participant during the testing session. Morae Recorder software was running on the participant’s laptop to capture keystrokes, and audio and video of the session. The moderator had a paper version of the test script to capture additional observations and notes during the testing session.
Some of the testing scenarios involved using one of the GIC email messages: Report Alert, Variant Update Alert, and Summary Alert. A study specific Microsoft Outlook mailbox was used to mimic the participants’ email. Folders were set up in the mailbox for each study participant, and each folder contained the set of three emails required for the tasks. Each participant was given a username and password to log into the application.
2.5 Usability Test Procedure
A usability test plan was developed and reviewed by the research team prior to testing. The test plan included details on the testing procedure, tasks, usability metrics, usability goals, and appendices of the scenarios, pre-test instructions, usability questionnaire, and post-test interview questions.
At each testing session, the moderator described the usability test procedure to the participant using scripted pre-test instructions to ensure all participants received the same introduction. The moderator described the “think aloud” process, asking the participants to share their thought process and expectations while completing the tasks [26, 27]. An example of “think aloud” was also demonstrated to participants. The participant was told that the purpose of the study was to evaluate the GIC and that the usability testing session was being recorded.
After the moderator discussed the pre-test instructions with the participant, they were introduced to the tool and began the task scenarios. They were asked to read each task aloud and inform the moderator when they completed the task. At the end of each task, the moderator asked the participant to grade the task (A→F) based on how easy/difficult it was to complete. Once the participant completed all the tasks, they were asked to complete a written questionnaire, the System Usability Scale (SUS) [28], to assess their overall feelings about GIC. The participants ranked their agreement with 10 statements based on a 5 point Likert-type scale to assess overall perceived usability. The moderator then administered a 6 question verbal post-test interview regarding their experience with the tool, recommendations for enhancements and expected value of the tool in their practice. For the remainder of the session, the moderator solicited additional feedback on the four main screens of the GIC. Participants were asked to suggest the intended function or meaning of certain interface elements (i.e., icons, links and labels) and interpret specific content on these screens.
2.6 Usability Metrics & Data Collection
In addition to the observations and comments captured during the testing session, usability measurements were recorded during the test or in the post-test analysis of the session video. Usability metrics are measurements collected to determine to what extent the usability goals have been met and to compare those measurements to those gathered from subsequent tests to assess whether design changes have made a difference. Task completion success rates, time-on-task, error rates, and assists by the moderator were collected for each participant during the testing session and coded during the post-test analysis.
Each scenario requested that the participant obtain specific data or complete specific actions that would be used in the course of the task. We recorded task success or failure based on the intended outcome as indicated in the final test plan.
Participants who were having trouble completing the tasks were given assists at the moderator’s discretion. An assist is defined as active direction by the moderator to the participant in order to help them achieve the task goal. Assists were only given when the participant was visibly frustrated or did something that would prevent completion of subsequent tasks. We captured the number of assists and included them as a metric in the analysis.
We measured the time for each task by noting the start and end times for each task (beginning when the participant finished reading the task scenario and ending when the participant either completed the task or indicated to the moderator that they believed they completed the task). When a participant required an assist, the time on task included the assist time. We also captured errors that occurred during the completion of the tasks. Errors can include those that would not have an impact on the final output of the task but would result in the task being completed less efficiently. Other errors, such as obtaining incorrect information, could result in task failure.
2.7 Data Analysis and Reporting
Each usability test session was recorded and analyzed using Morae software. The moderator and the observer reviewed each recording together to log the time on task, errors, assists and task completion for each participant. In addition, the moderator and observer marked specific quotes and usability issues that the participants encountered during the test. During multiple reviews of the Morae recordings and transcripts of the sessions, the usability expert identified usability issues based on observations of the participants during the task and their think-aloud comments. The transcripts of the usability tests were organized by task and participant, and quotes were identified that illustrated a user expectation, frustration or misinterpretation of content or functionality. The frequency of the issue, identified by the number of participants experiencing the problem, was captured. In addition, the usability heuristic related to each problem was identified. Finally, the issue was categorized based on how much of the system was affected. For example, a global issue affects multiple pages of the site and a local issue is specific to a particular interface element [24, 27].
The usability test report generated for the GIC development team included tables summarizing the usability metrics as well as a summary of the usability findings, organized by their scope and frequency with direct quotes from participants to support the analysis. In addition, a highlight video was generated with the consent of participants that included clips from the participant’s usability testing sessions that were used to illustrate some of the major findings.
2.8 Dissemination of Findings
The report was provided to all members of the Research team as well as members of the Quality Assurance and Development teams. The highlight video and major findings were presented and discussed at a meeting with the entire group.
3. Results
3.1 Participant Characteristics
The seven participants who participated from the two sites included two genetic counselors, one nurse practitioner, one nurse and three physicians (Table 1). Five out of the seven participants were introduced to the tool months previously through a demo presentation. None of the participants received formal training or had the opportunity to review the user guide before participating in a usability test.
Table 1.
Participants
| Clinic | Role | GeneInsight Clinic Exposure prior to Usability Test |
Internet Use |
|---|---|---|---|
| Site 1 | Physician | Demo | Daily |
| Site 1 | Genetic Counselor | Demo | Daily |
| Site 2 | Physician | Early demo | Daily |
| Site 2 | Physician | Early demo, meeting presentation | Daily |
| Site 2 | Research Nurse Coordinator | None | Daily |
| Site 2 | Nurse Practitioner | None | Daily |
| Site 2 | Genetic Counselor | Early demo, meeting presentation | Daily |
3.2 Usability Findings
Users were able to perform critical activities such as viewing a patient’s genetic test results and high level variant updates and searching patients on multiple dimensions. Participants rarely felt frustrated by the system although they did verbalize confusion or uncertainty during some of the tasks. Almost all observed usability issues fell under the following categories: icon inconsistencies, labeling and language issues, and placement and organization. Inconsistencies in the behavior and application of some icons with external conventions and among screens of the application led participants to misunderstand the process or status of the data presented. One participant expressed confusion about the meaning of the icons, “I’m not sure the red thing – high level alert? The green – lesser level? The ones without anything are insignificant variant changes?” Other issues included organization of the summary email and labels on the review buttons and for the reported probands and families counts that did not clearly indicate to some users what they were for or what was being communicated. One participant commented on the summary email, “There are so many highs…I'm trying to see what is urgent…they are all saying high alert and they are all high. I can't sit and open each one….” When asked about the reported probands and families counts, one participant incorrectly defined the counts, stating “I would think that would be a combo of reported in the lab and published....would not include other labs, just published data and the LMM lab.” Enhancements were incorporated into the current version of the software based on these usability findings. Participants were overall positive in their response to the interview questions. All participants said that they would recommend the tool to colleagues and that the tool would have a positive impact on their practice. One participant shared their thoughts on the tool, “It's nice to have updated access to the variant interpretation because it is important for patient management…it was really easy to use and self-explanatory.”
3.3 Usability Questionnaire
Overall scores from the post-test usability questionnaire (System Usability Scale) indicate an above average perceived ease of use. The overall average usability score was 89 out of 100 possible points across all participants. A score above an 80 places the application in the top 10% of scores [28–30].
3.4 Usability Metrics
Measurements captured during the usability tests show baseline results for the first version of the GIC and will be used for comparison with usability test results of subsequent revised versions. Geometric mean of time on task ranged from 8.17 seconds to 141.95 seconds. Additional measurements are summarized in Table 2.
Table 2.
Summary of Usability Test Results by Task
| Task Summary | |||
|---|---|---|---|
| Completion Rate (n=7) |
Average Grade (n=7) |
Error-Free Rate (n=7) |
|
| Task 1 – GIC Report Alert – locate patient with new report CRITICAL TASK |
100% | A− | 100% |
| Task 2 – View Test Report and ‘Mark Reviewed’ |
100% | A− | 100% |
| Task 3 – GIC Variant Alert – locate patient(s) with variant update CRITICAL TASK |
100% | A− | 100% |
| Task 4 – Locate unreviewed alert and change in variant interpretation CRITICAL TASK |
85.7% | A/A− | 71.4% |
| Task 5 – Locate overall report interpretation |
100% | A− | 71.4% |
| Task 6 – Locate number of reports and families with variant tested at lab |
14.3% | B+ * | 14.3% |
| Task 7 – Locate evidence for variant update |
100% | A | 100% |
| Task 8 – Mark variant reviewed | 57.1% | A * | 57.1% |
| Task 9 – Locate all of a patient’s variants. Locate reviewed variants info. |
57.1% | B+ | 42.9% |
| Task 10 – Locate variant history for reviewed variant |
85.7% | B+/B | 57.1% |
| Task 11 – Conduct patient search by variant |
85.7% | B | 71.4% |
| Task 12 – Conduct a search for patients with unreviewed information |
85.7% | B+/B | 85.7% |
| Task 13 – Locate alert on an incidental variant LOW PRIORITY TASK |
14.3% | A/A− | 14.3% |
| Task 14 – Review GIC Alert Summary |
100% | B | 100% |
These results are calculated on 6 participants rather than 7 because the moderator failed to solicit a grade from the participant or the participants’ workflow prevented the capture of task time.
Error-free rate and completion rate ranged from 14.3% to 100%. Six out of the 14 tasks (Tasks 1, 2, 3, 7 and 14) had a 100% completion and error-free rate. All participants were able to complete the critical tasks of viewing a new patient report and a high level variant update efficiently and without error. Locating the laboratory’s data on the number of reports and families (Task 6) as well as locating alerts on incidental variants (Task 13) had the lowest task completion rate and error-free rate (14.3% each); however, none of this information was previously provided to clinicians prior to GIC and the value of the incidental variant information is debatable, which will be explained in more detail in the discussion.
Average grades on all tasks, reflecting ease of the task, ranged from B to A. Grades were assigned by each participant at the completion of each task. The moderator asked the participant to assign a grade based on how easy/difficult the task was to accomplish. The highest average grade was given to the task of locating the evidence for a variant update, which was information of high value to the users. The two tasks that received the lowest grades included locating patients with the same variant after reviewing the variant in a single initial patient (Task 11) and reviewing the PGE alert summary email (Task 14).
Based on completion rate and error-free rate, task 6, 8, 9 and 13 showed the greatest room for improvement. Task 8 required the user to mark a variant reviewed by locating and using the button on the Individual Reported Variant Interpretation History page (Figure 2). Task 9 involved locating information on all of a patient’s variants, including updates that had already been reviewed.
4. Discussion
We assessed the usability of a tool for helping clinicians following patients who have had genetic testing. The clinicians identified a number of opportunities to improve the tool which were relatively straightforward to implement, including the addition of ordering provider to variant and report alerts and clarification of some language and labeling. Other modifications, such as the revisions to the icon scheme, took more development effort. Participants were enthusiastic about the possibility of receiving updates on variant information proactively, reducing their need to call the lab a day before a patient visit to quickly seek this information. One participant stated “this tool is important for any disease management where genotype comes into play. If we do not integrate this information with patient care we will never learn how and when to integrate genetic information into medicine.”
We learned that an interface that makes it very clear to the clinician what new knowledge is available and the potential significance of that information for patient and family treatment is important in displaying genetic data. Our findings suggest that clinicians, when reviewing genetic knowledge, like to have the evidence that supports a change in a variant’s significance and a clear understanding of exactly what has changed. It provides them with more confidence in making appropriate decisions for the patients and family. The usability findings help illuminate issues that may hinder the clinician from locating the relevant information as efficiently and effectively as possible. The lessons learned during the usability tests informed enhancements to the application that support user’s optimal use of the tool and will continue to inform future development.
As a result of the usability testing, the development team made several modifications to the application. The current release includes changes to the clinic emails and the interface that now specify the ordering provider. In the initial release authorized clinic users received alerts for all of the patients within their clinic, with no indication of which provider ordered each test. This was problematic for the busy clinicians who want to know immediately whether they need to be worrying about a variant update or particular test result on one of their patients. In the current release, the ordering providers are displayed for each case. In a future release, clinicians will be able to elect to receive alerts only for reports which list them as an ordering provider, as opposed to alerts for all providers in a clinic. In addition, the summary email was reorganized with high alerts at the top of the email. Participants felt that the high variant update alerts were the most important items and should appear first in the summary email. Also, the new report alerts in the summary emails were revised and are now labeled as medium rather than high level alerts, which allow the user to easily distinguish the variant updates of high importance.
To assist GIC users in locating those patients that have important information to be reviewed, icons were used in the search results and on the Tests page. In the initial version of GIC, inconsistencies in the behavior and application of some of the icons with external conventions and among screens of the application led participants to misunderstand the process or status of the data presented. Our findings suggested that the initial design tested with users did not provide the participants with a clear indication on the search page of whether a variant update existed and/or whether it had been reviewed by someone at the clinic. In designing applications that introduce a new workflow or new functionality, careful consideration should be taken to understand the current process it will be supporting and how the design could alleviate some of the mental effort required of the clinician, allowing the clinician to focus on the information important for treating their patients. In an environment where more than one person could potentially review one variant update, using the system to aid the clinician in recognizing those updates easily from the search results is important. As a result of this study, modifications were made to the icons used throughout the application in their design, placement and behavior. They were revised to more accurately and consistently reflect their meaning.
Additional modifications made to the application include small changes to the labels on the review buttons, and the counts of cases and families reported with a variant. Some language used in the application, such as the “reported and families counts”, was a point of confusion for some of the participants. This could be due, in part, to the fact that GIC was presenting this information in a more structured way than it was presented in written laboratory reports from the lab prior to the application. Clinicians were familiar with finding that type of information in the evidence for the change in variant category where additional context was provided. Participants during the usability test were not looking for a structured field for this and when asked about the counts they were not always sure whether the numbers reflected the lab counts or counts found in the literature or both. The findings from the usability test indicate that providing the right amount of detailed information and choosing the appropriate language is important, and could possibly reduce errors of interpretation. In this case, additional user research early in the development cycle may have been helpful. Providing this variant information in this format is new to the providers and therefore using familiar and clear language could help ease the transition and integrate the system into the providers’ current model. To achieve this, a label change was made to clarify that the reported and families count refers to those identified by the lab only.
Designing these applications to be scalable will be challenging but critical. Organizing and navigating this patient information so it can easily be interpreted by the clinician and shared with the patient will get more complex as the volume of testing increases and the number of variants per test increases. In this context it is important to identify and address aspects of the interface that can be distracting and not critical in supporting the user in accomplishing their main goal. In the version tested with users, incidental variants, which are not often reported by labs, are hidden by default on the Tests page and the user must click a link to reveal the incidental variant and any associated updates. Alerts can be associated with the incidental variants, but finding these alerts requires “un-hiding” the incidental variants. Some participants had trouble locating this information even though they were aware an update was available as indicated by the icon on the search page. The GIC was originally designed this way with the intention of reducing the risk that a clinician will misinterpret their significance. Especially since incidental variants are not clinically critical to convey, finding the balance between providing the clinician with all information versus limiting information to only that which is most clinically relevant is challenging. These findings have initiated a discussion within the GeneInsight team as to whether this functionality (low level alerts on incidental variants) should be removed from the application. As the volume of test results and variant updates increases over time, the time clinicians are able to spend reviewing variants and reports will decrease. Identifying and addressing any distracting elements of the interface may assist the clinicians in accomplishing the more critical tasks.
The subjective grade given by participants after each task suggests that their perception of the application is different than its actual performance in some cases. In instances such as this, the value of the tool in many ways may influence the clinician’s subjective assessment of its usability. People may be more willing to accept small problems when the benefit of the application is much greater than the time and effort required to use it and the cost of not using the tool is high [31, 32]. Also, as a result of the interface design, in some cases the user is unaware that they have obtained incorrect information or misinterpreted something. For this reason, taking the time to conduct usability tests and observe users can highlight some issues that may not have been articulated by users otherwise. This, in turn, allows designers and developers to understand underlying problems that users might encounter with the application to help prioritize their development and enhancement efforts [11]. A number of issues encountered during the usability test prompted additional discussions among the design team and also brought to light broader considerations that would be helpful to address to optimize its functionality and usefulness for personalized medicine. The larger study includes surveys and interviews with the users to discuss more in depth the utility of the application and its use in practice.
In addition to user requirements identified in the usability tests, business and regulatory requirements, especially for this type of application, must be considered during development and implementation as well. Concerns related to patient privacy and confidentiality, and technology requirements limit the extent to which certain designs and functionality is possible. It may not always be possible to respond to user requests as a result of these requirements. Therefore, it is challenging but essential for the development team to be aware of and manage these other elements as well, in order to successfully reach the goals for the GeneInsight Suite.
While usability testing is critical, combining it with other user research methods to understand additional facets of the user experience with the application is just as important. To understand the full picture of the utility, usefulness, and integration into clinical practice, conducting user research early and involving the user at each stage of the process is important for any new clinical application representing complex knowledge.
While the number of participants in this usability study was small, we studied a large proportion of the current users, and the usability issues identified provided significant insight into the user’s interactions with the GIC interface and most likely represent the majority of total issues [33]. Formative usability testing is useful to conduct on lower fidelity prototypes and changes incorporated before implementation of applications to save development and testing time and effort. The development team made many modifications as a result of the version one usability tests and the research team recently completed additional usability tests on the current version. As personalized medicine evolves, the ideal tool may require additional research and enhancements to support the needs of the clinicians.
5. Conclusions
Participants in usability studies encountered a number of usability issues with the first version of this novel application. However, they were impressed with the potential value of the tool in forwarding significant genetic variant updates immediately and electronically to the clinicians. In order to ensure effective use of the application, many of the issues identified during usability testing sessions were addressed in the most recent version of the software. This analysis will also inform future development as well. Usability evaluations are invaluable to the success of technology in an emerging area, especially in a complex domain such as genetics. With the potential volume and scope of personalized medicine, in addition to the burden already placed on clinicians to document and manage their patients’ information, designing tools that can alleviate mental load and automate an increasingly challenging process becomes more critical.
Highlights.
We evaluated an innovative application for communicating genetic results.
Application allows users to access knowledge updates to reported variants.
Optimizing accessibility and clinical relevance of genetic data is essential.
Incorporating user focused design can optimize value of application.
Providers felt the application had high value for delivering clinical care.
Supplementary Material
Acknowledgements
The National Library of Medicine, National Institutes of Health (RC1LM010526) provided financial support for the conduct of the research and preparation of this manuscript. The content is the responsibility of the authors and does not necessarily represent the official views of the National Library of Medicine or the National Institutes of Health.
We would also like to acknowledge Sara Samaha for assisting with the preparation of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The GeneInsight Clinic receives messages from GeneInsight Lab software. GeneInsight Lab is made available to outside laboratories. There is a fee associated with this service which supports the development and maintenance of the software.
References
- 1.Scheuner MT, de Vries H, Kim B, Meili RC, Olmstead SH, Teleki S. Are electronic health records ready for genomic medicine? Genet Med. 2009;11:510–517. doi: 10.1097/GIM.0b013e3181a53331. [DOI] [PubMed] [Google Scholar]
- 2.Hamburg MA, Collins FS. The path to personalized medicine. N Engl J Med. 2010;363:301–304. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 3.Lee C, Morton CC. Structural genomic variation and personalized medicine. N Engl J Med. 2008;358:740–741. doi: 10.1056/NEJMcibr0708452. [DOI] [PubMed] [Google Scholar]
- 4.Caleshu C, Day S, Rehm HL, Baxter S. Use and interpretation of genetic tests in cardiovascular genetics. Heart. 2010;96:1669–1675. doi: 10.1136/hrt.2009.190090. [DOI] [PubMed] [Google Scholar]
- 5.Genetic Testing Practice Guidelines: Translating Genetic Discoveries into Clinical Care. Genetics & Public Policy Center. 2008 [Google Scholar]
- 6.Aronson SJ, Clark EH, Babb LJ, Baxter S, Farwell LM, Funke BH, Hernandez AL, Joshi VA, Lyon E, Parthum AR, Russell FJ, Varugheese M, Venman TC, Rehm HL. The GeneInsight Suite: a platform to support laboratory and provider use of DNA-based genetic testing. Hum Mutat. 2011;32:532–536. doi: 10.1002/humu.21470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baars MJ, Henneman L, Ten Kate LP. Deficiency of knowledge of genetics and genetic tests among general practitioners, gynecologists, and pediatricians: a global problem. Genet Med. 2005;7:605–610. doi: 10.1097/01.gim.0000182895.28432.c7. [DOI] [PubMed] [Google Scholar]
- 8.Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 9.Baroudi J, Olson M, Ives B. An empirical study of the impact of user involvement on system usage and information satisfaction. Commun. ACM. 1986;29:232–238. [Google Scholar]
- 10.Rosenbaum S, Wilson CE, Jokela T, Rohn JA, Smith TB, Vredenburg K. Usability in Practice: user experience lifecycle - evolution and revolution, CHI '02 extended abstracts on Human factors in computing systems. Minneapolis, Minnesota, USA: ACM; 2002. pp. 898–903. [Google Scholar]
- 11.Gosbee JW. Conclusion: You need human factors engineering expertise to see design hazards that are hiding in "plain sight!". Jt Comm J Qual Saf. 2004;30:696–700. doi: 10.1016/s1549-3741(04)30083-3. [DOI] [PubMed] [Google Scholar]
- 12.Rogers ML, Patterson E, Chapman R, Render M. Usability Testing and the Relation of Clinical Information Systems to Patient Safety. In: Henriksen K, Battles JB, Marks ES, Lewin DI, editors. Advances in Patient Safety: From Research to Implementation. Volume 2. Rockville (MD): Concepts and Methodology; 2005. [PubMed] [Google Scholar]
- 13.Ash JS, Sittig DF, Poon EG, Guappone K, Campbell E, Dykstra RH. The extent and importance of unintended consequences related to computerized provider order entry. J Am Med Inform Assoc. 2007;14:415–423. doi: 10.1197/jamia.M2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bates DW, Kuperman GJ, Wang S, Gandhi T, Kittler A, Volk L, Spurr C, Khorasani R, Tanasijevic M, Middleton B. Ten commandments for effective clinical decision support: making the practice of evidence-based medicine a reality. J Am Med Inform Assoc. 2003;10:523–530. doi: 10.1197/jamia.M1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kushniruk AW, Patel VL. Cognitive and usability engineering methods for the evaluation of clinical information systems. J Biomed Inform. 2004;37:56–76. doi: 10.1016/j.jbi.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Armijo D, McDonnell C, Werner K. U.S.D.o.H.a.H. Services, editor. Rockville, MD: AHRQ Publications. 2009. Electronic health record usability: evaluation and use case framework. [Google Scholar]
- 17.Karsh BT, Weinger MB, Abbott PA, Wears RL. Health information technology: fallacies and sober realities. J Am Med Inform Assoc. 2010;17:617–623. doi: 10.1136/jamia.2010.005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schumacher R, Lowry S. In: NIST Guide to the Processes Approach for Improving the Usability of Electronic Health Records. U.S.D.o. Commerce, editor. National Institute of Standards and Technology; 2010. [Google Scholar]
- 19.Saleem JJ, Russ AL, Sanderson P, Johnson TR, Zhang J, Sittig DF. Current challenges and opportunities for better integration of human factors research with development of clinical information systems. Yearb Med Inform. 2009:48–58. [PubMed] [Google Scholar]
- 20.Smelcer J, Miller-Jacobs H, Kantrovich L. Usability of Electronic Medical Records. Journal of Usability Studies. 2009;4:70–84. [Google Scholar]
- 21.Aronson SJ, Clark EH, Varugheese M, Baxter S, Babb LJ, Rehm HL. Communicating New Knowledge on Previously Reported Genetic Variants. Genet Med. 2012 doi: 10.1038/gim.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jordan DM, Kiezun A, Baxter SM, Agarwala V, Green RC, Murray MF, Pugh T, Lebo MS, Rehm HL, Funke BH, Sunyaev SR. Development and validation of a computational method for assessment of missense variants in hypertrophic cardiomyopathy. Am J Hum Genet. 2011;88:183–192. doi: 10.1016/j.ajhg.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.GeneInsight Executive Summary. Partners HealthCare System, Inc.; 2010. [Google Scholar]
- 24.Nielsen J. Heuristic Evaluation. In: Nielsen J, Mack RL, editors. Usabililty Inspection Methods. New York, NY: John Wiley & Sons; 1994. [Google Scholar]
- 25.Morae, Okemos, MI: TechSmith Corporation; [Google Scholar]
- 26.Jaspers MW, Steen T, van den Bos C, Geenen M. The think aloud method: a guide to user interface design. Int J Med Inform. 2004;73:781–795. doi: 10.1016/j.ijmedinf.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Dumas JS, Redish J. A practical guide to usability testing, Rev. ed., Exeter, England: Intellect Books; Portland, Or. 1999. [Google Scholar]
- 28.Brooke J. SUS: A ―quick and dirty usability scale. In: Jordan PW, Thomas B, Weerdmeester BA, McClelland, editors. Usability Evaluation in Industry. London, UK: Taylor & Francis; 1996. pp. 189–194. [Google Scholar]
- 29.Bangor A, Kortum P, Miller J. Determining What Individual SUS Scores Mean: Adding an Adjective Rating Scale. Journal of Usability Studies. 2009;4:114–123. [Google Scholar]
- 30.Sauro J. Measuring Usability with the System Usability Scale (SUS) 2011 [Google Scholar]
- 31.Nielsen J, Levy J. Measuring usability: preference vs. performance. Commun. ACM. 1994;37:66–75. [Google Scholar]
- 32.Bailey R. Performance Vs. Preference. Human Factors and Ergonomics Society Annual Meeting Proceedings. 1993;37:282–286. [Google Scholar]
- 33.Virzi RA. Refining the test phase of usability evaluation: how many subjects is enough? Hum. Factors. 1992;34:457–468. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.