Abstract
Azotobacter vinelandii is a well-studied model system for nitrogen fixation in bacteria. Regulation of nitrogen fixation in A. vinelandii is independent of NtrB/NtrC, a conserved nitrogen regulatory system in proteobacteria. Previous work showed that an ntrC mutation in A. vinelandii resulted in a loss of induction of assimilatory nitrate and nitrite reductases encoded by the nasAB operon. In addition to NtrC, several other proteins, including NasT, a protein containing a potential RNA-binding domain ANTAR (AmiR and NasR transcription antitermination regulators), have been implicated in nasAB regulation. In this work, we characterize the sequence upstream of nasA and identify several DNA sequence elements, including two potential NtrC binding sites and a putative intrinsic transcriptional terminator upstream of nasA that are potentially involved in nasAB regulation. Our analyses confirm that the nasAB promoter, PnasA, is under NtrC control. However, unlike NtrC-regulated promoters in enteric bacteria, PnasA shows high activity in the presence of ammonium; in addition, the PnasA activity is altered in the nifA gene mutation background. We discuss the implication of these results on NtrC-mediated regulation in A. vinelandii. Our study provides direct evidence that induction of nasAB is regulated by NasT-mediated antitermination, which occurs within the leader region of the operon. The results also support the hypothesis that NasT binds the promoter proximal hairpin of nasAB for its regulatory function, which contributes to the understanding of the regulatory mechanism of ANTAR-containing antiterminators.
INTRODUCTION
Azotobacter vinelandii is a well-studied Gram-negative, aerobic diazotroph. Expression of nitrogen fixation genes in A. vinelandii is tightly regulated by NifA, a transcriptional activator under the control of NifL, an antiactivator protein that detects the cellular nitrogen level and redox state (15, 31). In addition, NifL is a receptor of the nitrogen signal from GlnK, a PII-like protein (24, 36, 37, 50, 57) that is modulated by the global nitrogen sensor protein GlnD (13, 39, 53), a bifunctional uridylyltransferase/UMP-removing (UTase/UR) enzyme (27, 37).
A. vinelandii encodes NtrB/NtrC (58), a conserved two-component regulatory system in proteobacteria involved in the control of nitrogen gene expression. In enteric bacteria, NtrB/NtrC, GlnK, and GlnD constitute a global nitrogen regulatory system (41). As a transcriptional activator, NtrC directly controls the expression of the genes related to nitrogen assimilation and metabolism. The activity of NtrC is regulated by NtrB (29), a histidine kinase under the direct control of the PII protein (45). When the cellular nitrogen status is low, NtrC is phosphorylated by NtrB (26, 28). Phosphorylated NtrC activates transcription of target nitrogen genes. In Klebsiella pneumoniae, NtrC tightly regulates the expression of nifLA (17, 40), which in turn regulates nif gene transcription in response to oxygen and the nitrogen signal from GlnK (8, 15, 33). However, in A. vinelandii, nifLA is expressed constitutively (7), independent of NtrB/NtrC (58).
Although ntrC is not involved in nif regulation in A. vinelandii, mutational analyses showed that a functional ntrC is required for synthesis of assimilatory nitrate/nitrite reductases (58), encoded by the nasAB operon (48). In addition to NtrC, nasAB expression requires σ54 (52, 58), an alternative sigma factor required by NtrC-regulated promoters. It was also shown that nasAB expression was repressed in the presence of ammonium and induced by the presence of nitrate or nitrite. These observations led to the hypothesis that NtrC in A. vinelandii regulates nasAB expression analogously to NtrC-mediated regulation in enteric bacteria (48). However, this hypothesis disagrees with an earlier observation that the promoter of nifLA from K. pneumoniae, which is regulated solely by NtrC, was highly expressed in A. vinelandii in the presence of high concentrations of ammonium (30).
An earlier study identified the gene products of a second operon, nasST, located 10 kb upstream of nasAB and involved in nasAB induction (21). The results of mutational analysis suggest that NasT plays a positive role, while NasS plays a negative role, in nasAB regulation. The predicted NasT protein contains a putative RNA-binding domain that is conserved in several antiterminators, suggesting that the protein may have antitermination functions (19, 35, 43, 56).
The results from earlier studies also suggest that nasB has a negative autoregulatory role in operon induction, which is evidenced by the finding that nasAB exerts high expression when nasB is mutated even in the absence of nitrate/nitrite induction (21, 48). Adding to the complexity of nasAB regulation is the possibility of competition for molybdenum between nitrogenase and nitrate reductase (NasB), which in turn affects nasAB regulation (22).
The aim of the present study was to elucidate the molecular mechanism of nasAB induction. We identified the promoter of nasAB and characterized its properties. Our results demonstrate that NasT-mediated antitermination plays the essential role in regulation of nasAB.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table S1 in the supplemental material. A. vinelandii UW136 and its derivatives were grown at 30°C in modified Burk's nitrogen-free salts medium (44) supplemented with 1% sucrose (BS). When needed, BS medium was supplemented with the following fixed nitrogen sources: ammonium acetate, 15 mM; urea, 10 mM; NaNO2, 5 mM; KNO3, 10 mM. Escherichia coli DH5α was grown on Luria-Bertani broth or agar medium at 37°C. Media were supplemented with antibiotics where appropriate: for A. vinelandii, carbenicillin (20 μg ml−1) and gentamicin (0.05 μg ml−1); for E. coli, carbenicillin (50 μg ml−1), chloramphenicol (34 μg ml−1), and gentamicin (15 μg ml−1).
Oligonucleotides.
The oligonucleotides used in this study were purchased from Integrated DNA Technologies, Inc. (Coralville, IA) (see Table S2 in the supplemental material).
Bioinformatics.
DNA secondary structure analysis was performed using Mfold (60); the putative σ54 binding site and NtrC binding sites were identified using PromScan (http://molbiol-tools.ca/promscan/). The sequence of the A. vinelandii genome is available at the website http://www.azotobacter.org/. A protein sequence BLAST search against the Conserved Domain Database (CDD) was performed at the NCBI website, http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi. The sequence alignment was created using the emma program of the EMBOSS (European Molecular Biology Open Software Suite) package and manipulated in GeneDoc (http://www.psc.edu/biomed/genedoc).
DNA manipulation.
Plasmid isolations were carried out using the GeneJET Plasmid miniprep kit (Fermentas, Glen Burnie, MD). Restriction enzyme digestions, ligations, cloning, and DNA electrophoresis were performed using standard protocols (51). DNA fragments were purified from agarose gels using the QIAquick gel extraction kit (Qiagen, Valencia, CA).
Transformations.
For general cloning, plasmids were transformed into chemically competent E. coli DH5α (51). A. vinelandii transformations were performed on competence medium as described previously (3).
Cloning the nasAB promoter and surrounding sequence.
A. vinelandii UW136 genomic DNA was isolated and purified as described previously (49). Purified DNA was digested with XhoI and separated by agarose gel electrophoresis. DNA fragments of approximately 7.8 kb were excised from the gel, purified, and ligated into the XhoI site of pBluescript II KS(+). After transformation into E. coli DH5α, positive clones were identified by colony PCR using primers P35 and P36 (see Table S2 in the supplemental material). The plasmid pWB30 was sequenced using primers P26 and P27 (see Table S2) to confirm the cloned sequence.
For colony PCR in this study, GoTaq Green Master Mix (Promega, Madison, WI) was used, and the PCR was carried out as follows: 92°C for 2 min, then 30 cycles (92°C for 1 min, 55°C for 1 min, and 68°C for 1 min for extension) for amplification, and 1 extension step at 72°C for 5 min.
Construction of the ntrC deletion mutant.
A 2.4-kb fragment containing ntrC was amplified from the A. vinelandii chromosome using primer pairs P1 and P2 (see Table S2 in the supplemental material) and cloned into the pGEM-T vector (Promega). The resulting plasmid was linearized with SacI, blunt-ended using T4 DNA polymerase, and self-ligated to yield pWB680. PCR amplification of this plasmid was performed using primer pair P3 and P4 (see Table S2), and the PCR product was digested with SacI and ligated with the SacI-Gm-SacI cassette from pTnMod-OGm, giving rise to pWB685. The orientation of the Gmr cassette with respect to ntrC in pWB685 was confirmed by sequencing. Plasmid pWB685 was transformed into A. vinelandii, and a carbenicillin-sensitive and gentamicin resistance transformant was selected on BS medium supplied with ammonium acetate (BSN). The ΔntrC::Gm allelic replacement mutation in the transformant was confirmed by colony PCR using the primer pair P1 and P2 (see Table S2).
Construction of lacZ fusion reporters for A. vinelandii.
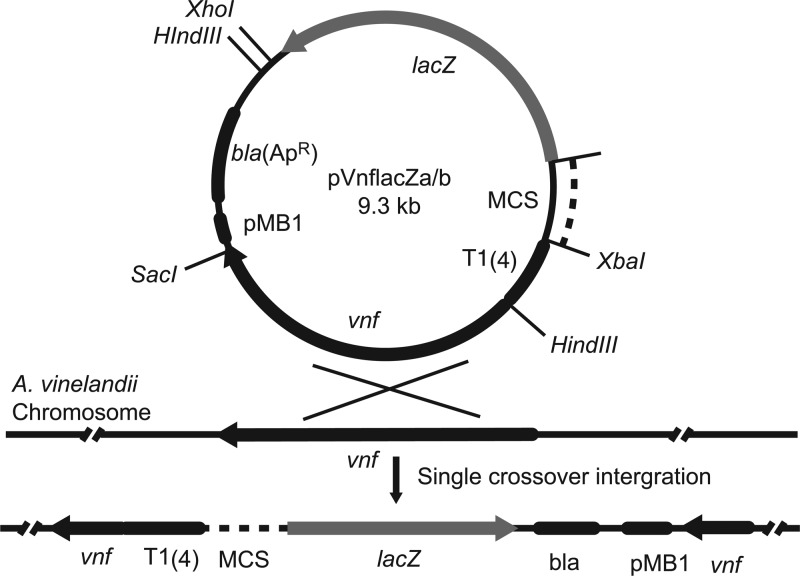
Transcriptional and translational lacZ fusion probes for A. vinelandii were constructed, as described below, such that the two plasmids differed primarily in the multiple cloning sites (MCSs) preceding the lacZ start codon (see Fig. 2).
Fig 2.
Map of the A. vinelandii lacZ reporter vectors. Each vector can be integrated into the vnf region of the A. vinelandii genome to provide stable single-copy expression data. Plasmid pVnflacZa is a translational fusion probe containing the XbaI-BcuI-BamHI cloning site, and pVnflacZb is a transcriptional fusion probe containing the XbaI-BcuI-BamHI-XbaI site followed by an SD sequence 8 nucleotides upstream of the lacZ. MCS, multiple cloning sites; pMB1, replicon region; T1(4), four tandem copies of the T1 terminator from the E. coli rrnB1 operon (42); vnf, vanadium(V)-containing nitrogenase gene (6).
To construct the backbone of the probes, the β-lactamase (bla) gene and the pMB1 replicon were amplified from the plasmid pBluescript II KS(+) using the primer pairs P16 and P17 (see Table S2 in the supplemental material). The PCR product was gel purified and self-ligated, giving rise to the plasmid pWhite. A 2.8-kb vnf sequence was amplified from the plasmid pJW1 using primer pairs P33 and P34 (see Table S2), digested with SacI and XhoI, and cloned into the SacI-SalI sites of pWhite, giving rise to the plasmid pWvnf.
For the translational fusion probe, a fragment containing four tandem copies of the E. coli transcriptional terminator rrnB1 was amplified from pPROBE-NT using primer pairs P8 and P9 (see Table S2 in the supplemental material) and cloned into the SphI site of pIC20H using blunt-end ligation to create pICT4. The 3-kb BamHI-lacZ-XhoI fragment was released from the plasmid pSUP102::Tn5-B21 using BamHI and XhoI and cloned into the BamHI-XhoI sites of pBluescript II KS(+), yielding the plasmid pBlue-lacZ. The XbaI-lacZ-XhoI fragment containing the MCS was removed from pBlue-lacZ and cloned into the XbaI-XhoI sites of pICT4, creating the probe cassette in pIC-lacZ. pIC-lacZ was digested by HindIII, and an ∼4-kb HindIII-rrnB1-lacZ-HindIII fragment was cloned into the HindIII site of pWvnf, leading to the probe plasmid pVnflacZa.
Plasmid pKT2lacZ was digested with BamHI and SalI, and the BamHI-lacZ-SalI fragment, containing a Shine-Dalgarno (SD) sequence, was cloned into the BamHI-XhoI site of pVnflacZa, leading to the transcriptional fusion probe pVnflacZb.
Successful insertion of foreign sequences into both probe plasmids was verified by PCR using primers P14 and P25 (see Table S2 in the supplemental material), which amplified the region between the rrnB1 terminators and the 70th bp in the lacZ open reading frame (ORF).
Construction of site-directed and deletion mutations.
Site-specific substitution and deletion mutations were constructed using the overlap extension method based on two rounds of PCR (25). In the first round, two fragments were amplified from the wild-type sequence using two pairs of primers: one of each complementary primer pair contained the desired mutation. In the second round, the two products of the first reaction were used as templates. The amplification was performed with two external primers to create a PCR product containing the desired mutation that was the fusion of two fragments from the first round of PCR. PCRs were carried out with Pfu DNA polymerase (Stratagene, La Jolla, CA) as follows: 95°C for 1 min, then 25 cycles of 92°C for 1 min, 55°C for 1 min, and 72°C for 1 min, followed by an extension step at 72°C for 5 min.
Construction of plasmids for expression analysis in E. coli MC1061.
nasT was amplified from pMAS20 using primer pairs P37 and P38 (see Table S2 in the supplemental material). The PCR product was digested with EcoRI and BglII and cloned into the EcoRI-BamHI site of the plasmid pDK6 to construct pDK6-T. Using a similar strategy, pDK6-S was assembled by amplifying nasS from pMAS20 using primer pairs P39 and P40 (see Table S2) and cloning into pDK6.
The lacZ reporter plasmid pBTW was engineered from plasmid pBT. The DNA fragment containing the chloramphenicol resistance gene, p15A replicon, and lacUV5 promoter was amplified from pBT using primer pairs P41 and P42 (see Table S2 in the supplemental material) and digested with NotI and XhoI. The resulting fragment was linked with the NotI-lacZ-XhoI cassette from the plasmid pblue-lacZ to generate pBTW. The tested DNA sequence can be cloned into the NotI-BamHI region of pBTW for expression analysis.
The DNA fragment containing the nasA leader extending into the 162nd nucleotide of nasA was amplified from the plasmid pWB552 using primer pairs P6 and P43 (see Table S2 in the supplemental material) and cloned into the NotI-BamHI region of pBTW to construct pBW555. Using a similar strategy, DNA fragments were amplified from plasmids pWB650, pWB664, and pWB668 and cloned into pBTW, respectively, to create pBW901, pBW902, and pBW910.
β-Galactosidase assay.
β-Galactosidase activity in E. coli was assayed as described by Miller in 1972 (51). β-Galactosidase activity in A. vinelandii was assayed as following: cultures (2 ml) were centrifuged (3,000 × g) and rinsed once with 1 ml Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 10 mM dithiothreitol), cell pellets were dissolved in 1 ml Z buffer, and 0.2 ml of cell solutions was used for β-galactosidase assays as described previously, and results were reported in Miller units (59).
RESULTS
Characterization of the sequence upstream of the nasA start codon.
Genome annotation indicated that the nasAB operon consists of four ORFs (54) instead of two as identified previously (48). For consistency, we refer to these 4 ORFs as the nasAB operon. One newly identified ORF (Avin23360) is located between nasA and nasB, and the other (Avin23340) is located downstream of nasB. These two ORFs are named nasC and nasH, respectively. Amino acid similarity analysis (BLASTP) suggested that the predicted proteins NasC and NasH are homologous to subunits of nitrite reductase (47.57% identity) and uroporphyrinogen-III methylase (52.77% identity), respectively, which might be involved in synthesis of the prosthetic group of nitrite reductase. Compared to nasA, which is 2,448 nt in length, both nasC and nasH are small (324 nt and 735 nt, respectively).
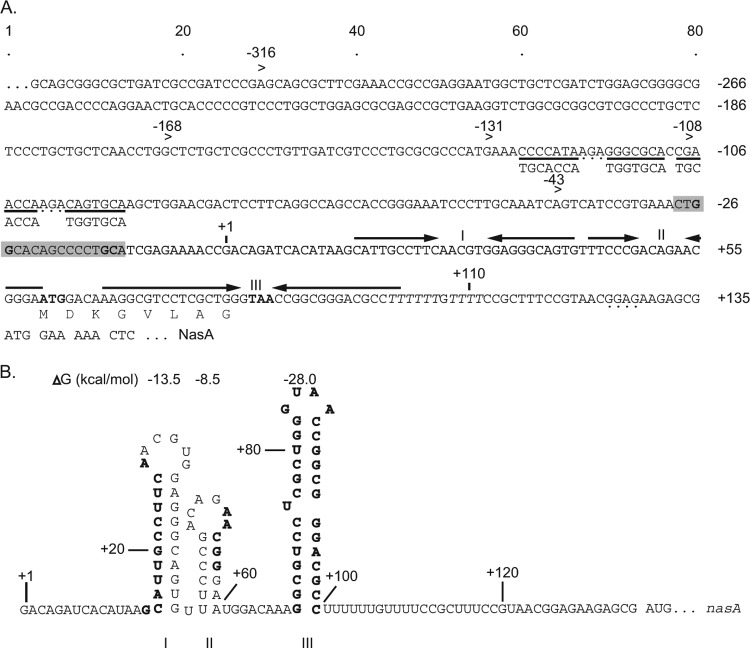
The sequence around the NasA start codon was determined originally by Maria Tortolero and verified by the A. vinelandii genome sequence (54). Using PromScan software (http://molbiol-tools.ca/promscan/), we identified a potential σ54 binding sequence (5′-CTGGCACAGCCCCTGCA-3′) (conserved nucleotides are underlined) (5) 146 nt upstream of the nasA start codon (Fig. 1A). Two regions of dyad symmetry homologous to the consensus NtrC binding sequence 5′-TGCACCNNNTGGTGCA-3′ (18) were identified 80 nt upstream of the σ54 binding site. The arrangement of the predicted σ54 and NtrC binding sites closely resembles the organization of NtrC-regulated promoters in enteric bacteria (32).
Fig 1.
(A) Nucleotide sequence from 477 bases upstream of nasA to the beginning of the NasA ORF. The predicted transcription start site is marked with +1; the putative σ54-binding sequence is shown in a light gray box with the conserved GG/GC bases in bold; the putative NtrC protein binding sites are underlined, with the consensus NtrC binding sequence shown underneath for comparison; inverted repeats are above the specific sequences with facing arrows; the start and stop codons of the putative octapeptide ORF in the leader region are in boldface. The predicted poly(T) sequence adjacent to hairpin III is italicized. The 8 codons of the putative octapeptide sequence are indicated, and the putative NasA Shine-Dalgarno (SD) sequence has a dashed underline. (B) Predicted secondary palindromic structures within the nasAB leader sequence. RNA sequence numbers are relative to the predicted transcription start site; the free energies (kcal/mol) of the predicted hairpins are indicated at the top of hairpins; the letters in boldface mark the sequences deleted in the mutant constructs.
Transcription initiation of σ54-dependent promoters usually begins 12 nt downstream of the σ54 binding site followed closely by the translation initiation region (4). However, the sequence between the predicted transcriptional initiation site and the nasA start codon is 135 nt, raising the possibility that additional promoters exist upstream of nasA. Bioinformatic analysis of this region did not identify other potential sigma factor binding sites. Another possibility is that the nasAB transcript has a long 5′ leader sequence with potential regulatory function. Posttranscriptional regulation of many bacterial operons occurs within the 5′ end leader transcript regions and is often used to fine-tune gene expression. Analysis in support of this revealed several potential regulatory features upstream of nasA.
RNA secondary structure prediction by Mfold (60) revealed three potential hairpin structures located between the putative σ54 binding site and the nasA start codon (Fig. 1A and B). Hairpin I has a 10-bp stem and a hexanucleotide AACGTG loop (ΔG = −13.5 kcal/mol), and an adjacent hairpin II has a 6-bp stem and a hexanucleotide ACAGAA loop (ΔG = −8.5 kcal/mol). These two hairpins are separated by one T nucleotide. Hairpin III has a high GC content (ΔG = −28 kcal/mol) and is situated immediately upstream of a poly(T) sequence 25 nt upstream of the nasA start codon. The combination of the hairpin and poly(T) sequence resembles an intrinsic transcriptional terminator structure. This suggested the possibility that antitermination is involved in nasAB regulation.
In addition to the three potential hairpin structures, we identified a small putative ORF encoding the octapeptide MDKGVLAG within this leader region. The small ORF begins at the last base of the middle hairpin II and ends within the loop region of the terminator hairpin (Fig. 1A).
Construction of LacZ probes specific for A. vinelandii.
A. vinelandii lacks β-galactosidase, enabling lacZ to be used as an expression reporter. Previously, lacZ was integrated into the genomic copy of nasA or nasB for expression studies (48). Using the reporter integrated into the genome of bacteria instead of within a plasmid in trans ensures the consistency of the gene copy number. However, integration of the reporter gene into the nasAB structural genes may disrupt normal operon expression. Previous work showed that lacZ integrated into nasB resulted in higher expression of nasB even in the absence of nitrate induction (21, 48). Based on these considerations, we designed lacZ reporters that can be integrated into the A. vinelandii genome, but outside the nasAB region, via homologous recombination. For convenience, we selected the vnf locus for reporter plasmid integration. The vnf genes encode an alternative nitrogenase that uses vanadium as the metal cofactor (6). In addition, vnf is not essential for bacterial viability, and its expression is inhibited in the presence of molybdenum (6), which was included in the medium used in these studies.
We constructed two reporter plasmids, pVnflacZa and pVnflacZb, for analysis of nasAB expression (Fig. 2). Plasmid pVnflacZa is a translational fusion vector used to determine the effects of mutations within the region identified upstream of nasA on translation of nasAB. This region with Shine-Dalgarno sequence was cloned into the MCS XbaI-BcuI-BamHI of pVnflacZa to form an in-frame translational fusion with the LacZ ORF. The lacZ sequence begins with ATGGATCC, where GGATCC is a BamHI restriction site. Plasmid pVnflacZb is a transcriptional fusion vector used to measure promoter activity. This plasmid contains the MCS site XbaI-BcuI-BamHI-XbaI followed by an SD sequence (5′-AGGAGGT-3′), located 8 nt upstream of the lacZ start codon. For both plasmids, a four tandem terminator rrnB1 sequence T1(4) was inserted upstream of MCSs to block background lacZ expression (42).
In addition to the ∼2.8-kb vnf sequence, each plasmid contains a pMB1 replicon for propagation in E. coli and a bla gene (encoding β-lactamase) for ampicillin or carbenicillin selection in E. coli and A. vinelandii. We found that the integrated plasmids were stably maintained within the A. vinelandii genome for more than 30 generations even in the absence of antibiotic selection (data not shown).
Time course expression of the translation fusion nasA′-′lacZ in A. vinelandii.
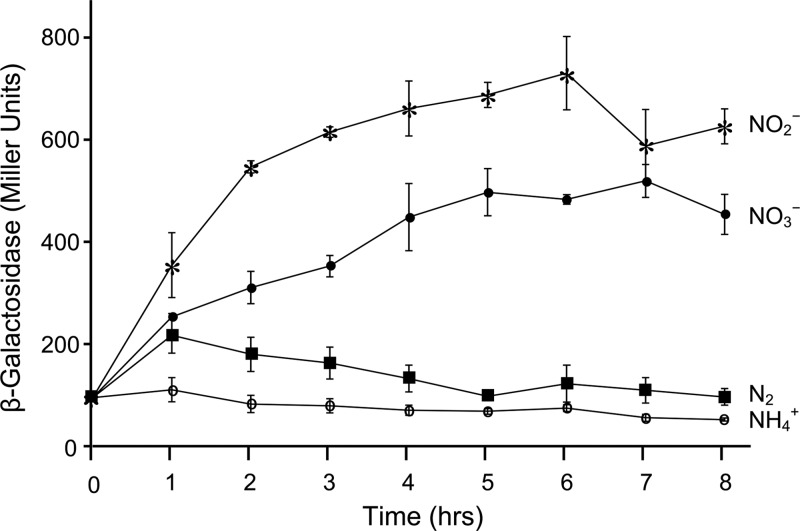
A 613-nt sequence starting with nucleotide 190 upstream of the potential NtrC binding sites and ending with the 162nd nucleotide of the nasA gene was selected for further analysis. This fragment was cloned into the XbaI-BamHI region of pVnflacZa, resulting in plasmid pWB552 with an in-frame fusion between nasA′ and ′lacZ. One transformant with pWB552 integrated in the genome in the vnf locus was selected for lacZ expression analysis. The transformant was grown in medium containing different nitrogen sources. At multiple time points, culture samples were harvested and analyzed for β-galactosidase activity (Fig. 3).
Fig 3.
Time course of the expression of the translational nasA′-′lacZ fusion integrated into the vnf region of the A. vinelandii genome. nasA′ in the fusion starts with the nucleotide 190 upstream of the potential NtrC binding site and ends with the 162nd nucleotide of the NasA ORF. Supplied nitrogen sources were nitrite (∗), nitrate (•), dinitrogen (▪), and ammonium (∘). Bacteria were grown on BS medium supplemented with the indicated nitrogen sources. At the times indicated, samples were taken and assayed for β-galactosidase expression.
In the presence of ammonium, the cellular β-galactosidase activity remained at background levels throughout growth. In contrast, in the presence of nitrate or nitrite, β-galactosidase activity increased over time and reached maximal levels after 6 h, in agreement with the expression profile of lacZ integrated into nasA published previously (38, 48, 58). In the absence of fixed nitrogen sources, A. vinelandii can utilize atmospheric N2 for growth. Under these conditions, β-galactosidase activity remained at background levels, indicating nasAB translation is low even in the absence of ammonium.
Induction of nasAB by nitrate and nitrite occurs via antitermination.
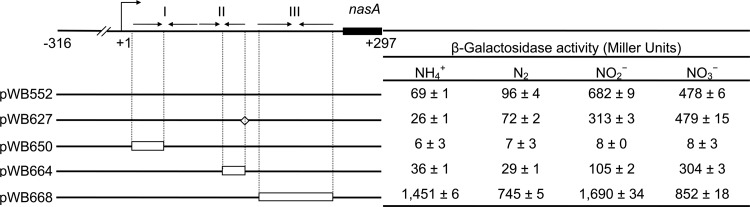
After confirming that the nasAB sequence selected is suitable for studies of nasAB regulation, we tested whether the predicted transcriptional terminator structure upstream of nasA is involved in operon regulation. We constructed a hairpin III deletion mutant, which has the entire hairpin III deleted (Fig. 1B and 4). The hairpin III deletion sequence was then cloned into the XbaI-BamHI region of pVnflacZa to construct a translation fusion. The new construct differed from pWB552 only by the loss of hairpin III. Analysis of β-galactosidase activity demonstrated that deletion of hairpin III resulted in increased lacZ expression under all conditions tested, indicating that hairpin III has a negative role in nasAB regulation. This result is consistent with the hypothesis that hairpin III and the poly(T) sequence immediately downstream of it constitute an intrinsic transcription terminator. However, the observation of high β-galactosidase activity in the presence of ammonium contrasts with the assumption that the promoter of nasAB is subjected to ammonium repression, which is mediated by NtrC. When introduced into a ntrC null mutant, the translational fusion showed trace amounts of expression under all nitrogen conditions tested, suggesting that the nasAB promoter is under NtrC regulation (Table 1).
Fig 4.
Effects of deletions and modifications within the leader sequence on nasA′-′lacZ expression (not drawn to scale). The top line represents the DNA sequence from nt −316 to + 297 (the 162nd nucleotide of nasA); the bent arrow and +1 represent the direction of transcription and initiation site; arrows facing one another represent the palindromic sequences comprising hairpins I, II, and III; empty rectangles represent deleted sequences; the open diamond represents the ATG→TAG nonsense mutation. Cultures were grown in BS medium supplemented with the indicated nitrogen sources, and β-galactosidase activity was measured 7 h after inoculation. All data represent means of three replicates ± standard deviations from a representative experiment.
Table 1.
Expression of the translational nasA′-′lacZ fusion in A. vinelandii grown with different nitrogen sources
| Relevant genotypic characteristic |
β-Galactosidase activity (Miller units)b after 7 h in BS medium supplemented with: |
||||
|---|---|---|---|---|---|
| nasA′a | A. vinelandii | NH4+ | N2 | NO2− | NO3− |
| Wild typec | Wild type | 69 ± 1 | 96 ± 4 | 682 ± 9 | 478 ± 6 |
| Wild type | nifA mutation | 27 ± 1 | 72 ± 2 | 980 ± 12 | 1,262 ± 18 |
| nasA′ with GG/GC→AA/AT substitution in the σ54 binding site | Wild type | <5 | 5 ± 2 | 7 ± 1 | 5 ± 1 |
| Wild type | ΔntrC::Gmr | <5 | <5 | 23 ± 1 | <5 |
| nasA′ with hairpin III deletion | ΔntrC::Gmr | <5 | <5 | <5 | <5 |
nasA′ refers to the sequence of nt −316 to +297, which includes the nasAB promoter PnasA and the beginning of the NasA ORF.
All data are mean values of triplicate samples ± standard deviations from a representative experiment.
Refers to the sequence of the nasA region used in the fusions.
The use of antitermination to regulate assimilatory nitrate reductase operon induction was identified previously in Klebsiella oxytoca (pneumoniae) M5al (34). The leader region of the assimilatory nitrate reductase operon nasFEDCBA (referred to as nasF) in K. oxytoca (pneumoniae) M5al features a factor-independent terminator and one promoter-proximal hairpin required for antitermination (34). The promoter-proximal hairpin is the potential binding site of antiterminator protein NasR and plays an important role in nitrate/nitrite-induced antitermination (11, 12). Although no sequence similarity between the nasF leader sequence and the sequence upstream of nasA was identified, the independence of potential hairpins I and II from hairpin III (Fig. 1B) resembles that of the promoter-proximal hairpin and terminator hairpin within the leader of nasF. To determine whether disruption of predicted hairpins I and II structures altered nasAB expression, we constructed two hairpin deletion mutants. These two constructs contain partial deletions within hairpins I and II, respectively (Fig. 1B and 4). Secondary structure analyses indicated that the partial deletion in each hairpin did not significantly alter the potential free energy and formation of the adjacent hairpins. Each of the individual sequences with deletions was cloned into the XbaI-BamHI region of pVnflacZa to construct translation fusions, analogous to the construction of the hairpin III deletion mutant. β-Galactosidase activity analysis showed that disruption of hairpin I abolished lacZ expression under all tested nitrogen conditions, while disruption of hairpin II reduced lacZ expression when induced by nitrite but did not reduce expression to background levels (Fig. 4). These results suggest that both hairpins I and II play a positive role in nasAB induction.
The loss of lacZ expression in the hairpin I deletion in the presence of nitrate or nitrite could be due to disruption of nitrate/nitrite-induced antitermination or to disruption of the integrity of the nasAB promoter, which prevents transcription initiation. A separate mutant with all three hairpins deleted was constructed (pWB643) (Fig. 5). The deletion of all three hairpins resulted in high β-galactosidase activity detected under all tested nitrogen conditions, indicating that the integrity of nasAB promoter was retained when all three hairpins were deleted. Thus, the lack of lacZ expression of the hairpin I deletion is most likely due to disruption of nitrate/nitrite-induced antitermination.
Fig 5.
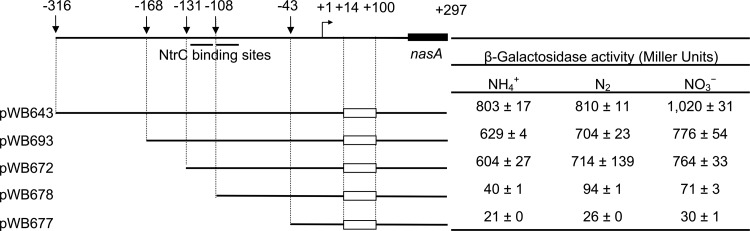
Effects of deletions upstream of the putative σ54 binding site on nasA′-′lacZ expression (not drawn to scale). pWB643 has all three hairpins (nt +14 to +100) within the leader region deleted. All other deletion mutants were constructed based on the sequence around the nasA promoter region in the plasmid pWB643. The top line represents the DNA sequence from nt −316 to + 297 (the 162nd nucleotide of nasA); the bent arrow and +1 represent the direction and initiation site of transcription; empty rectangles represent three-hairpin deletions within the leader region upstream of nasA; vertical arrows indicate the 5′ ends (−316, −168, −131, −108, and −43) of tested sequence mutants. β-Galactosidase activity assays were performed as described in the legend for Fig. 4.
For several bacterial operons involved in amino acid biosynthesis, translation of small ORFs located within the 5′ leader mRNA region is involved in ribosome-mediated antitermination (20, 23). To test whether the putative ORF located within the nasAB leader region behaved in an analogous manner, the start codon (ATG) of the 8-amino-acid peptide ORF was converted into a TAG stop codon, eliminating potential translation of this small ORF. The construct with the mutated leader region was cloned into pVnflacZa and introduced into A. vinelandii. β-Galactosidase assays showed that this mutation had lacZ expression similar to that of the wild-type control (Fig. 4), indicating that this putative small ORF had no role in nasAB regulation.
nasAB has an NtrC-dependent promoter.
To further define the position of the nasAB promoter(s), we constructed a series of 5′ end deletions upstream of the potential σ54 binding region in pWB643 and cloned them into the MCS of pVnflacZa (Fig. 1A and 5). β-Galactosidase assays showed that deletions ending at nucleotides −168 and −131, both of which maintained the putative NtrC binding sites, exhibited levels of β-galactosidase activity that were slightly lower than, but similar to, those of the control pWB643. A deletion that removed one putative NtrC binding site upstream of base −108 resulted in a 90% reduction in β-galactosidase activity, suggesting this sequence is directly involved in promoter function. Deletion to nucleotide −43, which removes the other putative NtrC binding site plus some downstream sequences, caused a 95% reduction in β-galactosidase activity compared to the full-length control. This result is similar to the deletion analysis of the nifLA promoter in K. pneumoniae, where deletions reaching nucleotide −28 led to a 93% reduction of promoter activity (16). In addition, replacement of the conserved dinucleotide pairs GG/GC with AA/AT in the potential σ54 binding site reduced β-galactosidase activity to background levels under all tested nitrogen conditions (Table 1), suggesting that the nasAB promoter requires σ54 for transcription. Taken together, we conclude that expression of nasAB is driven by an NtrC-regulated promoter, PnasA.
NasT in A. vinelandii is homologous to RNA-binding antiterminators AmiR and EutV.
NasT (GenBank accession no. CAA58582) consists of two domains (21): an N-terminal domain (amino acids 5 to 117), which is homologous to the REC (receiver) domain of the response regulator of two-component regulatory systems (43), and a C-terminal domain (amino acids 141 to 184), which is homologous to an RNA-binding domain, ANTAR (35, 56). The overall structure of NasT is homologous to that of AmiR (47) and EutV (14) (Fig. 6), two RNA-binding antiterminators identified in Pseudomonas aeruginosa and Enterococcus faecalis, respectively. Protein sequence homology analysis using the water program of EMBOSS indicated that NasT and AmiR share 47.4% similarity and 24.7% identity, while NasT and EutV share 58.3% similarity and 28.9% identity.
Fig 6.
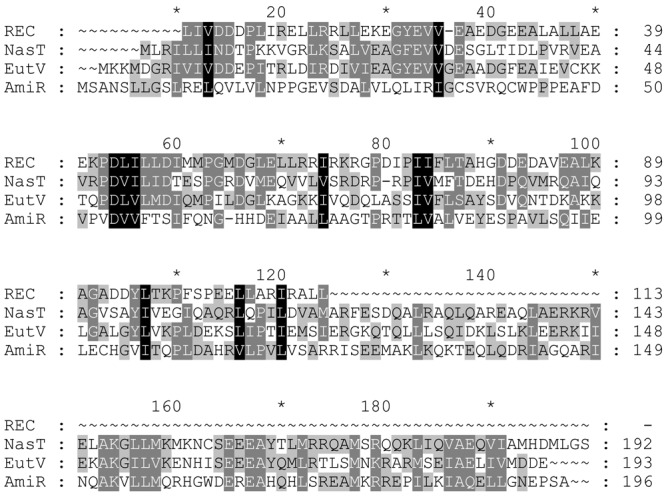
Sequence alignment of NasT, EutV, AmiR, and REC domain. The REC domain sequence (CDD no. cd00156) is from the Conserved Domain Database (CDD) at NCBI (National Center for Biotechnology Information). Protein sequences were obtained from NCBI protein databases: NasT (GenBank accession no. CAA58582), EutV (GenBank accession no. ZP_03948869) from Enterococcus faecalis TX0104, and AmiR (GenBank accession no. CAA32023) from Pseudomonas aeruginosa. − and ∼, gap in alignment; black background, conservation of nucleotides in all compared sequences; gray background, conservation in some compared sequences.
NasT is required for nasAB antitermination.
We constructed a bacterial heterologous expression system to test whether NasT functions within the leader region of nasAB. The system is comprised of a protein-expressing plasmid pDK6, a newly constructed lacZ reporter plasmid, and the E. coli strain MC1061, in which the lac operon has been deleted (10). pDK6 features a tac promoter upstream of an MCS and a lacIq gene, which prevents tac from being activated in the absence of the inducer isopropyl-β-d-thiogalactoside (IPTG) (1). For our purposes, we cloned nasT into the EcoRI-BamHI region of the pDK6 MCS. The lacZ reporter plasmid was derived from pBT, a component of the BacterioMatch II two-hybrid system (Stratagene). pBT is a low-copy-number plasmid in E. coli and features a lacUV5 promoter upstream of the λ cI gene. We replaced the cI gene in pBT with the lacZ reporter cassette, which contains an XbaI-BamHI cloning site preceding lacZ, resulting in the reporter plasmid pBTW. lacZ in the reporter cassette starts with ATGGATCC, where GGATCC is a BamHI restriction site. We cloned the nasAB leader sequence extending into the 162nd nucleotide of nasA into the XbaI-BamHI site of pBTW, resulting in the plasmid pBW555, which contains the translational fusion lacUV5-nasA′-′lacZ. In addition, we constructed three mutants with deletions at different hairpins of the leader region for expression comparison (Fig. 7).
Fig 7.
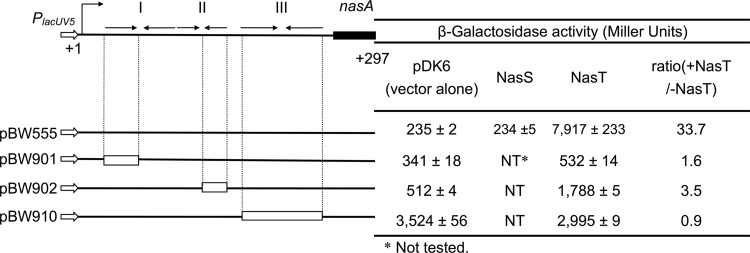
Analysis of NasT functions within the leader region of nasAB. The top line represents the DNA sequence from nt +1 to +297 (the 162nd nucleotide of nasA); the bent arrow and +1 represent the initiation site and direction of transcription; arrows facing one another represent the palindromic sequences comprising hairpins I, II, and III. (Bottom left) nasA′ fragments in the lacUV5-nasA′-′lacZ cassettes. The hollow arrows represent the lacUV5 promoter; the empty rectangles represent the deletion within the leader regions. (Bottom right) Results of expression analyses of nasA′-′lacZ with/without coexpressed NasS or NasT. β-Galactosidase activity was measured after 3 h of growth in LB medium. All data are presented as the mean values of triplicate samples ± standard deviations from a representative experiment.
β-Galactosidase expression assays showed that lacUV5-nasA′-′lacZ in E. coli has a basal level of expression, and deletion of the transcriptional terminator hairpin increased the level of β-galactosidase activity 15-fold, confirming the negative role of the transcriptional terminator in E. coli. In the presence of coexpressed NasT, the level of β-galactosidase activity increased more than 30-fold, suggesting that NasT is synthesized as a positive regulator. The positive role of NasT was compromised when the integrity of hairpin I or II was disrupted: partial deletion of hairpin I reduced β-galactosidase activity to the basal level, while partial deletion of hairpin II reduced β-galactosidase activity by 87%. The results are consistent with in vivo analyses (Fig. 4). We conclude that NasT acts within the leader region of nasAB and that its function requires hairpins I and II.
The activity of the nasAB promoter is altered in the nifA mutation background.
Expression analysis of the translational nasA′-′lacZ fusion showed that nasA′-′lacZ integrated into the genomes of the strain UW1 (with the nifA mutation) and the wild-type strain UW136 had a similar expression pattern under the conditions tested (Table 1). However, β-galactosidase activity levels induced by nitrate or nitrite in UW1 were much higher than in UW136. This expression difference led to the hypothesis that PnasA has lower activity in a functional nif system.
The DNA sequence −316 to +16 that contained PnasA and upstream sequences but lacked the three hairpin sequences was inserted into the MCS upstream of lacZ in the transcriptional reporter pVnflacZb, giving rise to the transcriptional PnasA-lacZ fusion. In UW136, PnasA-lacZ showed similar expression levels under all tested conditions and was not expressed in an ntrC mutant background (Table 2), similar to the expression profiles of the hairpin III deletion or three-hairpin-deletion derivatives (Fig. 4 and 5). However, in UW1, PnasA-lacZ showed a very different expression profile. Although the transcriptional fusion in UW1 showed expression levels similar to those in UW136 when supplied with ammonium, replacement of ammonium by other nitrogen sources led to 4-fold or higher levels of lacZ expression, suggesting the activity of PnasA is under partial repression in the presence of ammonium. When atmospheric N2 was used as the sole nitrogen source, UW1 did not grow, as evidenced by the lack of an increase in cell number, but PnasA-lacZ maintained strong expression. Urea is a nitrogen source that can be used by A. vinelandii without repressing nitrogen fixation (59). The results from this experiment lead to the conclusion that the activity of PnasA is altered in the nifA mutation background.
Table 2.
Expression of the transcriptional PnasA-lacZ fusion in A. vinelandii grown with different nitrogen sourcesa
| Relevant genotypic characteristic | β-Galactosidase activity (Miller units)b after 7 h of growth in BS medium supplemented with: |
||||
|---|---|---|---|---|---|
| NH4+ | N2 | Urea | NO2− | NO3− | |
| Wild type | 594 ± 15 | 307 ± 14 | 324 ± 10 | 439 ± 10 | 394 ± 25 |
| ΔntrC::Gmr | <5 | 8 ± 1 | <5 | <5 | <5 |
| nifA mutation | 559 ± 17 | 5,328 ± 30 | 3,470 ± 34 | 2,087 ± 142 | 2,677 ± 32 |
PnasA refers to the sequence of nt −316 to +12 around the nasAB promoter.
All data are mean values of triplicate samples ± standard deviations from a representative experiment.
DISCUSSION
Our study showed that the reported lack of expression of nasAB, the assimilatory nitrate reductase operon in A. vinelandii, in the presence of ammonium was not due to the repression of the promoter PnasA but to transcriptional termination within the nasAB leader region. The arrangement of NtrC and σ54 binding sites of PnasA is similar to that of NtrC-regulated promoters in enteric bacteria (32). However, the high activity of PnasA in the presence of ammonium distinguishes it from NtrC-regulated promoters in enteric bacteria (41). We attribute this unusual activity of PnasA to NtrC-mediated regulation in A. vinelandii. Although we cannot exclude the possibility that other regulatory factors might also be involved in nasAB activation, the abolishment of nasAB promoter activation in an ntrC mutant suggests the role of other potential regulatory factors is secondary to that of NtrC. Relaxed NtrC regulation in the presence of ammonium also explains the previous observation that the promoter of nifL from K. pneumoniae, which is regulated solely by NtrC, also was highly expressed in A. vinelandii in the presence of high concentrations of ammonium (30).
Although PnasA showed similar activity levels in the presence of ammonium, nitrogen, urea, nitrite, or nitrate in the wild-type strain UW136, the promoter showed much higher activity (4- to 17-fold higher) in UW1 when ammonium was removed or replaced with other nitrogen sources (Table 2). Since UW1 and UW136 differ only in nifA function, it is possible that NifA directly regulates PnasA. However, no potential NifA binding sequence TGT-N10-ACA (9) was identified within PnasA or its upstream region, suggesting that the regulatory role of nifA on the regulation of PnasA might be indirect. Activity analysis of both nitrogenase and PnasA in future studies may bring new insight into nitrogen regulation in A. vinelandii.
The increase of PnasA activity in UW1 in the absence of ammonium resembles the relief of ammonium repression on NtrC-regulated promoters in enteric bacteria. We speculate that the ammonium from the medium or nitrogen fixation increases cellular nitrogen status, resulting in the reduced activity of NtrC. It was demonstrated previously that GlnK in A. vinelandii transduces the nitrogen signal from GlnD to NifL (36, 37, 50, 57). It is possible that GlnK also transduces a nitrogen signal to NtrBC, a cascade similar to the general nitrogen regulatory system (GlnD→GlnK→NtrBC) in enteric bacteria (41).
Antitermination mediated by NasT plays an essential role in nasAB induction. The functional and structural features of NasT suggest that it belongs to the same family of regulators as AmiR and EutV. The direct ANTAR-RNA interaction has been previously confirmed in AmiR (46), EutV (19), and NasR (11) by means of electrophoretic mobility shift assays. Unlike the REC-ANTAR structure, the N terminus of NasR in K. oxytoca (pneumoniae) M5al is a nitrate- and nitrite-sensing (NIT) domain, and the binding of ligand at NIT triggers the antitermination function of NasR (11, 55).
The mechanism of ANTAR-mediated antitermination is still largely unknown. Alignment of multiple RNAs targeted by EutV shows a 13-nt shared conserved sequence (AGCAANGRRGCUY) (19); this sequence overlaps with the left stem of the terminator hairpin and the right stem of a putative low-stability antitermination hairpin (2). This conserved feature led to the hypothesis that binding of EutV to the conserved sequence increases the stability of the antiterminator structure, preventing the formation of a terminator hairpin (2).
The putative EutV binding sequence is not identified in the leader region of nasAB. The results of our deletion mutational analyses indicate that hairpins I and II, which are separated from the terminator hairpin, are potential binding sites of NasT. Similarly, the potential binding site of NasR is a promoter-proximal hairpin separated from the terminator (12). Thus, the model of ANTAR-related antitermination reconciling all these findings remains to be defined in future studies.
In the absence of nitrate and nitrite induction, the translational nasA′-′lacZ fusion showed low levels of expression (Fig. 4). Although low, these expression levels are still higher than the levels produced by the hairpin I deletion mutant, suggesting that read-through of the terminator may occur at a low level. We speculate that low levels of nasAB expression in the absence of nitrate and/or nitrite might be related to the regulatory role of NasB. Previous analysis suggested that NasB has a negative role on nasAB expression, and deletion of nasB resulted in high expression of nasA even without nitrate/nitrite induction (21, 48). How NasB is involved in nasAB operon regulation remains to be determined.
In summary, identification of NasT-mediated antitermination in nasAB regulation expands our understanding of nitrate/nitrite reductase regulation in bacteria and ANTAR-containing antiterminators. The study also sheds additional insights into nitrogen regulation in A. vinelandii and indicates that it is much more complicated than previously thought. nasAB represents the first NtrC-regulated operon in A. vinelandii that has been studied in detail, and it will be of interest to learn how other NtrC-regulated operons in this bacterium regulate their expression in a substrate-specific manner.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bentley A. Fane and Yeou-Cherng Bor for discussions and critical review of the manuscript.
Footnotes
Published ahead of print 6 July 2012
This article is dedicated to the memory of Christina Kennedy (1945–2009).
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Amann E, Brosius J, Ptashne M. 1983. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene 25:167–178 [DOI] [PubMed] [Google Scholar]
- 2. Baker KA, Perego M. 2011. Transcription antitermination by a phosphorylated response regulator and cobalamin-dependent termination at a B12 riboswitch contribute to ethanolamine utilization in Enterococcus faecalis. J. Bacteriol. 193:2575–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bali A, Blanco G, Hill S, Kennedy C. 1992. Excretion of ammonium by a nifL mutant of Azotobacter vinelandii fixing nitrogen. Appl. Environ. Microbiol. 58:1711–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barrios H, Valderrama B, Morett E. 1999. Compilation and analysis of sigma(54)-dependent promoter sequences. Nucleic Acids Res. 27:4305–4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beynon J, Cannon M, Buchanan-Wollaston V, Cannon F. 1983. The nif promoters of Klebsiella pneumoniae have a characteristic primary structure. Cell 34:665–671 [DOI] [PubMed] [Google Scholar]
- 6. Bishop PE, Jarlenski DM, Hetherington DR. 1980. Evidence for an alternative nitrogen fixation system in Azotobacter vinelandii. Proc. Natl. Acad. Sci. U. S. A. 77:7342–7346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blanco G, Drummond M, Woodley P, Kennedy C. 1993. Sequence and molecular analysis of the nifL gene of Azotobacter vinelandii. Mol. Microbiol. 9:869–879 [DOI] [PubMed] [Google Scholar]
- 8. Buchanan-Wollaston V, Cannon MC, Beynon JL, Cannon FC. 1981. Role of the nifA gene product in the regulation of nif expression in Klebsiella pneumoniae. Nature 294:776–778 [DOI] [PubMed] [Google Scholar]
- 9. Buck M, Miller S, Drummond M, Dixon R. 1986. Upstream activator sequences are present in the promoters of nitrogen fixation genes. Nature 320:374–378 [Google Scholar]
- 10. Casadaban MJ, Cohen SN. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179–207 [DOI] [PubMed] [Google Scholar]
- 11. Chai W, Stewart V. 1998. NasR, a novel RNA-binding protein, mediates nitrate-responsive transcription antitermination of the Klebsiella oxytoca M5al nasF operon leader in vitro. J. Mol. Biol. 283:339–351 [DOI] [PubMed] [Google Scholar]
- 12. Chai W, Stewart V. 1999. RNA sequence requirements for NasR-mediated, nitrate-responsive transcription antitermination of the Klebsiella oxytoca M5al nasF operon leader. J. Mol. Biol. 292:203–216 [DOI] [PubMed] [Google Scholar]
- 13. Contreras A, et al. 1991. The product of the nitrogen fixation regulatory gene nfrX of Azotobacter vinelandii is functionally and structurally homologous to the uridylyltransferase encoded by glnD in enteric bacteria. J. Bacteriol. 173:7741–7749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Del Papa MF, Perego M. 2008. Ethanolamine activates a sensor histidine kinase regulating its utilization in Enterococcus faecalis. J. Bacteriol. 190:7147–7156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dixon R, Kahn D. 2004. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2:621–631 [DOI] [PubMed] [Google Scholar]
- 16. Drummond M, Clements J, Merrick M, Dixon R. 1983. Positive control and autogenous regulation of the nifLA promoter in Klebsiella pneumoniae. Nature 301:302–307 [DOI] [PubMed] [Google Scholar]
- 17. Espin G, Alvarez-Morales A, Cannon F, Dixon R, Merrick M. 1982. Cloning of the glnA, ntrB and ntrC genes of Klebsiella pneumoniae and studies of their role in regulation of the nitrogen fixation (nif) gene cluster. Mol. Gen. Genet. 186:518–524 [DOI] [PubMed] [Google Scholar]
- 18. Ferro-Luzzi Ames G, Nikaido K. 1985. Nitrogen regulation in Salmonella typhimurium. Identification of an ntrC protein-binding site and definition of a consensus binding sequence. EMBO J. 4:539–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fox KA, et al. 2009. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc. Natl. Acad. Sci. U. S. A. 106:4435–4440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gollnick P, Babitzke P. 2002. Transcription attenuation. Biochim. Biophys. Acta 1577:240–250 [DOI] [PubMed] [Google Scholar]
- 21. Gutierrez JC, Ramos F, Ortner L, Tortolero M. 1995. nasST, two genes involved in the induction of the assimilatory nitrite-nitrate reductase operon (nasAB) of Azotobacter vinelandii. Mol. Microbiol. 18:579–591 [DOI] [PubMed] [Google Scholar]
- 22. Gutierrez JC, Santero E, Tortolero M. 1997. Ammonium repression of the nitrite-nitrate (nasAB) assimilatory operon of Azotobacter vinelandii is enhanced in mutants expressing the nifO gene at high levels. Mol. Gen. Genet. 255:172–179 [DOI] [PubMed] [Google Scholar]
- 23. Henkin TM, Yanofsky C. 2002. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. Bioessays 24:700–707 [DOI] [PubMed] [Google Scholar]
- 24. Hill S, Austin S, Eydmann T, Jones T, Dixon R. 1996. Azotobacter vinelandii NIFL is a flavoprotein that modulates transcriptional activation of nitrogen-fixation genes via a redox-sensitive switch. Proc. Natl. Acad. Sci. U. S. A. 93:2143–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59 [DOI] [PubMed] [Google Scholar]
- 26. Jiang P, Ninfa AJ. 1999. Regulation of autophosphorylation of Escherichia coli nitrogen regulator II by the PII signal transduction protein. J. Bacteriol. 181:1906–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang P, Peliska JA, Ninfa AJ. 1998. Enzymological characterization of the signal-transducing uridylyltransferase/uridylyl-removing enzyme (EC 2.7.7.59) of Escherichia coli and its interaction with the PII protein. Biochemistry 37:12782–12794 [DOI] [PubMed] [Google Scholar]
- 28. Kamberov ES, Atkinson MR, Ninfa AJ. 1995. The Escherichia coli PII signal transduction protein is activated upon binding 2-ketoglutarate and ATP. J. Biol. Chem. 270:17797–17807 [DOI] [PubMed] [Google Scholar]
- 29. Keener J, Kustu S. 1988. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc. Natl. Acad. Sci. U. S. A. 85:4976–4980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kennedy C, Drummond MH. 1985. The use of cloned nif regulatory elements from Klebsiella pneumoniae to examine nif regulation in Azotobacter vinelandii. J. Gen. Microbiol. 131:1787–1795 [Google Scholar]
- 31. Kennedy C, Robson RL. 1983. Activation of nif gene expression in Azotobacter by the nifA gene product of Klebsiella pneumoniae. Nature 301:626–628 [DOI] [PubMed] [Google Scholar]
- 32. Kustu S, North AK, Weiss DS. 1991. Prokaryotic transcriptional enhancers and enhancer-binding proteins. Trends Biochem. Sci. 16:397–402 [DOI] [PubMed] [Google Scholar]
- 33. Leonardo JM, Goldberg RB. 1980. Regulation of nitrogen metabolism in glutamine auxotrophs of Klebsiella pneumoniae. J. Bacteriol. 142:99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin JT, Stewart V. 1996. Nitrate and nitrite-mediated transcription antitermination control of nasF (nitrate assimilation) operon expression in Klebsiella pneumoniae M5al. J. Mol. Biol. 256:423–435 [DOI] [PubMed] [Google Scholar]
- 35. Lin JT, Stewart V. 1998. Nitrate assimilation by bacteria. Adv. Microb. Physiol. 39:1–30, 379 [DOI] [PubMed] [Google Scholar]
- 36. Little R, Colombo V, Leech A, Dixon R. 2002. Direct interaction of the NifL regulatory protein with the GlnK signal transducer enables the Azotobacter vinelandii NifL-NifA regulatory system to respond to conditions replete for nitrogen. J. Biol. Chem. 277:15472–15481 [DOI] [PubMed] [Google Scholar]
- 37. Little R, Reyes-Ramirez F, Zhang Y, van Heeswijk WC, Dixon R. 2000. Signal transduction to the Azotobacter vinelandii NIFL-NIFA regulatory system is influenced directly by interaction with 2-oxoglutarate and the PII regulatory protein. EMBO J. 19:6041–6050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Luque F, Santero E, Medina JR, Tortolero M. 1987. An effective mutagenic method in Azotobacter vinelandii. Microbiologia 3:45–49 [PubMed] [Google Scholar]
- 39. Meletzus D, Rudnick P, Doetsch N, Green A, Kennedy C. 1998. Characterization of the glnK-amtB operon of Azotobacter vinelandii. J. Bacteriol. 180:3260–3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Merrick MJ. 1983. Nitrogen control of the nif regulon in Klebsiella pneumoniae: involvement of the ntrA gene and analogies between ntrC and nifA. EMBO J. 2:39–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Merrick MJ, Edwards RA. 1995. Nitrogen control in bacteria. Microbiol. Rev. 59:604–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miller WG, Leveau JHJ, Lindow SE. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant Microbe Interact. 13:1243–1250 [DOI] [PubMed] [Google Scholar]
- 43. Morth JP, Feng V, Perry LJ, Svergun DI, Tucker PA. 2004. The crystal and solution structure of a putative transcriptional antiterminator from Mycobacterium tuberculosis. Structure 12:1595–1605 [DOI] [PubMed] [Google Scholar]
- 44. Newton JW, Wilson PW, Burris RH. 1953. Direct demonstration of ammonia as an intermediate in nitrogen fixation by Azotobacter. J. Biol. Chem. 204:445–451 [PubMed] [Google Scholar]
- 45. Ninfa AJ, Magasanik B. 1986. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 83:5909–5913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Norman RA, Poh CL, Pearl LH, O'Hara BP, Drew RE. 2000. Steric hindrance regulation of the Pseudomonas aeruginosa amidase operon. J. Biol. Chem. 275:30660–30667 [DOI] [PubMed] [Google Scholar]
- 47. O'Hara BP, et al. 1999. Crystal structure and induction mechanism of AmiC-AmiR: a ligand-regulated transcription antitermination complex. EMBO J. 18:5175–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ramos F, Blanco G, Gutiérrez JC, Luque F, Tortolero M. 1993. Identification of an operon involved in the assimilatory nitrate-reducing system of Azotobacter vinelandii. Mol. Microbiol. 8:1145–1153 [DOI] [PubMed] [Google Scholar]
- 49. Robson RL, et al. 1984. Genome size and complexity in Azotobacter chroococcum. J. Gen. Microbiol. 130:1603–1612 [DOI] [PubMed] [Google Scholar]
- 50. Rudnick P, Kunz C, Gunatilaka MK, Hines ER, Kennedy C. 2002. Role of GlnK in NifL-mediated regulation of NifA activity in Azotobacter vinelandii. J. Bacteriol. 184:812–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 52. Santero E, Luque F, Medina JR, Tortolero M. 1986. Isolation of ntrA-like mutants of Azotobacter vinelandii. J. Bacteriol. 166:541–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Santero E, Toukdarian A, Humphrey R, Kennedy C. 1988. Identification and characterization of two nitrogen fixation regulatory regions, nifA and nfrX, in Azotobacter vinelandii and Azotobacter chroococcum. Mol. Microbiol. 2:303–314 [DOI] [PubMed] [Google Scholar]
- 54. Setubal JC, et al. 2009. Genome sequence of Azotobacter vinelandii, an obligate aerobe specialized to support diverse anaerobic metabolic processes. J. Bacteriol. 191:4534–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shu CJ, Ulrich LE, Zhulin IB. 2003. The NIT domain: a predicted nitrate-responsive module in bacterial sensory receptors. Trends Biochem. Sci. 28:121–124 [DOI] [PubMed] [Google Scholar]
- 56. Shu CJ, Zhulin IB. 2002. ANTAR: an RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem. Sci. 27:3–5 [DOI] [PubMed] [Google Scholar]
- 57. Soderback E, et al. 1998. The redox- and fixed nitrogen-responsive regulatory protein NIFL from Azotobacter vinelandii comprises discrete flavin and nucleotide-binding domains. Mol. Microbiol. 28:179–192 [DOI] [PubMed] [Google Scholar]
- 58. Toukdarian A, Kennedy C. 1986. Regulation of nitrogen metabolism in Azotobacter vinelandii: isolation of ntr and glnA genes and construction of ntr mutants. EMBO J. 5:399–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Walmsley J, Kennedy C. 1991. Temperature-dependent regulation by molybdenum and vanadium of expression of the structural genes encoding three nitrogenases in Azotobacter vinelandii. Appl. Environ. Microbiol. 57:622–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.