Abstract
Shiga toxin-producing Escherichia coli (STEC) strains are important food-borne pathogens capable of causing hemolytic-uremic syndrome. STEC O157:H7 strains cause the majority of severe disease in the United States; however, there is a growing concern for the amount and severity of illness attributable to non-O157 STEC. Recently, the Food Safety and Inspection Service (FSIS) published the intent to regulate the presence of STEC belonging to serogroups O26, O45, O103, O111, O121, and O145 in nonintact beef products. To ensure the effective control of these bacteria, sensitive and specific tests for their detection will be needed. In this study, we identified single nucleotide polymorphisms (SNPs) in the O-antigen gene cluster that could be used to detect STEC strains of the above-described serogroups. Using comparative DNA sequence analysis, we identified 22 potentially informative SNPs among 164 STEC and non-STEC strains of the above-described serogroups and designed matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF) assays to test the STEC allele frequencies in an independent panel of bacterial strains. We found at least one SNP that was specific to each serogroup and also differentiated between STEC and non-STEC strains. Differences in the DNA sequence of the O-antigen gene cluster corresponded well with differences in the virulence gene profiles and provided evidence of different lineages for STEC and non-STEC strains. The SNPs discovered in this study can be used to develop tests that will not only accurately identify O26, O45, O103, O111, O121, and O145 strains but also predict whether strains detected in the above-described serogroups contain Shiga toxin-encoding genes.
INTRODUCTION
Escherichia coli is a diverse bacterial species whose members include beneficial commensals and overt pathogens capable of causing intestinal, as well as extraintestinal, disease (39–42). E. coli strains that cause intestinal disease can be grouped into several different categories based on pathogenic features, including enteropathogenic (EPEC), enteroaggregative (EAEC), enteroinvasive (EIEC), enterotoxigenic (ETEC), and Shiga toxin-producing (STEC) E. coli (46). Enterohemorrhagic E. coli (EHEC) strains comprise a subgroup within the STEC that cause hemorrhagic colitis and severe disease in humans (35, 38). EHEC strains are thought to have evolved from an EPEC progenitor and to have acquired Shiga toxins through phage insertions (54). Shiga toxins (encoded by stx1 and stx2) are among the virulence factors associated with severe disease and illness (6, 11, 58). EPEC strains do not contain Shiga toxin-encoding genes; however, they do contain other genes that are responsible for attachment to the epithelium and also genes that may be associated with particular clinical manifestations (1). EPEC strains can be divided into typical and atypical based on the presence or absence of bundle-forming pili (bfpA) (61). Atypical EPEC (aEPEC) strains lack the bfpA gene, are typically found in developed countries, and have been linked to diarrhea in several studies (2, 45, 47). Other examples of genes that have been linked to virulence or adhesion include eae, a gene that encodes intimin, an outer membrane protein that facilitates attachment to the epithelium (48); efa1 (lifA), a gene that encodes the protein lymphostatin, which has adhesive properties and is associated with diarrhea in atypical EPEC strains (3, 45); nleB, the non-locus of enterocyte effacement-borne effector B gene, which is associated with diarrhea in atypical EPEC strains (45); and ehxA, a gene located on a plasmid commonly found in EHEC strains and which encodes a hemolysin referred to as enterohemolysin (52).
E. coli O157:H7 strains are strongly linked to human illness in the United States and in other countries and are responsible for the majority of outbreaks in the United States (51, 57, 60). However, non-O157:H7 strains have also been linked to outbreaks (14, 44, 53) and illness (41) in the United States, and in some countries, non-O157:H7 strains are more commonly linked to illness (5, 9, 34). Cattle have been identified as reservoirs for STEC, and consequently, raw milk and undercooked ground beef have been implicated as sources of human infection (20, 55, 56). The Food Safety and Inspection Service (FSIS) recently announced that in 2012, STEC serogroups O26, O45, O103, O111, O121, and O145 would be declared adulterants in nonintact raw beef and mandatory testing for these serogroups will be implemented (27a). The association between STEC serogroups found in cattle and those causing human illness is complex, because only a small number of the STEC serogroups found in cattle have been associated with human disease (4, 10, 12). In addition, many strains of E. coli in cattle have the same O antigens as the STEC strains targeted for regulation but lack the genes encoding Shiga toxins and other virulence factors and have not been associated with disease in humans. Tests are needed that will not only identify the targeted STEC serogroups but also differentiate between strains of these serogroups that contain STEC-associated virulence factors and those that do not.
A study conducted by the Centers for Disease Control and Prevention (CDC) found that there were 940 lab-confirmed cases of non-O157 STEC human infection in the United States from 1983 to 2002 (15). The majority of the strains (71%) belonged to one of six major serogroups, including O111, O26, O103, O145, O45, and O121. Current research is focusing on these six serogroups to better understand their epidemiology in humans and cattle (12), develop accurate testing procedures (30), and establish intervention strategies to prevent contamination of ground beef (4). Studies to learn about the epidemiology of these six serogroups in humans are challenging because clinical laboratories are increasingly using strategies to diagnose STEC infections that do not yield an isolate for further characterization. There is also a poor understanding of the prevalence of non-O157 STEC in cattle and nonintact raw beef. Conducting studies to obtain prevalence estimates for non-O157 STEC in cattle and nonintact raw beef are complex because of a lack of standardized isolation and culturing methods. A primary step in studying and understanding the potential impact of non-O157 STEC strains in cattle and nonintact raw beef will be to develop accurate tests to identify specific O serogroups that are affordable, easy to implement in a variety of lab settings, easy to duplicate both within and between labs, and comparable across study populations.
One of the potential targets for identifying specific E. coli O serogroups is the O-antigen gene cluster. The O antigen is part of the lipopolysaccharide present on the outer membrane of E. coli and other Gram-negative bacteria. E. coli has over 181 O serogroups based on the structural variability of the O antigen (25). The variability is the result of differences in sugar composition and the sequences and linkages of these sugars (65). The O-antigen gene cluster consists of genes that are well conserved between O serogroups and also genes that differ across serogroups. These O-serogroup-specific genes are responsible for the synthesis and assembly of the O antigen (62, 63).
The objective of this study was to identify single nucleotide polymorphisms (SNPs) within the O-antigen gene cluster of non-O157 E. coli serogroups O26, O45, O103, O111, O121, and O145. Reported here are newly described nucleotide polymorphisms in the O-antigen gene cluster of these O serogroups that can differentiate between STEC and non-STEC strains within a serogroup. Differences in the genetic sequence of the O-antigen gene cluster corresponded well with differences in the virulence gene profiles and provide evidence of separate clustering for the majority of STEC and non-STEC strains. The SNPs discovered in this study can be used to develop tests that will specifically identify STEC strains within serogroups O26, O45, O103, O111, O121, and O145.
MATERIALS AND METHODS
Strain collection.
A set of 164 non-O157 strains isolated from various sources and geographic areas were used for SNP discovery (see Table S1 in the supplemental material). These included 64 O26, 47 O111, 23 O103, 6 O145, 12 O45, and 12 O121 strains. The strains originated from a variety of sources, including humans, cattle, sheep, goats, swine, turkey, chickens, dogs, whitetail deer, flies, and ecological surfaces, and from the following states in the United States: California, Colorado, Delaware, Florida, Iowa, Idaho, Indiana, Kansas, Louisiana, Nebraska, New Hampshire, New Jersey, Oklahoma, South Dakota, Tennessee, Texas, Utah, Wisconsin, and Washington. There were also strains from Switzerland, Germany, Australia, England, Kenya, Peru, Italy, Brazil, Uruguay, Denmark, Canada, Cuba, and Japan.
DNA isolation.
A single representative colony from each strain was inoculated into 10 ml of Luria broth and incubated overnight at 37°C on a shaker. Genomic DNA was extracted using the Qiagen Genomic-tip 100/G columns (Valencia, CA) and techniques previously described (21). The DNA concentration was determined with a spectrophotometer (NanoDrop Technologies, Wilmington, DE) and diluted with Tris-EDTA (for 50 ml TE: 500 μl 1 M Tris [pH 8], 10 μl 0.5 M EDTA, and 49.5 ml molecular-grade water) to a 5-ng/μl working solution.
PCR, DNA sequencing, and analysis.
Identification of SNPs was accomplished by PCR of short fragments of DNA followed by Sanger sequencing. The O-antigen gene cluster (Fig. 1) was amplified for all of the strains using six to nine different PCR primer pairs that collectively targeted the entire length of the gene cluster (see Table S2 in the supplemental material). Each of the PCR fragments was, on average, 1,500 to 2,000 bp in length, and fragments overlapped by approximately 500 bp. Amplification reaction mixtures contained 3.2 μl deoxynucleoside triphosphates (dNTPs) (1.25 mM; Promega, Madison, WI), 2 μl 10× buffer (HotStarTaq DNA polymerase kit; Qiagen), 0.1 μl HotStarTaq DNA polymerase (5 U/μl; Qiagen), 0.2 μl each primer (30 μM), and 1 μl template DNA (5 ng/μl) in a 20-μl reaction volume. Amplifications were performed with an S1000 or Peltier Dyad Bio-Rad thermocycler (Hercules, CA) under the following conditions: an initial denaturation step at 95°C for 15 min, followed by 30 s at 94°C, 30 s at 52°C, and 2 min at 72°C for 35 cycles and a final extension step at 72°C for 10 min. Amplification was verified by running PCR products on a 1% agarose gel containing ethidium bromide.
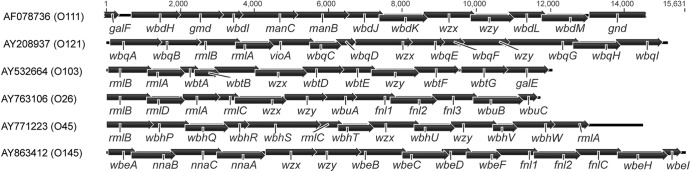
Fig 1.
Schematic representation of the O-antigen gene clusters from E. coli serotypes O26, O45, O103, O111, O121, and O145. GenBank accession numbers are indicated on the left followed by the E. coli serotype in parentheses. Base pair length is located above the gene clusters.
DNA exonuclease digestions for Sanger sequencing were set up using a modified 2.0 BigDye protocol (Applied Biosystems, Carlsbad, CA) (59). For the initial reaction, 5.5 μl of the PCR product and 7 μl exonuclease I (ExoI) (0.1 U/μl) were added to each well. Reactions were run on a Peltier Dyad Bio-Rad thermocycler under the following conditions: hold at 37°C for 1 h, followed by 20 min at 65°C. Following the digestion in the thermocycler, 23 μl 100% ethyl alcohol (EtOH) was added to each well and centrifuged at 3,200 rpm for 30 min at room temperature. The plate was inverted and centrifuged briefly at 500 rpm and allowed to dry at room temperature for at least 30 min. The sequencing reaction mixture contained 0.25 μl BigDye, 1.75 μl 5× buffer, and 0.11 μl primer (30 μM), for a final volume of 5 μl per well. Reactions were run on a Peltier Dyad Bio-Rad thermocycler under the following conditions: for 25 cycles, 30 s at 96°C, 2.5-s ramp to 96°C, 10 s at 96°C, 2.5-s ramp to 50°C, 5 s at 50°C, 2.5-s ramp to 60°C, and 4 min at 60°C. The sequencing reaction DNA contents were precipitated and washed with the following: addition of 22 μl 70% isopropanol, centrifugation for 30 min at 3,200 rpm, a brief centrifugation of the inverted plate at 500 rpm, addition of 22 μl 70% EtOH, centrifugation for 30 min at 3,200 rpm, another brief centrifugation of the inverted plate at 500 rpm, and a minimum air dry of at least 10 min. The plates were stored at −20°C prior to sequencing. DNA sequences were determined using a 3730xl DNA analyzer (Applied Biosystems).
DNA sequences were analyzed and contigs were assembled using Geneious Pro version 5.3.6 (Biomatters Ltd., Auckland, New Zealand). Assembled contigs of the O-antigen gene clusters (Fig. 1) were aligned and compared to reference sequences and each other using the Clustal alignment feature in Geneious and unweighted-pair group method with arithmetic mean (UPGMA) trees generated from the results. Bootstrap values were calculated for the branches using 1,000 pseudoalignments. The strains were grouped into clusters on the dendrogram; however, clusters do not have a statistical significance and were grouped merely for the convenience of presenting the results and discussion.
PCR of virulence genes.
PCR detection of the stx1, stx2, eae, and ehxA genes was performed with a multiplex PCR assay (52). Amplification of the bfpA, efa1 (lifA), and nleB genes was accomplished using previously described primers and protocols (45, 61). The Clermont typing procedure was used to divide the strains into phylogroups (22). The Clermont procedure is a multiplex PCR for amplification of the chuA and yjaA genes and the TspE4C2.2 fragment. Strains containing chuA and yjaA are classified as B2, strains containing TspE4C2.2 are classified as B1, strains containing chuA are classified as D, and strains that do not contain any of the three targets are classified as A. Amplification of the espK gene was performed on several O26 strains to determine if these strains were EHEC derivatives. Amplification was performed using two previously published primer sets, one set targeting the 3′ end and another set targeting the 5′ end, to ensure that strains with a truncated gene were detected (13).
STEC allele frequency of SNPs.
Polymorphisms were genotyped by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) genotyping (Sequenom, Inc., San Diego, CA). MALDI-TOF assay and multiplexing design was conducted with MassARRAY assay design software as recommended by the manufacturer (Sequenom, Inc.) (see Table S2 in the supplemental material). Up to 36 polymorphisms were accepted for each multiplex, and the assays were conducted with iPLEX Gold chemistry on a MassARRAY genotyping system per instructions of the manufacturer (Sequenom, Inc.). “High confidence” genotype calls by the Genotyper software were accepted as correct. “Aggressive” calls were inspected manually and verified as needed by Sanger sequencing or replicate MALDI-TOF assays. The frequency of the STEC alleles for the SNPs was determined for a panel of bacterial strains that were independent from the sequencing panel. This included 192 O157:H7, 4 O157:non-H7, 4 O55:H7, 2 O55:H6, 106 O26, 40 O45, 109 O103, 127 O111, 33 O121, 34 O145, and 22 other STEC strains; 175 O-antigen standards; 61 Salmonella strains; and 37 other bacteria (see Table S1 in the supplemental material). The strains originated from a variety of sources, including antelope, cattle, dogs, poultry, flies, goats, guinea pigs, humans, sheep, and swine, and from several geographic areas, including the United States, Canada, Chile, Denmark, England, France, Germany, Italy, Mexico, Panama, Scotland, and Sri Lanka.
Statistical tests.
Fisher's exact test was used to examine unconditional associations between the STEC-associated allele and O serogroup (P < 0.05) (STATA IC release 11.0). STEC-associated allele and O serogroup were analyzed as binary variables. Sensitivity and specificity estimates were calculated with exact 95% confidence intervals for each of the MALDI-TOF assays. The sensitivity estimate determined the ability of the assay to detect the STEC allele in an STEC strain, and the specificity determined the ability of the assay to not detect the STEC allele in a non-STEC strain. The calculations were based on a binary outcome of the STEC or non-STEC-associated allele. All strains with the STEC-associated allele were considered STEC, whereas non-STEC was any strain with the alternate allele, an indecisive allele call, or no allele call. The results of the assays were compared to three different known gene combinations: those containing stx alone, stx with eae, and stx with eae and ehxA.
Nucleotide sequence accession numbers.
Nucleotide sequences determined in this study (see Table S1 in the supplemental material) have been submitted to GenBank under accession numbers JN871613 to JN871676 (O26 strains), JN859197 to JN859208 (O45 strains), JN859209 to JN859220 (O121 strains), JN862595 to JN862617 (O103 strains), JN887644 to JN887690 (O111 strains), and JN850039 to JN850044 (O145 strains).
RESULTS
O26.
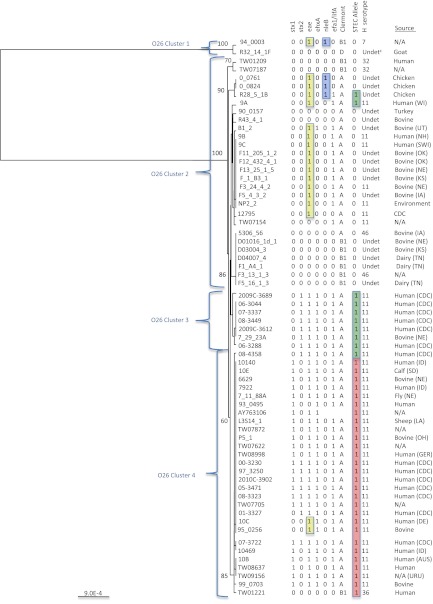
An 11,530-bp region of the O-antigen gene cluster from 64 O26 strains was sequenced and aligned with a reference sequence for E. coli O26:H11 (AY763106 [26]). A total of 174 polymorphisms were identified, of which 14.4% (25/174) were nonsynonymous. The majority (86.2%; 150/174) of the polymorphisms were found in only two strains (94_0003 and R32_14_1F [see Table S3 in the supplemental material]). For the most part, the polymorphisms from the two strains were in a 2-kb region of the gene cluster that included rmlB, rmlD, and the first 200 bp of rmlA. The most similar sequence by BLASTN was to a region of an E. coli O35 O-antigen gene cluster at 97%, while the sequence identity to O26 was 93%. A UPGMA tree generated from the alignment illustrated that sequence differences in the O-antigen gene cluster displayed distinct lineage differences, with the majority of the O26 STEC and O26 non-STEC strains clustered separately (Fig. 2). Clusters 1 and 2 contained non-STEC and EPEC strains, and clusters 3 and 4 contained mostly STEC strains. A number of the O26 cluster 1 and 2 strains contained nleB and eae. Cluster 3 strains contained stx2, eae, and ehxA, while cluster 4 strains contained stx1, stx2, or stx1 and stx2 along with eae and ehxA. Interestingly, two of the three non-STEC strains that fall into cluster 4 also contained ehxA and eae.
Fig 2.
UPGMA tree of the O-antigen gene clusters from 65 O26 Escherichia coli strains. Bootstrap values are presented on the corresponding branches, and clusters are in parentheses. EPEC strains are highlighted in yellow, strains with the nleB gene are highlighted in blue, highlighted in green are strains with the T STEC allele for polymorphism wzx 953 G→T, and highlighted in red are strains with the G and A STEC alleles for polymorphisms rmlA 30 G→T and fnl1 88 G→A. The unit of measure for the scale bar is the number of nucleotide substitutions per site. SWI, Switzerland; DEU, Germany; AUS, Australia; URU, Uruguay; N/A, not available. Two-letter abbreviations are postal abbreviations for U.S. states.
Three nucleotide polymorphisms were identified that differentiated O26 STEC strains from O26 non-STEC strains (Table 1). The polymorphisms are described by the gene in which they occur, the base pair position within that gene, and the allele change (non-STEC allele → STEC allele). The three polymorphisms were rmlA 30 G→T, wzx 953 T→G, and fnl1 88 G→A, with the last two polymorphisms resulting in amino acid coding changes. All three polymorphisms were found in genes that are not specific to the O26 serogroup. The T and A STEC-associated alleles of polymorphisms rmlA 30 G→T and fnl1 88 G→A were found in the same strains and captured the majority of the O26 STEC strains; however, wzx 953 T→G was included to capture a small subset of strains that contained only stx2. A combination of either rmlA 30 G→T or fnl1 88 G→A with wzx 953 T→G captured all of the O26 strains in our sequencing panel that contained stx1, stx2, or both stx1 and stx2. There were three strains included with the rmlA 30 G→T and fnl1 88 G→A STEC-associated alleles (highlighted in red) and two strains included with the wzx 953 T→G STEC-associated allele (highlighted in green) that did not contain stx (Fig. 2). Strains R28_5_1B, 10C, and 95_0256 contained espK and are most likely EHEC derivatives.
Table 1.
Gene and amino acid data for the single nucleotide polymorphisms identified in the O-antigen gene clusters of non-O157 Escherichia coli
| Serogroup | Gene | bp position | Polymorphisma | AAb position | AA change | Association (P value)c |
|---|---|---|---|---|---|---|
| O26d | rm1A | 30 | G→T | 10 | Synonymous | <0.001 |
| wzx | 953 | T→G | 318 | Phe→Leu | <0.001 | |
| fnl1 | 88 | G→A | 30 | Ala→Thr | <0.001 | |
| O45e | rmlB | 966 | T→C | 303 | Synonymous | <0.001 |
| wbhQ | 721 | C→A | 241 | Leu→Ile | <0.001 | |
| wbhU | 241 | G→A | 80 | Val→Ile | <0.001 | |
| wzy | 752 | T→C | 251 | Val→Ala | <0.001 | |
| wzy | 906 | T→C | 302 | Synonymous | <0.001 | |
| wbhW | 21 | C→T | 7 | Synonymous | <0.001 | |
| wbhW | 997 | T→G | 333 | Stop codon→Glu | <0.001 | |
| Intergenic | 13299l | C→A | NAk | NA | <0.001 | |
| Intergenic | 13340l | A deletionf | NA | NA | NA | |
| Intergenic | 13534l | A→C | NA | NA | <0.001 | |
| O103g | wbtD | 937 | C→T | 313 | His→Tyr | <0.001 |
| O111h | Intergenic | 492f | G→T | NA | NA | <0.001 |
| wbdH | 1006 | G→A | 336 | Val→Ile | <0.001 | |
| wbdK | 687 | C→T | 229 | Synonymous | <0.001 | |
| wzx | 1128 | A→T | 376 | Synonymous | <0.001 | |
| O121i | vioA | 313 | C→T | 105 | Pro→Ser | <0.001 |
| wbqE | 437 | C→T | 146 | Ala→Val | <0.001 | |
| wbqI | 582 | G→A | 194 | Synonymous | <0.001 | |
| O145j | wzy | 37 | A→C | 13 | Ile→Leu | <0.001 |
Non-STEC→STEC.
AA, amino acid.
Fisher's exact test P value testing the association between the STEC-associated allele and O serogoup.
GenBank AY763106 used as a reference.
GenBank AY771223 used as a reference.
Deletion found in the two non-STEC that do not share the other 9 SNPs.
GenBank AY532664 used as a reference.
GenBank AF078736 used as a reference.
GenBank AY208937 used as a reference.
GenBank AY647260 used as a reference.
NA, not applicable.
Position within O-antigen gene cluster.
MALDI-TOF assays were used to determine the STEC allele frequencies of the three O26 polymorphisms in a panel of 1,045 strains, including 93 O26 STEC strains. The rmlA 30 T, wzx 953 G, and fnl1 88 A STEC-associated alleles were found to be significantly associated (P < 0.001) with the O26 serogroup. The sensitivity and specificity estimates were based on the use of either rmlA 30 G→T or fnl1 88 G→A in conjunction with wzx 953 T→G for the O26 strains. The sensitivity and specificity estimates were similar regardless of whether stx alone, stx with eae, or stx with eae and ehxA was included as the classifier (Table 2).
Table 2.
Comparison of sensitivity and specificity estimates across different categories of virulence for the 21 assays targeting the single nucleotide polymorphisms in six O serogroups
| Assay |
stx alonea |
stx with eaeb |
stx with eae and ehxAc |
|||
|---|---|---|---|---|---|---|
| % Sed (95% CIf) | % Spe (95% CI) | % Se (95% CI) | % Sp (95% CI) | % Se (95% CI) | % Sp (95% CI) | |
| O26 | ||||||
| rmlA 30 and wzx 953 | 96.8 (90.4, 99.2) | 56.2 (53.0, 59.4) | 98.9 (93.4, 99.9) | 56.2 (52.9, 59.3) | 98.9 (93.3, 99.9) | 55.9 (52.7, 59.1) |
| fml1 88 and wzx 953 | 96.8 (90.4, 99.2) | 87.3 (84.9, 89.3) | 96.8 (90.3, 99.2) | 87.2 (84.8, 89.2) | 96.7 (90.1, 99.2) | 87.0 (84.6, 89.0) |
| O45 | ||||||
| rmlB 966 | 100.0 (88.8, 100.0) | 95.4 (93.9, 96.5) | 100.0 (88.6, 100.0) | 95.3 (93.8, 96.4) | 100.0 (88.6, 100.0) | 95.3 (93.8, 96.4) |
| wbhQ 721 | 100.0 (88.8, 100.0) | 96.5 (95.2, 97.5) | 100.0 (88.6, 100.0) | 96.4 (95.1, 97.4) | 100.0 (88.6, 100.0) | 96.4 (95.1, 97.4) |
| wbhU 241 | 97.4 (84.9, 99.9) | 93.6 (92.0, 95.0) | 100.0 (88.6, 100.0) | 93.7 (92.0, 95.0) | 100.0 (88.6, 100.0) | 93.7 (92.0, 95.0) |
| wzy 752 | 97.4 (84.9, 99.9) | 97.8 (96.7, 98.6) | 100.0 (88.6, 100.0) | 97.8 (96.7, 98.6) | 100.0 (88.6, 100.0) | 97.8 (96.7, 98.6) |
| wzy 906 | 97.4 (84.9, 100.0) | 97.2 (95.9, 98.0) | 100 (88.6, 100.0) | 97.2 (95.9, 98.0) | 100.0 (88.6, 100.0) | 97.2 (95.9, 98.0) |
| wbhW 21 | 97.4 (84.9, 99.9) | 96.7 (95.4, 97.6) | 100.0 (88.6, 100.0) | 96.7 (95.4, 97.7) | 100.0 (88.6, 100.0) | 96.7 (95.4, 97.7) |
| wbhW 997 | 97.4 (84.9, 99.9) | 97.0 (95.7, 97.9) | 100.0 (88.6, 100.0) | 96.8 (95.5, 97.7) | 100.0 (88.6, 100.0) | 96.8 (95.5, 97.7) |
| Interg 13299 | 100.0 (88.8, 100.0) | 95.9 (94.5, 97.0) | 100.0 (88.6, 100.0) | 95.8 (94.4, 96.9) | 100.0 (88.6, 100.0) | 95.8 (94.4, 96.9) |
| Inter 13534 | 97.4 (84.9, 99.9) | 96.9 (95.6, 97.8) | 100.0 (88.6, 100.0) | 96.9 (95.6, 97.8) | 100.0 (88.6, 100.0) | 96.9 (95.6, 97.8) |
| O103 | ||||||
| wbtD 937 | 75.2 (65.7, 82.9) | 99.8 (99.2, 100.0) | 75.2 (65.7, 82.9) | 99.8 (99.2, 100.0) | 75.2 (65.7, 82.9) | 99.8 (99.2, 100.0) |
| O111 | ||||||
| Inter 492 | 97.4 (92.1, 99.3) | 87.4 (85.1, 89.4) | 97.1 (91.3, 99.3) | 86.4 (84.1, 88.5) | 96.9 (90.5, 99.2) | 85.6 (83.2, 87.7) |
| wbdH 1006 | 97.4 (92.1, 99.3) | 44.9 (41.7, 48.2) | 97.2 (91.4, 99.3) | 44.6 (41.4, 47.8) | 96.9 (90.7, 99.2) | 44.1 (41.0, 47.3) |
| wbdK 687 | 95.9 (90.1, 98.5) | 90.3 (88.2, 92.1) | 95.5 (89.2, 98.3) | 89.3 (87.1, 91.2) | 95.0 (88.3, 98.2) | 88.5 (86.2, 90.4) |
| wzx 1128 | 97.5 (92.3, 99.3) | 88.1 (85.9, 90.1) | 97.2 (91.5, 99.3) | 87.1 (84.8, 89.1) | 97.0 (90.8, 99.2) | 86.3 (83.9, 88.4) |
| O121 | ||||||
| vioA 313 | 93.3 (76.5, 98.8) | 94. (92.8, 95.6) | 100.0 (85.0, 100.0) | 94.4 (92.8, 95.6) | 100.0 (85.0, 100.0) | 94.4 (92.8, 95.6) |
| wbqE 437 | 90.0 (72.3, 97.4) | 93.4 (91.7, 94.8) | 96.4 (79.8, 99.8) | 93.4 (91.7, 94.8) | 96.4 (79.8, 99.8) | 93.4 (91.7, 94.8) |
| wbqI 582 | 90.0 (72.3, 97.4) | 93.6 (91.9, 95.0) | 96.4 (79.8, 99.8) | 93.6 (92.0, 95.0) | 96.4 (79.8, 99.8) | 93.6 (92.0, 95.0) |
| O145 | ||||||
| wzy 37 | 94.1 (78.9, 99.0) | 98.1 (97.1, 98.8) | 94.1 (78.9, 99.0) | 98.1 (97.1, 98.8) | 93.8 (77.8, 98.9) | 97.9 (96.9, 98.7) |
All strains containing stx1, stx2, or both were considered virulent (true positive).
All strains containing either stx1, stx2, or both and also containing eae were considered virulent.
All strains containing either stx1, stx2, or both and also containing both eae and ehxA were considered virulent.
Se, sensitivity.
Sp, specificity.
CI, confidence interval.
Inter, intergenic.
O45.
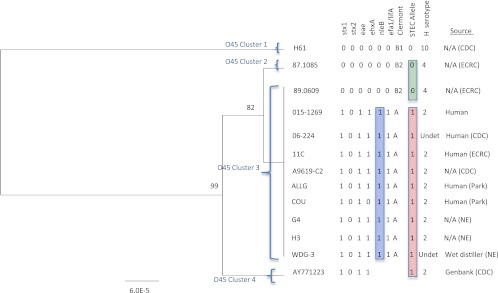
A 14,483-bp region of the O-antigen gene cluster from 12 O45 strains was sequenced and aligned with a reference sequence for E. coli O45:H2 (AY771223 [23]). A total of 20 polymorphisms were identified, of which 5 were synonymous, 2 were deletions, and 15 were found only in strain H61 (see Table S3 in the supplemental material). A UPGMA tree generated from the alignment illustrated that sequence differences in the O-antigen gene cluster displayed lineage differences, with the majority of O45 STEC and O45 non-STEC strains clustered separately (Fig. 3). Cluster 1 contained non-STEC strain H61, and cluster 2 contained non-STEC strain 87.1085, which is diverging from the strains in cluster 3. The O45 cluster 1 and 2 strains did not contain any of the virulence genes. The STEC strains were in clusters 3 and 4, and all the strains in cluster 3, have identical genetic sequences. Interestingly, one non-STEC strain (89.0609) also shared this sequence but had the same virulence gene profile and Clermont typing classification as non-STEC strain 87.1085 (cluster 2). The Clermont typing procedure classified the cluster 1 strain as B1 and the cluster 2 strain as B2. All of the STEC strains in cluster 3 contained eae, nleB, and efa1 (lifA) and were classified as A by the Clermont typing procedure, and all but one strain contained ehxA.
Fig 3.
UPGMA tree of the O-antigen gene clusters from 13 O45 Escherichia coli strains. Bootstrap values are presented on the corresponding branches, and clusters are represented in parentheses. Strains with the nleB gene are highlighted in blue, highlighted in green are strains with the A base deletion in polymorphism intergenic 13340, and highlighted in red are strains with the C, A, A, T, C, C, T, G, and A STEC-associated alleles in the other nine identified polymorphisms. The unit of measure for the scale bar is the number of nucleotide substitutions per site. N/A, not available; ECRC, Escherichia coli Reference Center.
Ten polymorphisms were identified that differentiated O45 STEC strains from O45 non-STEC strains. The 10 polymorphisms were rmlB 966 T→C, wbhQ 721 C→A, wbhU 241 G→A, wzy 752 T→C, wzy 906 T→C, wbhW 21 C→T, wbhW 997 T→G, intergenic 13299 C→A, intergenic 13340 A base insertion, and intergenic 13534 A→C (Table 1). Polymorphisms wbhQ 721 C→A, wbhU 241 G→A, wzy 752 T→C, and wbhW 997 T→G resulted in amino acid coding changes (Table 1). The genes containing polymorphisms that were specific to the O45 serogroup include wbhQ, wbhU, and wbhW. A combination of any one of the nine other polymorphisms with intergenic 13340 A base insertion divided the O45 strains in our sequencing panel into one group that contained strains with stx and a second group containing strains that did not contain stx.
MALDI-TOF assays were used to determine the STEC allele frequencies of nine of the O45 polymorphisms in a panel of 1,096 strains, including 39 O45 STEC strains. The nine STEC-associated alleles from the O45 polymorphisms were significantly (P < 0.001) associated with the O45 serogroup. The sensitivity estimates were slightly improved for wbhU 241 G→A, wzy 752 T→C, wzy 906 T→C, wbhW 21 C→T, wbhW 997 T→G, and intergenic 13534 A→C when eae and ehxA were included as classifiers with stx. The specificity estimates were similar regardless of whether stx alone, stx with eae, or stx with eae and ehxA was included as the classifier (Table 2).
O103.
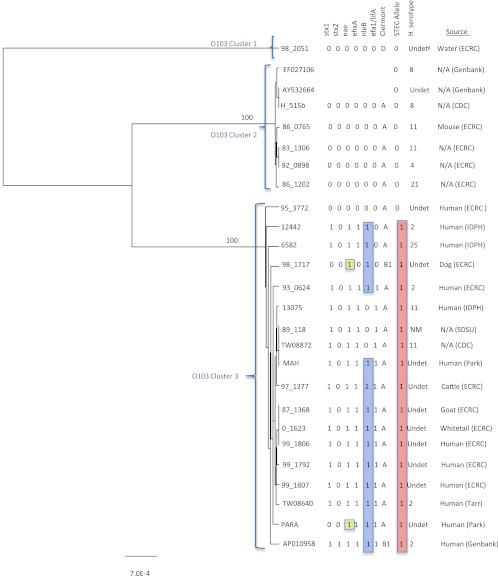
An 11,881-bp region of the O-antigen gene cluster from 23 O103 strains was sequenced and aligned with three E. coli O103 reference sequences (AY532664 [33], EF027106 [unpublished data], and AP010958 [50]). A total of 202 polymorphisms were identified, of which 40 were nonsynonymous and 1 was a deletion (see Table S3 in the supplemental material). A UPGMA tree generated from the alignment illustrated that O-antigen gene cluster sequences displayed distinct lineage differences, with the majority of O103 STEC and O103 non-STEC strains clustered separately (Fig. 4). Clusters 1 and 2 contained non-STEC strains, and cluster 3 contained all the STEC strains and three non-STEC strains. Strains in clusters 1 and 2 did not contain any of the virulence genes. The majority of the STEC strains in cluster 3 contained eae, ehxA, nleB, and efa1 (lifA), and two of the three non-STEC strains found in cluster 3 also contained eae.
Fig 4.
UPGMA tree of the O-antigen gene clusters from 26 O103 Escherichia coli strains. Bootstrap values are presented on the corresponding branches, and clusters are in parentheses. EPEC strains are highlighted in yellow, strains with the nleB gene are highlighted in blue, and strains highlighted in red have the T STEC allele for polymorphism wbtD 937 C→T. The unit of measure for the scale bar is the number of nucleotide substitutions per site. IDPH, Idaho Department of Public Health; SDSU, South Dakota State University.
One polymorphism was identified that differentiated O103 STEC strains from O103 non-STEC strains (Table 1). The wbtD 937 C→T polymorphism resulted in an amino acid coding change and is gene specific to the O103 serogroup. The polymorphism captured all O103 strains in our sequencing panel that contained stx (all of our O103 strains contain only stx1); however, it also included two strains that did not contain stx. The MALDI-TOF assay was used to determine the STEC allele frequency of the wbtD 937 C→T polymorphism in a panel of 1,084 strains, including 105 O103 STEC strains. The wbtD 937 T STEC-associated allele was significantly (P < 0.001) associated with the O103 serogroup. The sensitivity and specificity estimates were exactly the same regardless of whether stx alone, stx with eae, or stx with eae and ehxA was included as the classifier (Table 2).
O111.
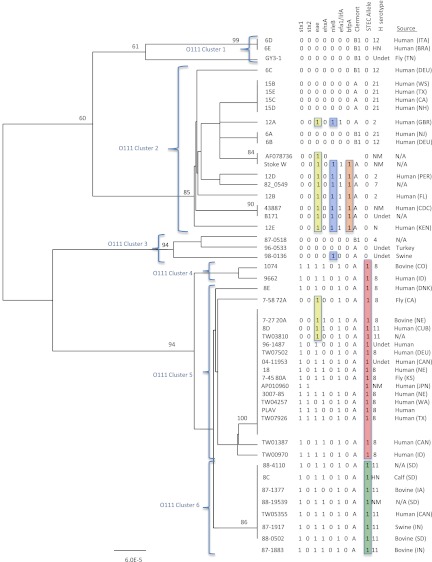
A 14,514-bp region of the O-antigen gene cluster from 47 O111 strains was sequenced and aligned with two E. coli O111 reference sequences (AF078736 [62] and AP010960 [50]). A total of 54 polymorphisms were identified, of which 22 were nonsynonymous and one was a deletion (see Table S3 in the supplemental material). A highly polymorphic region was observed at reference AF078736 nucleotide positions 3156 and 3165 in an intergenic region. At position 3156, 23 (85%) of the strains had more than 10 T's, while the remaining strains had no more than 9 T's. The majority (21/23; 91.3%) of the strains with more than 10 T's were STEC strains, while the majority (24/26; 92.3%) of the strains with no more than 9 T's were non-STEC. The same observation was made at position 3165, where the majority (23/26; 88.5%) of non-STEC strains only had three G's and the majority (20/23; 87%) of STEC strains had more than three. A UPGMA tree generated from the alignment illustrated that sequence differences in the O-antigen gene cluster displayed distinct lineage differences, with the majority of STEC and non-STEC strains clustered separately (Fig. 5). Clusters 1, 2, and 3 contained non-STEC strains, and cluster 2 primarily contained EPEC strains. The EPEC strains were the only non-STEC strains to contain nleB, efa1 (lifA), and bfpA. Clusters 4, 5, and 6 contained the STEC strains, and the clusters appeared to be associated with the H serogroup. The majority of the STEC strains in clusters 4, 5, and 6 contained eae, ehxA, and efa1 (lifA), and all four of the non-STEC strains in cluster 5 contained eae.
Fig 5.
UPGMA tree of the O-antigen gene clusters from 49 O111 Escherichia coli strains. Bootstrap values are presented on the corresponding branches, and clusters are represented in parentheses. EPEC strains are highlighted in yellow, strains with nleB are highlighted in blue, and strains with bfpA are highlighted in orange. Strains highlighted in red have the T, A, T, and T STEC-associated alleles for polymorphisms intergenic 492 G→T, wbdH 1006 G→A, wbdK 687 C→T, and wzx 1128 A→T; strains highlighted in green also have the STEC-associated alleles but are a cluster of O111:H11 strains. The unit of measure for the scale bar is the number of nucleotide substitutions per site. ITA, Italy; BRA, Brazil; DEU, Germany; GBR, England; PER, Peru; KEN, Kenya; DNK, Denmark; CAN, Canada; CUB, Cuba; JPN, Japan.
Four polymorphisms were identified that differentiated O111 STEC strains from O111 non-STEC strains. The four polymorphisms were intergenic 492 G→T, wbdH 1006 G→A, wbdK 687 C→T, and wzx 1128 A→T. The wbdH 1006 G→A polymorphism was the only O111 polymorphism that resulted in an amino acid coding change (Table 1). The genes containing polymorphisms that were specific to the O111 serogroup include wbdH and wbdK. The STEC-associated alleles of the four polymorphisms were found in the same O111 strains as genotyped in this study; however, only one of the polymorphisms is needed to differentiate the strains that contained stx. The polymorphisms captured all of the O111 strains in our sequencing panel with stx1 or both stx1 and stx2 (there were no strains with only stx2); however, there were four strains with the STEC-associated alleles that did not contain stx (Fig. 5).
MALDI-TOF assays were used to determine the STEC allele frequencies of the four O111 STEC-associated polymorphisms in a panel of 1,063 strains, including 116 O111 STEC strains. The intergenic T, wbdH 1006 A, wbdK 687 T, and wzx 1128 T STEC-associated alleles were significantly (P < 0.001) associated with the O111 serogroup. The sensitivity and specificity estimates were similar regardless of whether stx alone, stx with eae, or stx with eae and ehxA was included as the classifier (Table 2).
O121.
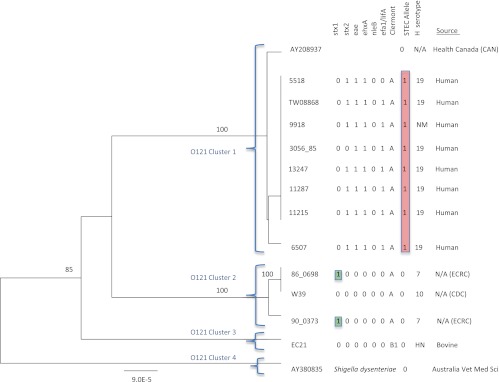
A 14,999-bp region of the O-antigen gene cluster from 12 O121 strains was sequenced and aligned with a reference sequence for E. coli O121 (AY208937 [31]) and a Shigella dysenteriae reference sequence (AY380835 [29]). A total of 43 polymorphisms were identified (see Table S3 in the supplemental material). A UPGMA tree generated from the alignment illustrated that sequence differences in the O-antigen gene cluster displayed lineage differences, with the majority of STEC and non-STEC strains clustered separately (Fig. 6). Clusters 1 and 2 contained both STEC and non-STEC strains, and cluster 3 contained a non-STEC strain. Cluster 4 contained Shigella dysenteriae 7, which had an O-antigen gene cluster that is closely related to the O-antigen gene cluster in the O121 strains. The strains in cluster 1 contained eae, ehxA, and efa1 (lifA). Interestingly, neither the STEC nor the non-STEC strains in cluster 2 or 3 contained any of the other virulence genes. The clusters also appeared to be associated with stx; strains in cluster 1 contained stx2, whereas strains in cluster 2 contained stx1.
Fig 6.
UPGMA tree of the O-antigen gene clusters from 13 O121 Escherichia coli strains and one Shigella dysenteriae strain. Bootstrap values are presented on the corresponding branches, and clusters are represented in parentheses. Highlighted in red are strains that have the T, T, and A STEC-associated alleles for polymorphisms vioA 313 C→T, wbaE 437 C→T, and wbaI 582 G→A. Highlighted in green are strains with stx1 but none of the other virulence factors. The unit of measure for the scale bar is the number of nucleotide substitutions per site.
Three nucleotide polymorphisms were identified that differentiated O121 STEC strains from O121 non-STEC strains (Table 1). The three polymorphisms were vioA 313 C→T, wbqE 437 C→T, and wbqI 582 G→A, with the first two polymorphisms resulting in an amino acid coding change. All three polymorphisms were contained in genes that were specific to the O121 serogroup. The STEC-associated alleles for the three polymorphisms were found in the same O121 strains as genotyped in this study; however, only one of the polymorphisms would be needed to differentiate the strains with stx. The polymorphisms captured all of the O121 strains in the sequencing panel containing only stx2; however, the STEC-associated allele does not capture the strains containing only stx1 (Fig. 6).
MALDI-TOF assays were used to determine the STEC allele frequencies of the three O121 polymorphisms in a panel of 1,094 strains, including 30 O121 STEC strains. The three STEC-associated alleles from the O121 polymorphisms were significantly (P < 0.001) associated with the O121 serogroup. The sensitivity estimates were improved when eae and ehxA were included as classifiers with stx. The specificity estimates were the same regardless of whether stx alone, stx with eae, or stx with eae and ehxA was used as the classifier (Table 2).
O145.
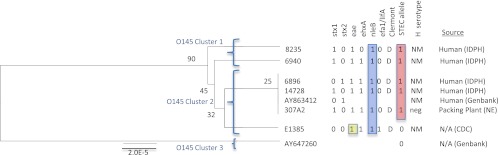
A 15,556-bp region of the O-antigen gene cluster from six O145 strains was sequenced and aligned with two E. coli O145 reference sequences (AY863412 [32] and AY647260 [28]). A total of eight polymorphisms were identified, of which one was a deletion (see Table S3 in the supplemental material). A UPGMA tree generated from the alignment illustrated that sequence differences in the O-antigen gene cluster displayed lineage differences, with the majority of STEC and non-STEC strains clustered separately (Fig. 7). Cluster 1 contained STEC strains, and cluster 2 contained both STEC and non-STEC strains. All of the strains in cluster 1 contained eae and nleB, and one strain contained ehxA. The strains in cluster 2 also contained eae, ehxA, and nleB, and in addition, one strain also contained efa1 (lifA).
Fig 7.
UPGMA tree of the O-antigen gene clusters from eight O145 Escherichia coli strains. Bootstrap values are presented on the corresponding branches, and clusters are represented in parentheses. EPEC strains are highlighted in yellow, strains with nleB are highlighted in blue, and strains highlighted in red have the C STEC-associated allele for polymorphism wxy 37 A→C. The unit of measure for the scale bar is the number of nucleotide substitutions per site.
One polymorphism was identified that differentiated O145 STEC strains from O145 non-STEC strains. The wzy 37 A→C polymorphism resulted in an amino acid coding change, was gene specific to the O145 serogroup, and captured all the O145 strains in our sequencing panel that contained stx (Table 1). The MALDI-TOF assay was used to determine the STEC allele frequencies of the wzy 37 A→C polymorphism in a panel of 1,102 strains, including 34 O145 STEC strains. The wzy 37 C STEC-associated allele was significantly (P < 0.001) associated with the O145 serogroup. The sensitivity and specificity estimates were similar regardless of whether stx alone, stx with eae, or stx with eae and ehxA was included as the classifier (Table 2).
DISCUSSION
Recent evidence suggests that non-O157 strains are responsible for an increasing number of human infections and that the role of these strains in causing diarrheal disease may be just as important as that of O157 strains (15). Obtaining true prevalence estimates for non-O157 infections is difficult due to the lack of mandatory characterization of isolates, loss of isolates due to the use of nonculture diagnostics, and also the lack of standardized testing procedures. The aim of this study was to compare the O-antigen gene cluster sequences of O26, O45, O103, O111, O121, and O145 E. coli strains to explore possible nucleotide associations with serogroups and pathotypes.
We found that there was a substantial amount of O-antigen gene cluster variation both within and between O serogroups. The degree of variation detected within and between O-antigen genes of E. coli was not unexpected due to lateral gene transfer events that are often the result of environmental pressures and the necessity to adapt for survival (36, 49). A complete genome comparison study between an E. coli O157:H7 strain isolated from the Sakai outbreak and a benign lab strain of E. coli, K-12 MG1655, found that the two strains shared a 4.1-Mb sequence that was believed to be the E. coli backbone; the remaining 1.4 Mb consisted of O157:H7-specific sequences that appeared to be foreign DNA acquired through lateral gene transfer (36). Another genome comparison study on O26, O111, and O103 E. coli strains found a large number of strain-specific genes; however, virulence genes were well conserved between strains (49). The conservation of virulence genes between strains may be explained by the results of another study which has shown through phylogenetic analysis that pathogenic strains of E. coli have evolved and acquired virulence plasmids in parallel (54). Dendrogram analyses of our strains also support this hypothesis. We found that the majority of the non-STEC strains had a different lineage than the STEC strains within the O serogroups (Fig. 2 to 7); however, STEC strains were more closely related to non-STEC strains within the same O serogroup than to STEC strains in the other O serogroups (data not shown).
Gene differences in the O-antigen gene clusters of E. coli strains have been used to create PCR-based assays for detection of specific non-O157 strains (24, 28). PCR assays were developed that were specific to the O26 and O103 serogroups and also were able to detect O26 and O103 strains in apple juice (24). One of the major differences between previous PCR-based assay targets and the polymorphisms found in this study was the ability to differentiate between STEC and non-STEC strains. Many strains of non-O157 E. coli are not considered pathogenic, and it will be important for the meat industry when responding to the mandatory testing requirements beginning in 2012 to be able to differentiate between regulated disease-causing non-O157 STEC strains and non-STEC strains that only share the same O-antigen genes.
We identified polymorphisms in a collection of E. coli strains from each of the six O-serogroups that were not only specific to the O serogroup but also, to a great extent, unique to STEC strains. There were several false negatives and false positives associated with the identified STEC-associated alleles for the strains sequenced in our collection; these included five false positives for the O26 strains, two false positives for the O103 strains, four false positives for the O111 strains, and two false negatives and one false positive for the O121 strains. However, 100% sensitivity or specificity for any STEC-associated allele would be highly unlikely due to the evolutionary nature of E. coli and lateral gene transfer. It is interesting to note that the two false-negative O121 strains (which contained stx1 but did not have the STEC-associated allele) did not contain eae or ehxA and were positive for the H7 antigen. This is in contrast to the majority of O121 STEC strains, which are positive for stx2 and the H19 antigen. All but one of the false-positive strains (i.e., had the STEC-associated allele but did not contain stx) in our sequencing panel contained eae.
Research is ongoing, and questions still remain unanswered as to what genes are necessary to confer virulence in humans (3, 5, 67). Research has shown that stx is associated with severe disease and hemolytic-uremic syndrome (HUS) (34), whereas eae is associated with attachment of the bacteria to the epithelium (1). Several studies have shown a significant association between stx2 and eae and severe clinical disease (11, 27, 64). The O121 strains sequenced in this study may have recently acquired stx1 or lost eae and ehxA. Another study conducted in Germany found that EPEC strains associated with bloody diarrhea and cases of HUS were closely related to STEC strains and clustered in the same multilocus sequence typing (MLST) clonal complexes (7). The authors hypothesized that these EPEC strains were originally STEC strains that lost stx once infection was established in the patients. Other studies have suggested that STEC strains evolved from EPEC strains by acquiring stx (54, 66). In the absence of genes encoding alternate mechanisms for colonizing hosts, STEC strains that do not contain eae or ehxA may be reduced in virulence and less clinically important. These strains may have once had the ability to cause severe disease but due to the loss of attachment genes may now be unable to sustain infection in a human host. The differences in the H antigen between the stx1 and stx2 strains also support the alternative hypothesis that an stx1 phage infected a particular lineage of O121 strains.
The five false-positive O26 strains (which have the STEC allele but do not contain stx) are also of particular interest because four of them contain eae (Fig. 1). Research has been conducted on the ability of O26 aEPEC strains to acquire stx-carrying phages and O26 EHEC strains to lose stx in vitro (8). It has been found that stx-carrying bacteriophages facilitate the bidirectional conversion between O26 aEPEC and EHEC pathotypes. Bugarel et al. identified espK as a unique genetic marker for EHEC and EHEC derivative strains (EHEC strains that have lost stx) (18). The espK gene was present in three of the four false-positive strains that contained eae. Two of these strains were found in cluster 4 and had the STEC alleles for polymorphisms rmlA 30 G→T and fnl1 88 G→A. The third strain was found in cluster 2 and contained the STEC allele for polymorphism wzx 953 G→T. Interestingly, this strain also contained nleB, whereas the other false-positive strain in cluster 2 did not contain espK or nleB. The finding of espK in the O26 false-positives would indicate that these were EHEC strains that lost their Shiga toxin-encoding genes.
Overall, the MALDI-TOF assays used to determine the frequency of the STEC alleles proved to accurately detect STEC strains within serogroups in this study (Table 2). The panel of bacterial strains used to validate the MALDI-TOF assays was independent from the bacterial strains used in the sequencing panel. Due to the complex relationship between the presence of virulence genes and the ability to cause human infection, we estimated the sensitivity and specificity for each of the assays using three different virulence gene classification groups. The first estimate classified all strains with stx alone as a true positive, the second estimate classified all strains that contained stx with eae as a true positive, and the third estimate classified all strains that contained stx with both eae and ehxA as a true positive. The different virulence gene classification groups only affected the sensitivity and specificity estimates for the O serogroups with a large diversity of strains, which included the O26, O45, O111, and O121 strains. Sensitivity and specificity estimates for classification with stx alone and stx with eae did not vary dramatically for the O111 and O26 groups because the majority of the strains with stx also contained eae. The O121 group and several of the assays in the O45 group showed an increased sensitivity for stx with eae and stx with eae and ehxA classifications because of strains with stx that did not contain eae or ehxA and did not have the STEC-associated allele (Table 2).
Overall, the sensitivity estimates of the 21 assays were high, except for the assay targeting the O103 wbtD 937 C→T polymorphism (75.2%) (Table 2). The low sensitivity was a result of 25 O103:H25 strains that did not have the STEC-associated allele and were classified as false negatives. Interestingly, two of the O103:H25 strains, one of which was included in the SNP discovery set, did have the STEC-associated allele. Additional sequencing of O103:H25 strains will be needed to determine whether an alternate or additional SNP is needed to incorporate O103:H25 STEC strains.
The majority of the assays had a high specificity, except for the O26 rmlA 30 G→T (with wzx 953 T→G) (56.2%) and O111 wbdH 1006 G→A (44.9%) assays (Table 2). The low specificity estimates were a result of the large number of false positives. A large number of non-O111 or non-O26 E. coli strains as well as Salmonella strains contained the O26 or O111 STEC-associated alleles for these two assays. One of the explanations for the large number of false positives may be the close relationship between E. coli and Salmonella. The polymorphisms in these two assays may be contained within a region that is highly conserved both between E. coli serogroups and between E. coli and Salmonella. Other studies have also found a high degree of similarity in the genetic sequences of the O antigens in E. coli and Salmonella enterica (37, 43).
Thirty-five (21.3%) of the strains sequenced in this study did not contain stx but did contain eae and were classified as EPEC, and the majority were O26 or O111 strains. Dendrogram analysis revealed some interesting patterns in regard to potential virulence factors and EPEC versus STEC strains (Fig. 2 and 5). The role of these strains in human illness is not fully understood; however, from the dendrogram analysis it appears that distinct lineages have evolved, perhaps through environmental or selective pressures to promote survival of the bacteria. In the O111 and O26 dendrograms, there was a distinct lineage separation, with the majority of the STEC and EPEC strains clustered separately. The majority of the O111 EPEC strains were typical and contained nleB (Fig. 5), but the O26 EPEC strains were atypical and only a few contained nleB (Fig. 2). In contrast to nleB, which was found only in the EPEC strains in the O26 and O111 serogroups, efa1 (lifA) was found in the majority of both the EPEC and STEC strains. nleB and efa1 (lifA) were found on the same pathogenicity island (OI-122); however, other studies have also reported variation in the carriage of these genes between EPEC strains and the existence of two main variants of the OI-122 pathogenicity island (3). It was also interesting to note that nleB was found in the STEC strains of the O45, O103, and O145 serogroups (Fig. 3, 4, and 7) and was absent in all of the O121 strains (Fig. 6). Different selective pressures and evolutionary niches may be responsible for differences in the carriage of nleB in O26 and O111 EPEC strains versus O45, O103, and O145 STEC strains. In contrast to the results found in this study, Bugarel et al. detected nleB in EHEC strains belonging to the O26:H11, O103:H2, O111:H8, O121:H19, and O145:H28 serotypes (16, 17). The primary reason for the contrasting results is most likely the different sample populations. Bugarel et al. investigated strains from the National Reference Laboratory for E. coli at the Federal Institute for Risk Assessment in Berlin, Germany, and the French Food Safety Agency in Maisons-Alfort, France. The strains investigated in this study were primarily from sources in the United States. It is not unusual to see differences in strain carriage across geographic regions or across source demographics.
Another group of interest is highlighted in green in the O111 dendrogram (Fig. 5). Strains in this group had identical O-antigen gene sequences and virulence gene profiles and are mostly serotype O111:H11. It is interesting that the majority of these strains grouped separately from the other O111 STEC strains, because the H11 antigen strains are primarily found in bovine hosts and do not typically cause disease in humans. The STEC serotype O111:H8 strains highlighted in red (Fig. 5) are more commonly associated with human illness (14, 19). These STEC O111:H11 strains contained the same polymorphisms as the O111:H8 STEC strains but also had two additional unique polymorphisms.
The presence of particular virulence genes does not appear to be host specific. In the O111 serogroup, nleB was found mostly in strains derived from humans, whereas in the O26 serogroup, nleB was found in environmental strains (any strain from a nonhuman source). In the O111 and O26 serogroups, ehxA was found in strains originating from both human and environmental sources. However, in order to further explore relationships between the genes and potential host-specific factors, a larger and more diverse sample of strains is needed. The majority of the O26 EPEC strains were environmental, and the majority of both the O111 EPEC and STEC strains were from human sources. This may be the reason we did not see any host-specific gene profiles and why all the typical EPEC strains were O111 strains. Other studies have found that typical EPEC strains are most commonly isolated from human sources and not found in bovine sources (68).
The polymorphisms discovered in this study were unique, because not only do they differentiate between the six O serogroups but also they are associated with STEC strains. Recent outbreaks of non-O157 STEC in the United States and Europe have increased awareness of these strains and highlighted the need to develop accurate tests for identification. Discussions on public health and prevention have led to the development of regulations in the meat industry concerning testing for certain non-O157 STEC strains. In order to prevent unnecessary loss of nonintact raw beef products and revenue, it will be essential to have tests available that are both fast and accurate. The polymorphisms presented in this study can be used to develop tests that should facilitate the identification of O26, O45, O103, O111, O121, and O145 STEC strains. The methods applied in this study can also be used to identify potential STEC-associated alleles in other non-O157 STEC serogroups. Sequencing of additional strains in the six serogroups presented in this article as well as additional serogroups will allow us to have a greater confidence in our sensitivity estimates, further understand the evolution of these strains, and potentially answer questions regarding the role of particular virulence genes in pathogenicity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sandy Fryda-Bradley, Renee Godtel, and Steve Simcox for their technical assistance and Joan Rosch for secretarial support.
The use of product and company names is necessary to accurately report the methods and results; however, the USDA neither guarantees nor warrants the standard of the products, and the use of the names by the USDA implies no approval of the product to the exclusion of others that may also be suitable.
Footnotes
Published ahead of print 13 July 2012
Supplemental material for this article can be found at http://aem.asm.org/.
REFERENCES
- 1. Afset JE, et al. 2008. Phylogenetic backgrounds and virulence profiles of atypical enteropathogenic Escherichia coli strains from a case-control study using multilocus sequence typing and DNA microarray analysis. J. Clin. Microbiol. 46:2280–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Afset JE, Bevanger L, Romundstad P, Bergh K. 2004. Association of atypical enteropathogenic Escherichia coli (EPEC) with prolonged diarrhoea. J. Med. Microbiol. 53:1137–1144 [DOI] [PubMed] [Google Scholar]
- 3. Afset JE, et al. 2006. Identification of virulence genes linked with diarrhea due to atypical enteropathogenic Escherichia coli by DNA microarray analysis and PCR. J. Clin. Microbiol. 44:3703–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arthur TM, Barkocy-Gallagher GA, Rivera-Betancourt M, Koohmaraie M. 2002. Prevalence and characterization of non-O157 Shiga toxin-producing Escherichia coli on carcasses in commercial beef cattle processing plants. Appl. Environ. Microbiol. 68:4847–4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aslani MM, Bouzari S. 2009. Characterization of virulence genes of non-O157 Shiga toxin-producing Escherichia coli isolates from two provinces of Iran. Jpn. J. Infect. Dis. 62:16–19 [PubMed] [Google Scholar]
- 6. Banatvala N, et al. 2001. The United States National Prospective Hemolytic Uremic Syndrome Study: microbiologic, serologic, clinical, and epidemiologic findings. J. Infect. Dis. 183:1063–1070 [DOI] [PubMed] [Google Scholar]
- 7. Bielaszewska M, et al. 2008. Shiga toxin-negative attaching and effacing Escherichia coli: distinct clinical associations with bacterial phylogeny and virulence traits and inferred in-host pathogen evolution. Clin. Infect. Dis. 47:208–217 [DOI] [PubMed] [Google Scholar]
- 8. Bielaszewska M, et al. 2007. Shiga toxin gene loss and transfer in vitro and in vivo during enterohemorrhagic Escherichia coli O26 infection in humans. Appl. Environ. Microbiol. 73:3144–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blanco JE, et al. 2004. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from human patients: prevalence in Lugo, Spain, from 1992 through 1999. J. Clin. Microbiol. 42:311–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blanco M, et al. 1997. Distribution and characterization of faecal verotoxin-producing Escherichia coli (VTEC) isolated from healthy cattle. Vet. Microbiol. 54:309–319 [DOI] [PubMed] [Google Scholar]
- 11. Boerlin P, et al. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bosilevac JM, Koohmaraie M. 2011. Prevalence and characterization of non-O157 Shiga toxin-producing Escherichia coli isolates from commercial ground beef in the United States. Appl. Environ. Microbiol. 77:2103–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brandal LT, et al. 2012. Norwegian sheep are an important reservoir for human-pathogenic Escherichia coli O26:H11. Appl. Environ. Microbiol. 78:4083–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brooks JT, et al. 2004. Outbreak of Shiga toxin-producing Escherichia coli O111:H8 infections among attendees of a high school cheerleading camp. Clin. Infect. Dis. 38:190–198 [DOI] [PubMed] [Google Scholar]
- 15. Brooks JT, et al. 2005. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983–2002. J. Infect. Dis. 192:1422–1429 [DOI] [PubMed] [Google Scholar]
- 16. Bugarel M, Beutin L, Fach P. 2010. Low-density macroarray targeting non-locus of enterocyte effacement effectors (nle genes) and major virulence factors of Shiga toxin-producing Escherichia coli (STEC): a new approach for molecular risk assessment of STEC isolates. Appl. Environ. Microbiol. 76:203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bugarel M, Beutin L, Martin A, Gill A, Fach P. 2010. Micro-array for the identification of Shiga toxin-producing Escherichia coli (STEC) seropathotypes associated with hemorrhagic colitis and hemolytic uremic syndrome in humans. Int. J. Food Microbiol. 142:318–329 [DOI] [PubMed] [Google Scholar]
- 18. Bugarel M, Beutin L, Scheutz F, Loukiadis E, Fach P. 2011. Identification of genetic markers for differentiation of Shiga toxin-producing, enteropathogenic, and avirulent strains of Escherichia coli O26. Appl. Environ. Microbiol. 77:2275–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. CDC. 2000. Escherichia coli O111:H8 outbreak among teenage campers—Texas, 1999. MMWR Morb. Mortal. Wkly. Rep. 49:321–324 [PubMed] [Google Scholar]
- 20. CDC. 1993. Update: multistate outbreak of Escherichia coli O157:H7 infections from hamburgers—western United States, 1992–1993. MMWR Morb. Mortal. Wkly. Rep. 42:258–263 [PubMed] [Google Scholar]
- 21. Clawson ML, et al. 2009. Phylogenetic classification of Escherichia coli O157:H7 strains of human and bovine origin using a novel set of nucleotide polymorphisms. Genome Biol. 10:R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. DebRoy C, Fratamico PM, Roberts E, Davis MA, Liu Y. 2005. Development of PCR assays targeting genes in O-antigen gene clusters for detection and identification of Escherichia coli O45 and O55 serogroups. Appl. Environ. Microbiol. 71:4919–4924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DebRoy C, et al. 2004. Detection of Escherichia coli serogroups O26 and O113 by PCR amplification of the wzx and wzy genes. Appl. Environ. Microbiol. 70:1830–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. D'Souza JM, Wang L, Reeves P. 2002. Sequence of the Escherichia coli O26 O-antigen gene cluster and identification of O26 specific genes. Gene 297:123–127 [DOI] [PubMed] [Google Scholar]
- 26. Durso LM, Bono JL, Keen JE. 2005. Molecular serotyping of Escherichia coli O26:H11. Appl. Environ. Microbiol. 71:4941–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ethelberg S, et al. 2004. Virulence factors for hemolytic uremic syndrome, Denmark. Emerg. Infect. Dis. 10:842–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a. Federal Register 2011. Shiga toxin-producing Escherichia coli in certain raw beef products. Docket no. FSIS-2012-0023. Fed. Regist. 76:58157–58165 [Google Scholar]
- 28. Feng L, et al. 2005. Structural and genetic characterization of enterohemorrhagic Escherichia coli O145 O-antigen and development of an O145 serogroup-specific PCR assay. J. Bacteriol. 187:758–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Feng L, et al. 2004. Structure of the Shigella dysenteriae 7 O-antigen gene cluster and identification of its antigen specific genes. Microb. Pathog. 36:109–115 [DOI] [PubMed] [Google Scholar]
- 30. Fratamico PM, et al. 2011. Detection by multiplex real-time polymerase chain reaction assays and isolation of Shiga toxin-producing Escherichia coli serogroups O26, O45, O103, O111, O121, and O145 in ground beef. Foodborne Pathog. Dis. 8:601–607 [DOI] [PubMed] [Google Scholar]
- 31. Fratamico PM, Briggs CE, Needle D, Chen CY, DebRoy C. 2003. Sequence of the Escherichia coli O121 O-antigen gene cluster and detection of enterohemorrhagic E. coli O121 by PCR amplification of the wzx and wzy genes. J. Clin. Microbiol. 41:3379–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fratamico PM, DebRoy C, Miyamoto T, Liu Y. 2009. PCR detection of enterohemorrhagic Escherichia coli O145 in food by targeting genes in the E. coli O145 O-antigen gene cluster and the Shiga toxin 1 and Shiga toxin 2 genes. Foodborne Pathog. Dis. 6:605–611 [DOI] [PubMed] [Google Scholar]
- 33. Fratamico PM, DebRoy C, Strobaugh TP, Jr, Chen CY. 2005. DNA sequence of the Escherichia coli O103 O-antigen gene cluster and detection of enterohemorrhagic E. coli O103 by PCR amplification of the wzx and wzy genes. Can. J. Microbiol. 51:515–522 [DOI] [PubMed] [Google Scholar]
- 34. Gerber A, Karch H, Allerberger F, Verweyen HM, Zimmerhackl LB. 2002. Clinical course and the role of Shiga toxin-producing Escherichia coli infection in the hemolytic-uremic syndrome in pediatric patients, 1997–2000, in Germany and Austria: a prospective study. J. Infect. Dis. 186:493–500 [DOI] [PubMed] [Google Scholar]
- 35. Griffin PM, Tauxe RV. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13:60–98 [DOI] [PubMed] [Google Scholar]
- 36. Hayashi T, et al. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11–22 [DOI] [PubMed] [Google Scholar]
- 37. Hu B, et al. 2010. Structural and genetic evidence for the close relationship between Escherichia coli O71 and Salmonella enterica O28 O-antigens. FEMS Immunol. Med. Microbiol. 59:161–169 [DOI] [PubMed] [Google Scholar]
- 38. Jure MA, et al. 1998. Association between hemolytic uremic syndrome and verotoxin-producing strains of E. coli. Rev. Latinoam. Microbiol. 40:1–8 [PubMed] [Google Scholar]
- 39. Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 40. Karmali MA, Steele BT, Petric M, Lim C. 1983. Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet i:619–620 [DOI] [PubMed] [Google Scholar]
- 41. Kim JP, et al. 2005. A case of hemolytic uremic syndrome with hemorrhagic colitis due to Escherichia coli O111 infection. Korean J. Gastroenterol. 45:365–368 [PubMed] [Google Scholar]
- 42. Liptakova A, et al. 2005. Hemolytic uremic syndrome caused by verotoxin-producing Escherichia coli O26. Case report. Folia Microbiol. (Praha) 50:95–98 [DOI] [PubMed] [Google Scholar]
- 43. Liu B, et al. 2010. Genetic and structural relationships of Salmonella O55 and Escherichia coli O103 O-antigens and identification of a 3-hydroxybutanoyltransferase gene involved in the synthesis of a Fuc3N derivative. Glycobiology 20:679–688 [DOI] [PubMed] [Google Scholar]
- 44. Misselwitz J, et al. 2003. Cluster of hemolytic-uremic syndrome caused by Shiga toxin-producing Escherichia coli O26:H11. Pediatr. Infect. Dis. J. 22:349–354 [DOI] [PubMed] [Google Scholar]
- 45. Narimatsu H, Ogata K, Makino Y, Ito K. 2010. Distribution of non-locus of enterocyte effacement pathogenic island-related genes in Escherichia coli carrying eae from patients with diarrhea and healthy individuals in Japan. J. Clin. Microbiol. 48:4107–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nguyen RN, Taylor LS, Tauschek M, Robins-Browne RM. 2006. Atypical enteropathogenic Escherichia coli infection and prolonged diarrhea in children. Emerg. Infect. Dis. 12:597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ogierman MA, Paton AW, Paton JC. 2000. Up-regulation of both intimin and eae-independent adherence of Shiga toxigenic Escherichia coli O157 by ler and phenotypic impact of a naturally occurring ler mutation. Infect. Immun. 68:5344–5353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ogura Y, et al. 2007. Extensive genomic diversity and selective conservation of virulence-determinants in enterohemorrhagic Escherichia coli strains of O157 and non-O157 serotypes. Genome Biol. 8:R138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ogura Y, et al. 2009. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 106:17939–17944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pai CH, Gordon R, Sims HV, Bryan LE. 1984. Sporadic cases of hemorrhagic colitis associated with Escherichia coli O157:H7. Clinical, epidemiologic, and bacteriologic features. Ann. Intern. Med. 101:738–742 [DOI] [PubMed] [Google Scholar]
- 52. Paton AW, Paton JC. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36:598–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Piercefield EW, Bradley KK, Coffman RL, Mallonee SM. 2010. Hemolytic uremic syndrome after an Escherichia coli O111 outbreak. Arch. Intern. Med. 170:1656–1663 [DOI] [PubMed] [Google Scholar]
- 54. Reid SD, Herbelin CJ, Bumbaugh AC, Selander RK, Whittam TS. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64–67 [DOI] [PubMed] [Google Scholar]
- 55. Riley LW, et al. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681–685 [DOI] [PubMed] [Google Scholar]
- 56. Rodrigue DC, et al. 1995. A university outbreak of Escherichia coli O157:H7 infections associated with roast beef and an unusually benign clinical course. J. Infect. Dis. 172:1122–1125 [DOI] [PubMed] [Google Scholar]
- 57. Ryan CA, et al. 1986. Escherichia coli O157:H7 diarrhea in a nursing home: clinical, epidemiological, and pathological findings. J. Infect. Dis. 154:631–638 [DOI] [PubMed] [Google Scholar]
- 58. Siegler RL, et al. 2003. Response to Shiga toxin 1 and 2 in a baboon model of hemolytic uremic syndrome. Pediatr. Nephrol. 18:92–96 [DOI] [PubMed] [Google Scholar]
- 59. Smith TP, Godtel RA, Lee RT. 2000. PCR-based setup for high-throughput cDNA library sequencing on the ABI 3700 automated DNA sequencer. Biotechniques 29:698–700 [DOI] [PubMed] [Google Scholar]
- 60. Tarr PI. 1995. Escherichia coli O157:H7: clinical, diagnostic, and epidemiological aspects of human infection. Clin. Infect. Dis. 20:1–8; quiz, 9–10 [DOI] [PubMed] [Google Scholar]
- 61. Tennant SM, et al. 2009. Characterisation of atypical enteropathogenic E. coli strains of clinical origin. BMC Microbiol. 9:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang L, Curd H, Qu W, Reeves PR. 1998. Sequencing of Escherichia coli O111 O-antigen gene cluster and identification of O111-specific genes. J. Clin. Microbiol. 36:3182–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang L, Reeves PR. 1998. Organization of Escherichia coli O157 O-antigen gene cluster and identification of its specific genes. Infect. Immun. 66:3545–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Werber D, et al. 2003. Strong association between Shiga toxin-producing Escherichia coli O157 and virulence genes stx2 and eae as possible explanation for predominance of serogroup O157 in patients with haemolytic uraemic syndrome. Eur. J. Clin. Microbiol. Infect. Dis. 22:726–730 [DOI] [PubMed] [Google Scholar]
- 65. Whitfield C. 1995. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 3:178–185 [DOI] [PubMed] [Google Scholar]
- 66. Wick LM, Qi W, Lacher DW, Whittam TS. 2005. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J. Bacteriol. 187:1783–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wickham ME, et al. 2006. Bacterial genetic determinants of non-O157 STEC outbreaks and hemolytic-uremic syndrome after infection. J. Infect. Dis. 194:819–827 [DOI] [PubMed] [Google Scholar]
- 68. Yuste M, De La Fuente R, Ruiz-Santa-Quiteria JA, Cid D, Orden JA. 2006. Detection of the astA (EAST1) gene in attaching and effacing Escherichia coli from ruminants. J. Vet. Med. B Infect. Dis. Vet. Public Health 53:75–77 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.