Abstract
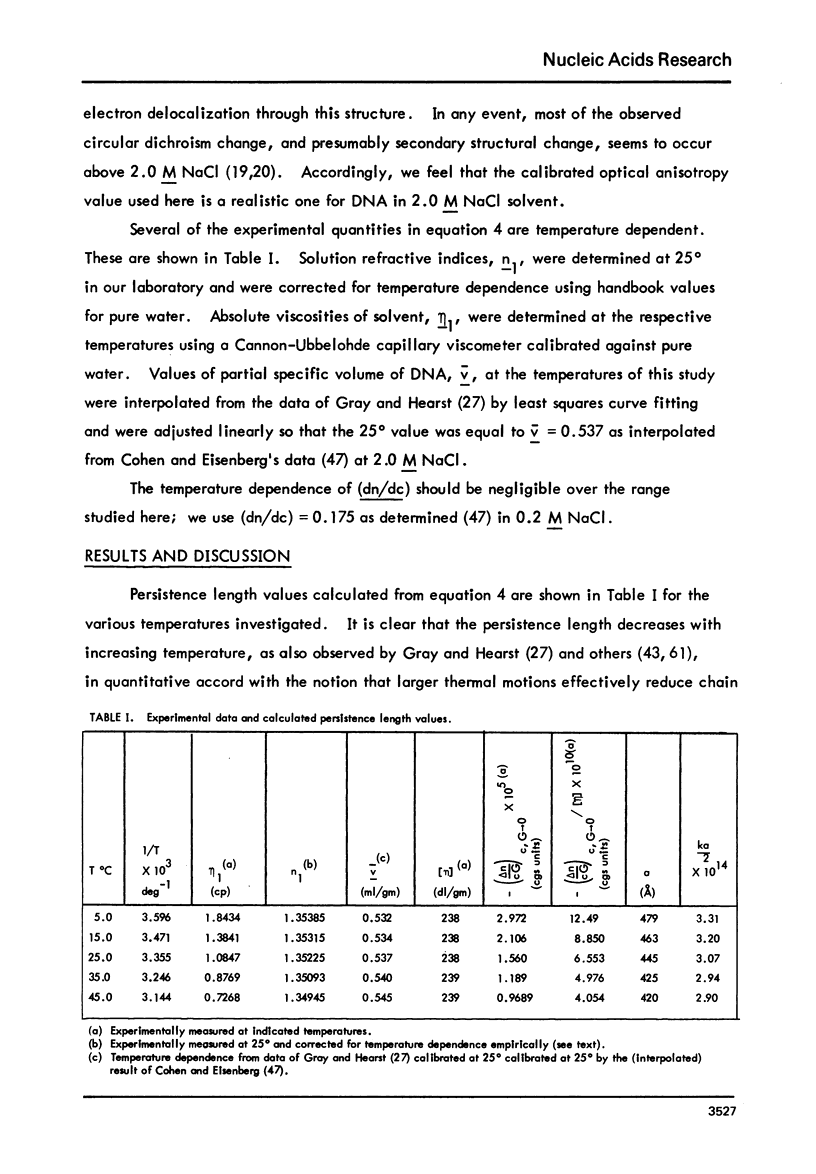
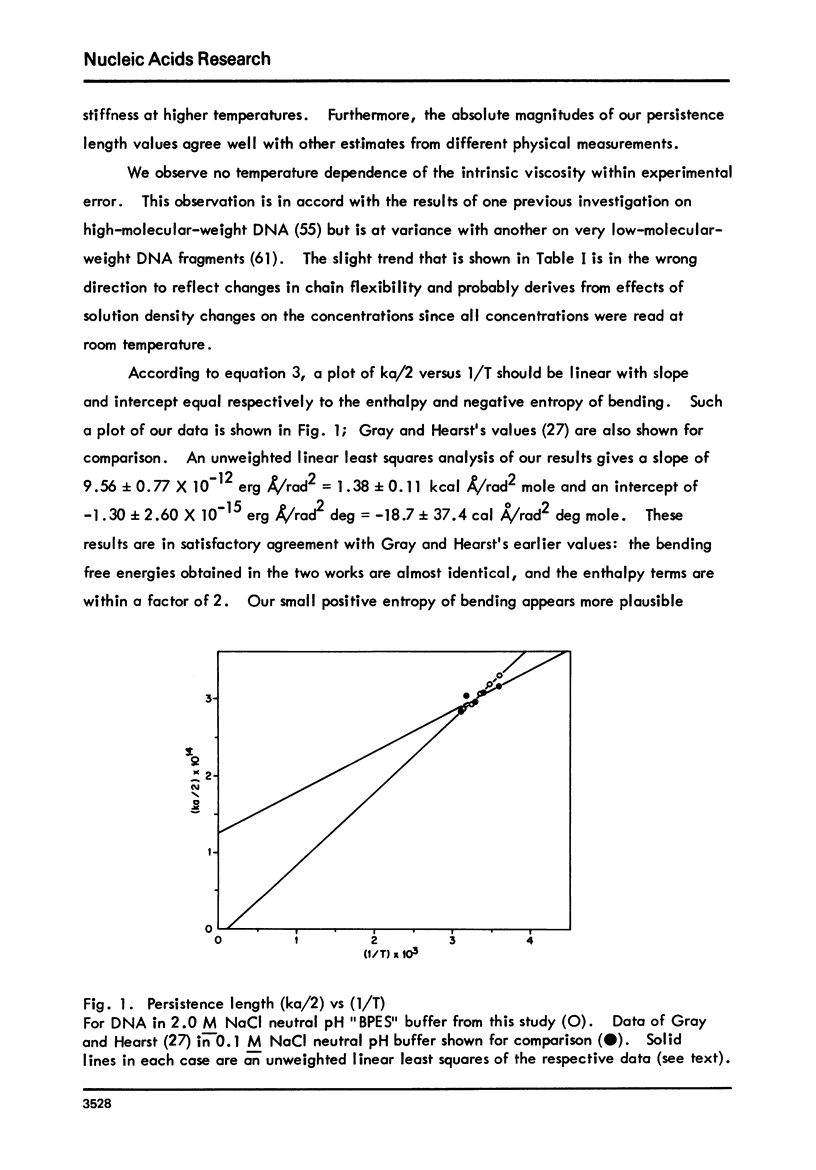
The persistence length of high-molecular-weight, monodisperse-bihelical DNA has been evaluated from low-shear flow birefingence and viscosity data at several temperatures in 2.0 M Nacl neutral pH buffer. At these solvent conditions, both the DNA and histone components of chromatin nu-bodies have structural features similar to those in the intact nucleohistone complex at low ionic strength. The theory of Landau and Lifshitz is used to relate the experimental result to the thermodynamic functions for bending 140 nucleotide pairs of DNA into a plausible model structure: per nu-body, delta Gb=43.8 +/- 5.3 kcal/mole, delta Hb= 45.7 +/- 3.7 kcal/mole, and delta Sb = 6.2 +/- 12.4 entropy units. This bending free energy is comparable to or less than that estimated to be required for a kinked DNA configuration and appears to be well within the range of estimated electrostatic free energies available from DNA-histone interactions in a nu-body assembly.
Full text
PDF














Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bruner R., Vinograd J. The evaluation of standard sedimentation coefficients of sodium RNA and sodium DNA from sedimentation velocity data in concentrated NaCl and CsCl solutions. Biochim Biophys Acta. 1965 Sep 6;108(1):18–29. doi: 10.1016/0005-2787(65)90104-8. [DOI] [PubMed] [Google Scholar]
- Cohen G., Eisenberg H. Conformation studies on the sodium and cesium salts of calf thymus deoxyribonucleic acid (DNA). Biopolymers. 1966 Apr-May;4(4):429–440. doi: 10.1002/bip.1966.360040404. [DOI] [PubMed] [Google Scholar]
- Cohen G., Eisenberg H. Deoxyribonucleate solutions: sedimentation in a density gradient, partial specific volumes, density and refractive index increments, and preferential interactions. Biopolymers. 1968;6(8):1077–1100. doi: 10.1002/bip.1968.360060805. [DOI] [PubMed] [Google Scholar]
- Crick F. H., Klug A. Kinky helix. Nature. 1975 Jun 12;255(5509):530–533. doi: 10.1038/255530a0. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Zimm B. H. Viscosity and sedimentation of the DNA from bacteriophages T2 and T7 and the relation to molecular weight. J Mol Biol. 1965 Jul;12(3):525–536. doi: 10.1016/s0022-2836(65)80310-2. [DOI] [PubMed] [Google Scholar]
- DAVISON P. F., FREIFELDER D., HEDE R., LEVINTHAL C. The structural unity of the DNA of T2 bacteriophage. Proc Natl Acad Sci U S A. 1961 Aug;47:1123–1129. doi: 10.1073/pnas.47.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C., Weintraub H. Chromosomal proteins and chromatin structure. Annu Rev Biochem. 1975;44:725–774. doi: 10.1146/annurev.bi.44.070175.003453. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Bellard M., Oudet P., Chambon P. Stability of nucleosomes in native and reconstituted chromatins. Nucleic Acids Res. 1976 Nov;3(11):3173–3192. doi: 10.1093/nar/3.11.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey J. E., Eisenberg H. The flexibility of low molecular weight double-stranded DNA as a function of length. I. Light scattering measurements and the estimation of persistence lengths from light scattering, sedimentation and viscosity. Biophys Chem. 1976 Sep;5(3):301–318. doi: 10.1016/0301-4622(76)80042-7. [DOI] [PubMed] [Google Scholar]
- Gralla J., Crothers D. M. Free energy of imperfect nucleic acid helices. II. Small hairpin loops. J Mol Biol. 1973 Feb 5;73(4):497–511. doi: 10.1016/0022-2836(73)90096-x. [DOI] [PubMed] [Google Scholar]
- Gray H. B., Jr, Hearst J. E. Flexibility of native DNA from the sedimentation behavior as a function of molecular weight and temperature. J Mol Biol. 1968 Jul 14;35(1):111–129. doi: 10.1016/s0022-2836(68)80041-5. [DOI] [PubMed] [Google Scholar]
- Harrington R. E. Flow birefringence of T2 bacteriophage DNA. Biopolymers. 1968;6(1):105–116. doi: 10.1002/bip.1968.360060109. [DOI] [PubMed] [Google Scholar]
- Harrington R. E. Optico-hydrodynamic properties of high molecular weight DNA from steady-state flow birefringence and viscosity at extremely low velocity gradients. Biopolymers. 1970 Feb;9(2):159–193. doi: 10.1002/bip.1970.360090204. [DOI] [PubMed] [Google Scholar]
- Harrington R. E. The flow birefringence of persistence length deoxyribonucleic acid. Hydrodynamic properties, optical anisotropy, and hydration shell anistropy. J Am Chem Soc. 1970 Nov 18;92(23):6957–6964. doi: 10.1021/ja00726a038. [DOI] [PubMed] [Google Scholar]
- Hirsh J., Schleif R. High resolution electron microscopic studies of genetic regulation. J Mol Biol. 1976 Dec;108(2):471–490. doi: 10.1016/s0022-2836(76)80131-3. [DOI] [PubMed] [Google Scholar]
- Houssier C., Fredericq E. Electro-optical properties of nucleic acids and nucleoproteins. I. Study of the gel-forming deoxyribonucleohistone. Biochim Biophys Acta. 1966 May 12;120(1):113–130. doi: 10.1016/0926-6585(66)90283-4. [DOI] [PubMed] [Google Scholar]
- Jolly D., Eisenberg H. Photon correlation spectroscopy, total intensity light scattering with laser radiation, and hydrodynamic studies of a well fractionated DNA sample. Biopolymers. 1976 Jan;15(1):61–95. doi: 10.1002/bip.1976.360150107. [DOI] [PubMed] [Google Scholar]
- KILKSON R., MAESTRE M. F. Structure of T-2 bacteriophage. Nature. 1962 Aug 4;195:494–495. doi: 10.1038/195494a0. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Thomas J. O. Chromatin structure; oligomers of the histones. Science. 1974 May 24;184(4139):865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- Kovacic R. T., van Holde K. E. Sedimentation of homogeneous double-strand DNA molecules. Biochemistry. 1977 Apr 5;16(7):1490–1498. doi: 10.1021/bi00626a038. [DOI] [PubMed] [Google Scholar]
- LEVINTHAL C., DAVISON P. F. Degradation of deoxyribonucleic acid under hydrodynamic shearing forces. J Mol Biol. 1961 Oct;3:674–683. doi: 10.1016/s0022-2836(61)80030-2. [DOI] [PubMed] [Google Scholar]
- MARVIN D. A., SPENCER M., WILKINS M. H., HAMILTON L. D. The molecular configuration of deoxyribonucleic acid. III. X-ray diffraction study of the C form of the lithium salt. J Mol Biol. 1961 Oct;3:547–565. doi: 10.1016/s0022-2836(61)80021-1. [DOI] [PubMed] [Google Scholar]
- Ohba Y. Structure of nucleohistone. I. Hydrodynamic behaviour. Biochim Biophys Acta. 1966 Jul 20;123(1):76–83. [PubMed] [Google Scholar]
- Olins A. L., Carlson R. D., Wright E. B., Olins D. E. Chromatin nu bodies: isolation, subfractionation and physical characterization. Nucleic Acids Res. 1976 Dec;3(12):3271–3291. doi: 10.1093/nar/3.12.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUBENSTEIN I., THOMAS C. A., Jr, HERSHEY A. D. The molecular weights of T2 bacteriophage DNA and its first and second breakage products. Proc Natl Acad Sci U S A. 1961 Aug;47:1113–1122. doi: 10.1073/pnas.47.8.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramm E. I., Vorob'ev V. I., Birshtein T. M., Bolotina I. A., Volkenshtein M. V. Circular dichroism of DNA and histones in the free state and in deoxyribonucleoprotein. Eur J Biochem. 1972 Feb 15;25(2):245–253. doi: 10.1111/j.1432-1033.1972.tb01690.x. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Woodbury C. P., Inman R. B. Characterization of rodlike RNA fragments. Biopolymers. 1975 Feb;14(2):393–408. doi: 10.1002/bip.1975.360140212. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Bourgeois S., Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J Mol Biol. 1970 Nov 14;53(3):401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Suzuki H., Bourgeois S. Lac repressor-operator interaction. I. Equilibrium studies. J Mol Biol. 1970 Feb 28;48(1):67–83. doi: 10.1016/0022-2836(70)90219-6. [DOI] [PubMed] [Google Scholar]
- Rinehart F. P., Hearst J. E. The ionic strength dependence of s o 20 w of phage DNA in NH 4 AC. Arch Biochem Biophys. 1972 Oct;152(2):712–722. doi: 10.1016/0003-9861(72)90267-6. [DOI] [PubMed] [Google Scholar]
- Rinehart F. P., Hearst J. E. The ionic strength dependence of the coil dimensions of viral DNA in NH 4 AC solutions. Arch Biochem Biophys. 1972 Oct;152(2):723–732. doi: 10.1016/0003-9861(72)90268-8. [DOI] [PubMed] [Google Scholar]
- Ross P. D., Scruggs R. L. Viscosity study of DNA. II. The effect of simple salt concentration on the viscosity of high molecular weight DNA and application of viscometry to the study of DNA isolated from T4 and T5 bacteriophage mutants. Biopolymers. 1968;6(8):1005–1018. doi: 10.1002/bip.1968.360060802. [DOI] [PubMed] [Google Scholar]
- Schellman J. A. Flexibility of DNA. Biopolymers. 1974 Jan;13(1):217–226. doi: 10.1002/bip.1974.360130115. [DOI] [PubMed] [Google Scholar]
- Sobell H. M., Tsai C. C., Gilbert S. G., Jain S. C., Sakore T. D. Organization of DNA in chromatin. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3068–3072. doi: 10.1073/pnas.73.9.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhai S. Energy bands and electronic delocalization in the sugar-phosphate backbone of DNA. Biopolymers. 1974;13(9):1739–1745. doi: 10.1002/bip.1974.360130906. [DOI] [PubMed] [Google Scholar]
- Van Holde K. E., Sahasrabuddhe C. G., Shaw B. R. A model for particulate structure in chromatin. Nucleic Acids Res. 1974 Nov;1(11):1579–1586. doi: 10.1093/nar/1.11.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Worcel A., Alberts B. A model for chromatin based upon two symmetrically paired half-nucleosomes. Cell. 1976 Nov;9(3):409–417. doi: 10.1016/0092-8674(76)90085-4. [DOI] [PubMed] [Google Scholar]
- Wilhelm F. X., Champagne M. H., Daune M. P. Conformation du DNA dans la nucléoprotéine. Eur J Biochem. 1970 Aug;15(2):321–330. doi: 10.1111/j.1432-1033.1970.tb01010.x. [DOI] [PubMed] [Google Scholar]



