Abstract
Scleractinian corals harbor microorganisms that form dynamic associations with the coral host and exhibit substantial genetic and ecological diversity. Microbial associates may provide defense against pathogens and serve as bioindicators of changing environmental conditions. Here we describe the bacterial assemblages associated with two of the most common and phylogenetically divergent reef-building corals in the Caribbean, Montastraea faveolata and Porites astreoides. Contrasting life history strategies and disease susceptibilities indicate potential differences in their microbiota and immune function that may in part drive changes in the composition of coral reef communities. The ribotype structure and diversity of coral-associated bacteria within the surface mucosal layer (SML) of healthy corals were assessed using denaturing gradient gel electrophoresis (DGGE) fingerprinting and 454 bar-coded pyrosequencing. Corals were sampled at disparate Caribbean locations representing various levels of anthropogenic impact. We demonstrate here that M. faveolata and P. astreoides harbor distinct, host-specific bacteria but that specificity varies by species and site. P. astreoides generally hosts a bacterial assemblage of low diversity that is largely dominated by one bacterial genus, Endozoicomonas, within the order Oceanospirillales. The bacterial assemblages associated with M. faveolata are significantly more diverse and exhibit higher specificity at the family level than P. astreoides assemblages. Both corals have more bacterial diversity and higher abundances of disease-related bacteria at sites closer to the mainland than at those furthest away. The most diverse bacterial taxa and highest relative abundance of disease-associated bacteria were seen for corals near St. Thomas, U.S. Virgin Islands (USVI) (2.5 km from shore), and the least diverse taxa and lowest relative abundance were seen for corals near our most pristine site in Belize (20 km from shore). We conclude that the two coral species studied harbor distinct bacterial assemblages within the SML, but the degree to which each species maintains specific microbial associations varies both within each site and across large spatial scales. The taxonomic scale (i.e., phylum versus genus) at which scientists examine coral-microbe associations, in addition to host-elicited factors and environmental fluctuations, must be considered carefully in future studies of the coral holobiont.
INTRODUCTION
Most eukaryotes are believed to associate with a diverse assemblage of microbial symbionts that aid in their development and health (88). In corals, bacterial assemblages found within the surface mucus layer (SML) are an important part of a larger holobiont that is composed of the coral (skeleton, tissues, and SML) along with a wide variety of other associated microbial taxa, including eukaryotes, archaea, viruses, and fungi (67). Coral microorganisms exhibit substantial genetic and ecological diversity and are believed to contribute to the overall health of the coral host (20, 51, 69, 70). In addition, these microbes may be ecologically important in that they appear to contribute to the ability of reef-building corals to adapt to and evolve under changing environmental conditions (64, 69). Just as humans are thought to have coevolved with their gut microorganisms (88), corals probably coevolved with their microbial symbionts, which likely fill a critical and beneficial role in coral colony immune function (90). Microorganisms are found throughout the coral holobiont and appear to be regulated in part by the coral host (6, 40). Recent evidence suggests that the diversity and types of coral-associated bacteria may also be tightly coupled with the clade of Symbiodinium spp. found within the coral host tissues (48, 51). Coral microbes are thought to benefit the host by providing nutritional by-products, protein, and nitrogenous compounds (46, 47, 87) and also by synthesizing essential vitamins (17). Microbial symbionts may also protect corals from disease by preventing opportunistic infections through the occupation of otherwise available niches and by producing antibacterial agents (41, 66, 72, 74). Shifts in coral microbial assemblages have been linked to bleaching (7, 59), thermal stress (48, 82), irradiance (54), disease (reviewed in reference 6), changes in dissolved organic nutrients (40, 76, 83), and shifting pH (83). Shifts in both microbial diversity and metabolism have also been related to the proximity of coral to human populations (19), demonstrating that geographic location may indirectly influence coral reef health through microbial mediation.
Although evidence suggests that congeneric coral species associate with similar microorganisms (51, 67), the metabolic functions and specificity of coral-microbe associations are less predictable than initially hypothesized (18, 19, 28, 42, 79). Coral microbiota may vary among reef locations that differ in water quality (38) and water depth (39). The operational taxonomic units (OTUs) of bacteria in the surface mucus layer of Montastraea faveolata assemblages vary on both the spatial (reef to reef) and temporal (month to month) scales, further suggesting that microbial assemblages are sensitive to surrounding environmental conditions (27). The coral microbiota is also believed to be thermally sensitive, losing protective antibacterial properties at sustained temperatures above 28 to 30°C (66). Bordering macroalgae might serve as bacterial vectors, as Halimeda macroalgae were observed to transfer white plague type II disease to exposed M. faveolata colonies (57). Thus, in addition to the host coral species, environmental factors such as water quality, location, depth, temperature, and other sessile organisms in close proximity may have significant effects on microbial diversity (4). Therefore, shifts in the coral microbiota could serve as bioindicators of environmental change and disease. Although we have made significant progress over the past decade, we still lack basic information about host specificity and the stability of healthy coral-associated microbial assemblages, including how and why assemblages change across large spatial scales and gradients of anthropogenic impact. We address these questions in part by investigating the bacteria associated with healthy coral colonies over large geographic scales and providing insight into overall reef resilience and the contribution of microorganisms to coral health. The better we understand healthy coral microbial assemblages and their spatial variability, the more efficient future studies can be in determining how and why corals are disturbed by infection and environmental stress.
The goal of the present study was to document the bacterial ribotype (16S rRNA gene) diversity associated with the SML of healthy Montastraea faveolata and Porites astreoides corals at four sites across the Caribbean: St. Thomas, U.S. Virgin Islands (USVI); Florida Keys inshore and offshore reefs; and Belize. These sites span the Caribbean Sea and are exposed to decreasing anthropogenic impacts and increasing distance from the mainland, in the order listed. This is the first study to compare structural bacterial diversity and species specificity among samples from such geographically disparate locations (sites were >1,100 km apart). Ribotype diversity was visualized by denaturing gradient gel electrophoresis (DGGE) analysis, in which band presence/absence provided molecular fingerprints for 26 colonies of each coral species. The bacterial assemblages associated with the SML of a representative subset of these colonies (n = 12) were analyzed using 454 bar-coded pyrosequencing of 16S rRNA gene amplicons.
MATERIALS AND METHODS
Sample collection.
Microbial samples were collected from the SML of apparently healthy M. faveolata and P. astreoides coral colonies on reefs adjacent to the following 4 sites: (i) MacLean Marine Science Center of the University of the Virgin Islands, St. Thomas, USVI (Flat Cay Reef, July 2009; n = 4 for M. faveolata and n = 4 for P. astreoides); (ii) Mote Marine Laboratory, Summerland Key, FL (inshore reef [Wonderland Reef], May 2009; n = 10 for M. faveolata and n = 12 for P. astreoides); (iii) Summerland Key, FL (offshore reef [Looe Key Reef], August 2009; n = 3 for M. faveolata and n = 3 for P. astreoides); and (iv) Carrie Bow Cay Field Station of the Smithsonian Institution, Belize (Southwater Cay Reef, August 2009; n = 7 for M. faveolata and n = 7 for P. astreoides). The four sampling sites increase in distance from the mainland as follows: St. Thomas, 2.5 km; Florida inshore reef, 5 km; Florida offshore reef, 12 km; and Belize, 20 km. The sampling sites span the Caribbean: the Florida Keys are ∼1,100 km from Belize and ∼1,800 km from St. Thomas, while Belize and St. Thomas are ∼2,500 km apart.
Collections were made by scuba diving to a 5- to 15-m depth and using sterile 5-ml syringes. A 5- by 5-cm area of mucus on the coral surface was gently agitated using the plastic tip of the syringe, which encourages sloughing of the viscous mucus and reduces aspiration of seawater (66). Sterile nitrile gloves were worn during collection to reduce human bacterial contamination, and syringes were capped after collection to prevent further seawater contamination. Mucus was collected from the upper surfaces of M. faveolata (n = 26) and P. astreoides (n = 26) colonies that were >1 m apart and not obviously interacting with any other coral, invertebrates, macroalgae, or benthic cyanobacteria. Syringes were placed in seawater-filled coolers and transported back to the laboratory (<3 h), where they were immediately processed for transport and subsequent culture-independent analyses. Syringes were placed tip down in test tube racks for ∼15 min to allow the mucus to settle to the bottom, and then 2 ml of concentrated mucus was transferred to cryovials and centrifuged at 10,000 × g for 10 min. The seawater supernatant was poured off and the remaining mucus pellet frozen at −20°C. Mucus pellets from M. faveolata and P. astreoides were transported to Auburn University and thawed at 4°C prior to DNA extraction using a Mobio Ultraclean microbial DNA isolation kit (Carlsbad, CA) according to the manufacturer's instructions, with an additional (10 min) heating step at 64°C to increase DNA yield. Extracted DNA was stored at −80°C until PCR amplification.
PCR amplification and DGGE protocol.
Universal bacterial primers 27F-GC (5′-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG CAG AGT TTG ATC MTG GCT CAG-3′) and 518R (5′-ATT ACC GCG GCT GCT GG-3′) were used to amplify the 16S rRNA gene, using genomic DNA extracted from coral mucus as the template. The forward primer was modified to incorporate a 40-bp GC clamp (underlined above) for resolution in a DGGE system (22, 55). These primers amplify a 491-bp section of the 16S rRNA gene of members of the domain Bacteria, including the highly variable V1-to-V3 region (3, 39). All PCRs were performed on a thermal cycler (Mastercycler ep gradient machine; Eppendorf, Hauppauge, NY), using reaction mixtures containing the following: 12.5 μl EconoTaq Plus Green 2× master mix (Lucigen, Middleton, WI) and 0.5 μl of each 20 μM primer in a final volume of 25 μl (adjusted with nuclease-free water). Strip tubes containing master mix and nuclease-free water were UV irradiated for 20 min prior to the addition of primers and DNA template under sterile conditions in a laminar flow hood to reduce contamination. The DNA template was added during an initial hot start of 3 min at 94°C, followed by a touchdown PCR protocol in which the annealing temperature was decreased from 65°C by 1°C every cycle until reaching a final temperature of 54°C, at which time 35 additional cycles of 94°C for 45 s, 54°C for 45 s, and 72°C for 1.5 min were performed; 1 final cycle of 94°C for 45 s, 54°C for 45 s, and 72°C for 7 min was followed by cooling to 4°C. PCR products were analyzed by agarose gel electrophoresis (1% [wt/vol] agarose), stained with ethidium bromide, and visualized using a UV transilluminator.
Samples were separated using a vertical gel electrophoresis apparatus (Hoefer model SE600; Hoefer Inc., San Francisco, CA) warmed with a tank heater (Lauda model M6a; Brinkmann Instruments, NY) for use as a DGGE system. PCR products were loaded onto an 8% acrylamide gel and run with 0.5× TAE buffer (Tris base, acetic acid, EDTA) and a 35 to 60% linear denaturing gradient of formamide and urea. Gels were electrophoresed in the DGGE system at 60°C, first for 15 min at 50 V and subsequently for 10 h at 100 V (or 1,000 V-h) (74). After electrophoresis, the gels were stained for 30 min with SYBR Gold nucleic acid stain at a 1:10,000 dilution ratio (Invitrogen, Carlsbad, CA) in TAE buffer, rinsed, and imaged using an AlphaImager HP gel documentation system (ProteinSimple, Santa Clara, CA). Images were saved as 8-bit TIFF files, and alignment, normalization, band class identification, and statistical analysis were performed using Bionumerics V. 5.0 (Applied Maths, Austin, TX).
Uniquely dominant and distinct bands were dabbed with a sterile pipette tip and placed directly into PCR strip tubes containing UV-sterilized nuclease-free water. Bands were reamplified with the previously described touchdown protocol by using the 27F-518R primer set without the GC clamp. PCR products were analyzed by agarose gel electrophoresis (1% [wt/vol] agarose), stained with ethidium bromide, and visualized using a UV transilluminator. Ammonium acetate-ethanol precipitation was performed, and the resulting product was amplified using a BigDye sequencing reaction mixture containing 1.0 μl of BigDye terminator, 1.5 μl of 5× buffer, 0.5 μl of 10 μM 27F, 4 μl of nuclease-free water, and 3 μl of template DNA. The resulting preparation was subjected to PCR under the following conditions: 95°C for 30 s, 50°C for 30 s, and 60°C for 4 min for 30 cycles. PCR products were purified using a BigDye XTerminator purification kit (Applied Biosciences) and shipped to the Smithsonian Institution Laboratories of Analytical Biology (Suitland, MD) for sequencing. The Smithsonian Institution performed high-throughput (96-well) Sanger sequencing on an ABI sequencer. Sequences were trimmed using CLC Genomics Workbench (CLC Bio, Cambridge, MA) and compared to the GenBank nr/nt database by BLASTn searches, and those sequences with >96% identity and expect values (E values) of <1 × 10−20 were accepted for downstream analysis.
DGGE analysis.
DGGE images were imported into Bionumerics V. 5.0 (Applied Maths) and subjected to a series of steps to allow multiple gel images to be compared reliably at one time: (i) each sample lane was identified, (ii) a background subtraction was applied, (iii) each lane was normalized to the reference standards run in each gel, and (iv) each band was identified and quantified. Sample comparison and band matching were initially optimized in Bionumerics, in which band classes were constructed based on optimal position tolerance and optimization settings. A bifurcating hierarchical dendrogram with a similarity matrix representing sample clusters was constructed for each coral species, using the Ward algorithm and Dice coefficients derived from the band alignment. A binary matrix based on band presence/absence was exported from Bionumerics, and singlet bands were removed. The binary matrix of DGGE profiles was converted to a distance matrix (Jaccard). Kruskal's nonmetric multidimensional scaling (nMDS) analysis and permutational multivariate analysis of variance (PERMANOVA) with pairwise comparisons were used to assess the multivariate relationships within and between sampling sites (50). The nMDS analysis was used to arrange multivariate data in a two-dimensional plane based on similarity coefficients between different samples. The Euclidean distances (we used the Jaccard index) between points in an nMDS plot are inversely proportional to the similarity of the samples. Kruskal's stress formula was used as an informal method of determining the appropriate number of dimensions (50). These data were analyzed using the metaMDS and Adonis utilities within the Vegan package in the R statistical package (31) and using PERMANOVA+ within the PRIMER 6.0 package (Primer-E Ltd.). Both software packages provided similar results, and the relationships among samples are represented in a plot of the first two dimensions of the nMDS results from PRIMER 6.0.
PCR and pyrosequencing preparation.
A 456-bp region of the 16S rRNA gene that includes the highly variable regions V3 and V4 was selected for bar-coded pyrosequencing using phosphorothioate primers (3). Phosphorothioate primers have a single phosphorothioate bond at the 3′ terminus that improves amplification of DNA sequences by DNA polymerases with proofreading activity (75). Two samples were selected from each coral species at each of three sites (St. Thomas, Florida inshore reef, and Belize; total = 12 samples). Samples were amplified using the bacterial forward primer 347F, which included the primer B adaptor for pyrosequencing and a unique 10-bp MID bar code on the 5′ end (5′-CGTATCGCCTCCCTCGCGCCATCAG-MID-GGAGGCAGCAGTRRGGAAT-3′), and the bacterial reverse primer 803R (5′-CTACCRGGGTATCTAATCC-3′). Bar code sequences can be found in Table S1 in the supplemental material. All PCRs were performed using an Eppendorf Master Cycler ep gradient thermal cycler and the following reagents: 25 μl Pfu (proofreading) DNA polymerase master mix, 2 μl of each 12.5 μM primer, and 3 to 4 μl DNA template (to generate consistent band intensities) in a final volume of 50 μl (adjusted with nuclease-free water). Amplifications were conducted under the following conditions: initial hot start of 2 min at 95°C followed by a touchdown PCR protocol in which the annealing temperature was decreased from 62°C by 1°C every cycle until reaching a touchdown temperature of 51°C, at which point 30 cycles of 95°C for 40 s, 51°C for 40 s, and 72°C for 1 min 15 s were performed; 1 final cycle of 95°C for 40 s, 51°C for 40 s, and 72°C for 10 min was followed by cooling to 4°C. PCR products were purified using ammonium acetate-ethanol precipitation, after which the concentration of each sample was quantified using a Nanodrop 1000 (ThermoScientific, Wilmington, DE) spectrophotometer. The 12 samples with unique bar codes were diluted to equimolar concentrations (16 ng/μl), pooled, and sequenced using a Roche 454 FLX sequencer with Titanium chemistry at Engencore (Columbia, SC).
Restriction fragment length polymorphism (RFLP) was used to type the Symbiodinium clade associated with each of the samples chosen for bar-coded pyrosequencing. Small-subunit rRNA (ssRNA) genes were amplified from total nucleic acid samples by using both the universal eukaryotic (SS5) and zooxanthella-specific (SS3Z) PCR primers (71). PCR products were digested with the TaqI restriction enzyme and compared with cultured standards to identify the Symbiodinium clade.
Pyrosequencing analysis.
For pyrosequencing analysis, sequences were trimmed using CLC Genomics Workbench (CLC Bio) with the following parameters, and any sequences not matching these criteria were excluded from downstream analysis: minimum quality score of 0.01 (99.9% quality), minimum sequence length of 200 bp, and no ambiguous bases in the sequence or mismatches in the primer sequence. Samples were imported and analyzed using the quantitative insights into microbial ecology (QIIME) pipeline (12). Sequences were then grouped into OTUs with an identity threshold of >97%. Using the Ribosomal Database Project (RDP) pipeline within QIIME, sequences were aligned with PyNast and grouped using a complete linkage clustering method. The sequences were clustered by uclust, which creates “seeds” of sequences that generate clusters based on percent identity. Finally, representative sequences from each OTU were selected, and taxonomic identity was assigned to each sequence by using the RDP taxonomic classifier at 90% confidence (84).
Sequences were analyzed using a number of descriptive and statistical methods within the QIIME pipeline. After bacterial libraries were rarefied so that sequencing efforts did not affect diversity comparisons, the following alpha diversity metrics were determined: total observed species (OTUs), predicted species (Chao1), and Shannon-Wiener diversity (H′). A beta diversity distance matrix was computed from the previously constructed OTU table based on the RDP taxonomic classifier. Weighted UniFrac distances were used to construct three-dimensional principal coordinate analysis (PCoA) plots. Jackknifed beta diversity metrics using 110 sequences per sample were used to directly measure the robustness of individual clusters in PCoA plots.
Sequences were further analyzed for diversity by two additional methods: the Blast2Go database and the SILVA comprehensive rRNA database (http://www.arb-silva.de/). These databases were searched using the Cornell Computation Biology Services Unit BioHPC Web computing resource with P-BLAST. Sequences with a >100-bp alignment, >96% identity, and E value of <1 × 10−20 were accepted for downstream analysis. QIIME bacterial sequence libraries (RDP results) were further analyzed using BLASTn searches against two different databases of 16S rRNA gene sequences, composed of sequences from (i) diseased coral-associated bacteria and (ii) healthy coral-associated bacteria (53). The number of significant hits that met the previously defined parameters was tallied, and an OTU distribution matrix was generated. The OTU community composition was analyzed with nMDS and PERMANOVA with pairwise comparisons in PRIMER 6.0 (Primer-E Ltd.) (see similar analyses in references 2 and 36). The 11 samples were located in the ordination space according to their pairwise dissimilarities (Jaccard index) of community composition.
Nucleotide sequence accession number.
All 16S rRNA gene sequences from the bar-coded pyrosequencing analysis have been archived in the NCBI sequence read archive (SRA) under study accession no. SRP009253.2.
RESULTS
DGGE analysis.
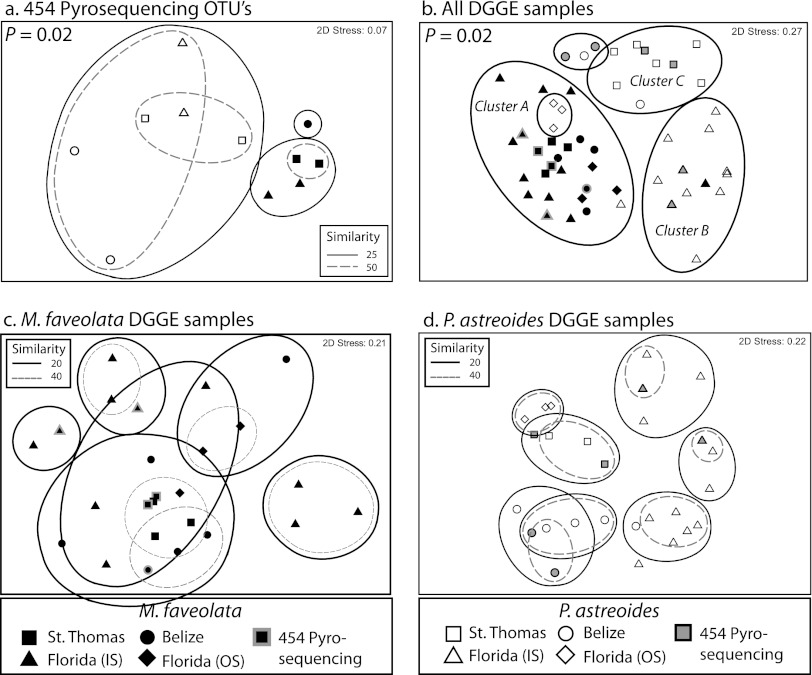
DGGE analysis of bacterial assemblages associated with the SML of M. faveolata (n = 26) and P. astreoides (n = 26) coral colonies revealed that the assemblages associated with M. faveolata generally clustered together, with no apparent pattern in terms of collection site, based on nMDS (Fig. 1b, cluster A, and c). Samples from P. astreoides clustered more consistently by site than by species (Fig. 1d). The P. astreoides samples collected from the Florida Keys offshore site at the Looe Key reef (n = 3) were distinct from those from all other sites and more similar to those from M. faveolata (Fig. 1b). Samples from P. astreoides at the Florida inshore site, St. Thomas, and Belize clustered independently and distinctly from M. faveolata samples (Fig. 1b, clusters B and C).
Fig 1.
nMDS plots of the first 2 dimensions based on Jaccard distances are shown for M. faveolata (filled symbols) and P. astreoides (open symbols) SML microbial assemblages from all field sites, i.e., St. Thomas, USVI (squares), Florida inshore reef (IS; triangles), Florida offshore reef (OS; diamonds), and Belize (circles). Emphasized shapes (grey outline or grey fill) indicate which samples were chosen for 16S rRNA gene pyrosequencing. (a) 16S rRNA gene pyrosequencing OTUs derived from the SILVA database (>97% identity). Contours represent PERMANOVA average similarity percentages. (b) Binary DGGE profiles (band presence/absence) for M. faveolata and P. astreoides. Contours are based on significant pairwise PERMANOVA results (P < 0.05). Cluster A represents the majority of M. faveolata samples, and clusters B and C represent P. astreoides samples from Florida (IS) and St. Thomas, respectively. (c and d) M. faveolata (c) and P. astreoides (d) binary DGGE banding patterns. Contours represent PERMANOVA average similarities within and between samples.
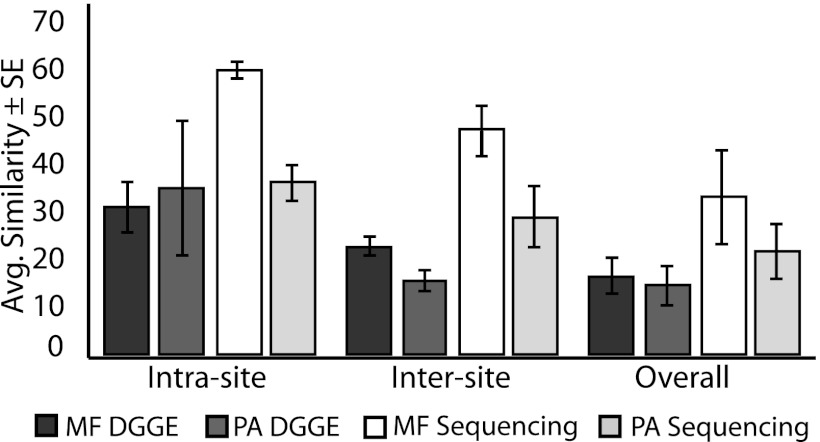
PERMANOVA confirmed the results of the nMDS analysis, in that bacterial assemblages varied significantly between coral species and among sampling sites (P < 0.01; F = 4.57 and 2.68, respectively) (Table 1). Pairwise PERMANOVA results suggested that the significant variation among sites was driven by P. astreoides-associated microbial assemblages rather than by those associated with M. faveolata (see Table S3 in the supplemental material). All site-by-site comparisons for P. astreoides were significantly different, but only two site-by-site comparisons differed significantly for M. faveolata (St. Thomas versus Florida [inshore site] [P = 0.01] and St. Thomas versus Florida [offshore site] [P = 0.04]) (see Table S3). Mean similarities among (intersite) and within (intrasite) sampling sites, derived from pairwise PERMANOVA results (see Table S3), further demonstrate that M. faveolata showed greater intersite similarity (22% ± 1.9% [mean ± standard error {SE}]) than P. astreoides (15% ± 2.1%), with less variability within a site (30% ± 5.1% versus 34% ± 13.8%) (Fig. 2).
Table 1.
Two-way PERMANOVA of binary DGGE profiles and 16S rRNA gene pyrosequencing samples, based on SILVA database-derived OTUs (>97% identity) and a Jaccard distance matrixa
| Study method and variable | df | SS | MS | F value | P(MC) |
|---|---|---|---|---|---|
| DGGE | |||||
| Coral | 1 | 13044 | 13044 | 4.5742 | <0.01 |
| Sites | 3 | 22,929 | 7,642.9 | 2.6801 | <0.01 |
| Coral × site | 3 | 21,967 | 7,322.3 | 2.5677 | <0.01 |
| Residual | 44 | 1.25E+05 | 2,851.7 | ||
| Total | 51 | 1.87E+05 | |||
| Pyrosequencing | |||||
| Coral | 1 | 6,750.20 | 6,750.20 | 3.86 | 0.02 |
| Site | 3 | 6,333.40 | 2,111.10 | 1.21 | 0.34 |
| Coral × site | 3 | 5,878.10 | 1,959.40 | 1.12 | 0.44 |
| Residual | 3 | 5,244.80 | 1,748.30 | ||
| Total | 10 | 25,949.00 |
The analysis was performed at the order level. See Table S3 in the supplemental material for pairwise comparison results. SS, sum of squares; MS, mean square. P values were based on the Monte Carlo statistic, P(MC), and on Jaccard distances. Values in bold are statistically significant.
Fig 2.
Average PERMANOVA similarities ± SE for M. faveolata (MF) and P. astreoides (PA) SML microbial samples within (intrasite) and between (intersite) sites and across all sites (overall). DGGE similarities are based on binary band presence/absence and Jaccard distances. 16S rRNA gene pyrosequencing similarities are based on OTUs derived from the SILVA database (>97% identity) and on Jaccard distances.
We excised and sequenced 43 clear, unique, and/or dominant bands from DGGE gels, and 26 yielded sequences that were of sufficiently high quality for taxon identification, with the remainder excluded mainly because of contamination from adjacent bands. The DGGE band sequences were affiliated with bacterial taxa in the following three phyla: (i) Cyanobacteria, including the genera Synechococcus (n = 7) and Prochlorococcus (n = 2); (ii) Proteobacteria, including the subclasses Alphaproteobacteria (n = 1) and Gammaproteobacteria, which include Edwardsiella (n = 1), Oceanospirillales (n = 13), and Escherichia coli (n = 1); and (iii) Actinobacteria, represented by one sequence.
454 pyrosequencing analysis.
A total of 8,547 high-quality pyrotag sequencing reads were obtained from a representative subset of samples (n = 12) chosen based on community patterns of the 52 initial samples assessed by DGGE analysis. One M. faveolata sample from Belize failed to provide any sequences, likely due an inaccurate initial NanoDrop spectrometry-determined concentration. Each of the remaining 11 samples contained between 397 and 1,375 reads (∼777 reads/sample), with average sequence lengths ranging from 212 to 436 bp per sample after primer and bar code removal (Table 2). The numbers of sequence hits per sample obtained using the SILVA and Blast2Go databases (>97% identity) were similar and are also reported in Table 2. In summary, samples from P. astreoides surface mucus had averages of 851 (QIIME) and 794 (SILVA) database hits per sample, while those from M. faveolata had averages of 735 (QIIME) and 611 (SILVA) database hits per sample. All P. astreoides samples were from colonies associated with clade A Symbiodinium, and M. faveolata colonies were associated with Symbiodinium clades A, B, and D (Table 2).
Table 2.
Bioinformatics analysis
| Coral species | Sitea | Symbiodinium sp. clade | No. of sequences with >96% identity |
% of sequences affiliated with sequences in databaseb |
||||
|---|---|---|---|---|---|---|---|---|
| RDP | SILVA | Blast2Go | Disease | BBD | Healthy | |||
| M. faveolata | STT | D | 1,014 | 819 | 821 | 67.9 | 4.9 | 4.1 |
| STT | D | 456 | 403 | 390 | 15.1 | 3.8 | 0.9 | |
| FLK | B | 949 | 704 | 650 | 33.2 | 12.6 | 2.9 | |
| FLK | B | 397 | 200 | 217 | 8.2 | 2.7 | 10.6 | |
| BLZ | A | 736 | 696 | 685 | 26.4 | 2.4 | 6.0 | |
| Avg ± SE | 735 ± 125 | 611 ± 114 | 606 ± 109 | 41.5 ± 10 | 4.4 ± 2 | 2.5 ± 2 | ||
| P. astreoides | STT | 531 | 490 | 495 | 1.5 | 0.5 | 0.0 | |
| STT | A | 688 | 569 | 576 | 6.8 | 3.4 | 0.1 | |
| FLK | A | 490 | 417 | 402 | 1.9 | 0.4 | 0.3 | |
| FLK | A | 1,102 | 1,056 | 1,043 | 4.1 | 0.5 | 5.6 | |
| BLZ | A | 1,375 | 1,348 | 1,344 | 3.4 | 0.0 | 1.6 | |
| BLZ | A | 809 | 770 | 788 | 0.5 | 0.1 | 2.2 | |
| Avg ± SE | 851 ± 154 | 794 ± 148 | 801 ± 148 | 3.1 ± 1 | 1.0 ± 1 | 1.0 ± 1 | ||
STT, St. Thomas, USVI; FLK, the Florida Keys; BLZ, Belize.
Percentage of sequences affiliated (>96% identity) with any sequence within databases (52) of potential disease-, black band disease (BBD)-, and healthy coral-associated bacteria.
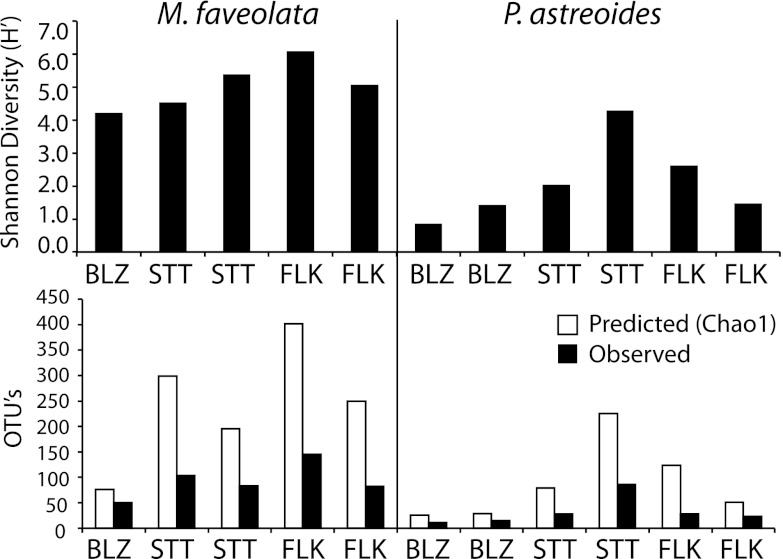
Bacterial diversity based on RDP results was highest for M. faveolata, with H′ values ranging from 4.2 to 6.1, which are comparable to the diversity values observed for the congeneric species Montastraea annularis (H′ = 2.8 to 5.0) in a similar study conducted by Barott and colleagues (4). However, Shannon H′ values were much lower for P. astreoides, at only 0.8 to 4.3 (Fig. 3). The highest average bacterial diversity was in Florida for M. faveolata (5.5 ± 0.5) and in St. Thomas for P. astreoides (3.1 ± 1.1) (Fig. 3). The largest range of OTUs, i.e., 50 to 145 OTUs, was seen for microbes in M. faveolata surface mucus, with a predicted (Chao1) range of 76 to 402 OTUs (Fig. 3). A much lower range of OTUs (11 to 85 OTUs) occurred in P. astreoides surface mucus, with a predicted (Chao1) range of 26 to 225 OTUs (Fig. 3).
Fig 3.
Alpha diversity metrics derived from 16S rRNA gene bar-coded pyrosequencing for bacteria associated with the surface mucus layer of M. faveolata and P. astreoides corals from St. Thomas, USVI (STT), the Florida Keys inshore site (FLK), and Belize (BLZ). (Top) Shannon-Weiner diversity of bacteria from each coral SML sample. (Bottom) Numbers of predicted (Chao1) and observed OTUs. OTUs were grouped at >97% similarity, based on RDP-classified results.
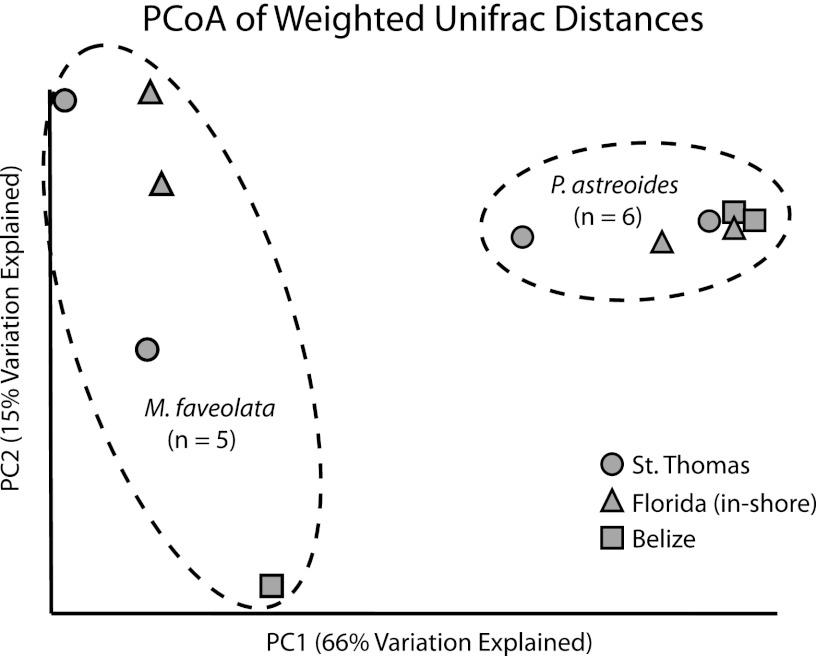
PCoA of the weighted UniFrac distances based on RDP results showed that M. faveolata and P. astreoides samples each harbored characteristic bacterial communities (Fig. 4). All of the P. astreoides samples clustered to the right of the graph, along the primary axis (66% of the variation) and away from the M. faveolata samples on the left. The P. astreoides samples also clustered in the second dimension (15% of the variation), but M. faveolata samples did not have a similar clustering in the second dimension (Fig. 4). The bacterial communities associated with both corals showed some spatial separation, but this was more evident in the nMDS results based on OTUs from the SILVA database results. Nonmetric MDS results showed that M. faveolata samples were more specific than P. astreoides samples, which is likely a result of the higher taxonomic resolution found with the SILVA database than with the RDP database (Fig. 1a versus Fig. 4). Thus, all of the results presented for coral-associated bacterial characterization are based on the SILVA database analysis rather than on RDP database analysis.
Fig 4.
Principal coordinate analysis (PCoA) of weighted UniFrac distances was performed based on RDP-classified results from 16S rRNA gene bar-coded pyrosequencing of M. faveolata (n = 5) and P. astreoides (n = 6) surface mucus layer bacterial assemblages from 3 sites in the Caribbean: St. Thomas, USVI, the Florida Keys (inshore), and Belize. Contours are based on significant coral species PERMANOVA results (P = 0.02).
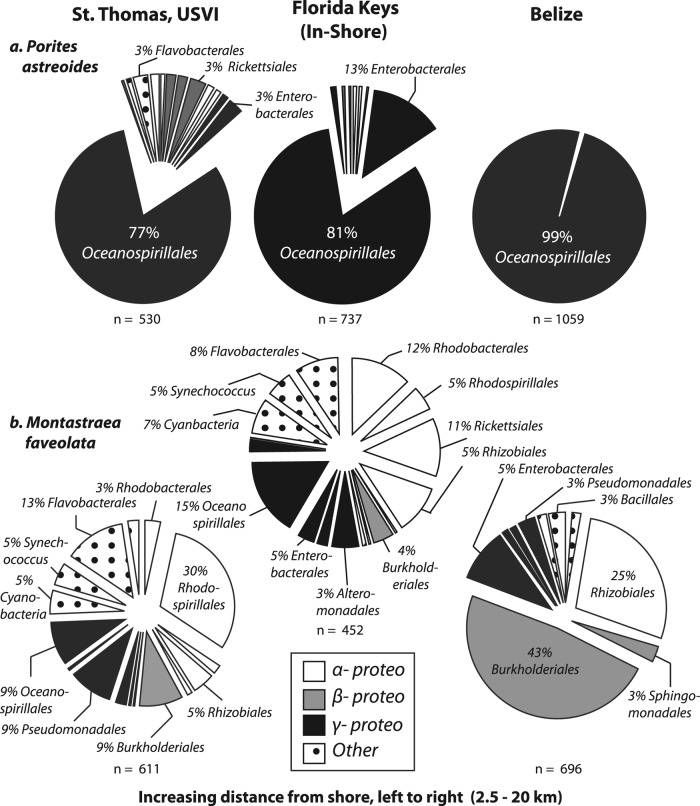
Coral-associated bacterial assemblages were dominated by sequences affiliated with the following classes or phyla: Alpha-, Beta-, and Gammaproteobacteria were the most dominant organisms, followed by members of the Bacteroidetes, Actinobacteria, Cyanobacteria, and Firmicutes. In general, the following five bacterial orders were common among both M. faveolata and P. astreoides pyrosequencing samples: (i) Haellaceae, a member of the Oceanospirillales, within the Gammaproteobacteria; (ii) Enterobacteriaceae, within the Gammaproteobacteria; (iii) Rhodospirillaceae, within the Alphaproteobacteria; (iv) Comamonadaceae, within the Burkholderiales and Betaproteobacteria groups; and (v) Flavobacteriaceae members of the Cytophaga-Flavobacteria-Bacteroides group (Table 3; Fig. 5). The most abundant sequences identified to the genus level from M. faveolata samples were sequences of Acinetobacter (3.5% ± 2.2% of total sequence reads), a member of the Pseudomonadales within the Gammaproteobacteria. Other common genera within M. faveolata mucus were Edwardsiella (3.3% ± 1.4%), Mesorhizobium (2.9% ± 1.8%), Flavobacterium (1.3% ± 0.3%), and Vibrio (1.1% ± 1.0%), with the Vibrio spp. including Vibrio vulnificus, Vibrio harveyi, Vibrio campbellii, and Vibrio alginolyticus. The dominant genus identified in P. astreoides samples was Endozoicomonas (85.1% ± 7.3%), within the order Oceanospirillales and the class Gammaproteobacteria. The average % Endozoicomonas spp. increased from 75.4% ± 16.8% in St. Thomas to 80.6% ± 14.4% in Florida (inshore site) and 99% ± 0.2% in Belize (Fig. 5). The next most abundant genera were Edwardsiella (4.4% ± 4.2%) and Plesiomonas (0.9% ± 0.8%), within the Enterobacteriales, and Cetobacterium, within the Fusobacteriales (0.5% ± 0.4%). Members of the Vibrionales made up only 0.3% ± 0.2% of the P. astreoides bacterial assemblages.
Table 3.
Ten most abundant bacterial groups for each coral species
| Coral species and bacterial groupa | % prevalence |
|---|---|
| P. astreoides | |
| Gammaproteobacteria, Oceanospirillales, Hahellaceae | 88 |
| Gammaproteobacteria, Enterobacteriaceae | 6 |
| Bacteroidetes, Flavobacteriaceae | 1 |
| Alphaproteobacteria, Rhodospirillaceae | 1 |
| Fusobacteria, Fusobacteriaceae | 1 |
| Betaproteobacteria, Comamonadaceae | 0.5 |
| Gammaproteobacteria,Alteromonadaceae | 0.4 |
| Gammaproteobacteria,Vibrionaceae | 0.4 |
| Firmicutes,Bacillaceae | 0.3 |
| Alphaproteobacteria,Phyllobacteriaceae | 0.2 |
| M. faveolata | |
| Flavobacteria | 17 |
| Alphaproteobacteria, Rhodospirillaceae | 17 |
| Betaproteobacteria, Comamonadaceae | 15 |
| Alphaproteobacteria,Rhodobacteraceae | 7 |
| Gammaproteobacteria, Hahellaceae | 7 |
| Alphaproteobacteria,Phyllobacteriaceae | 6 |
| Cyanobacteria,Synechococcus | 5 |
| Gammaproteobacteria, Enterobacteriaceae | 4 |
| Gammaproteobacteria,Moraxellaceae | 4 |
| Alphaproteobacteria,Methylobacteriaceae | 4 |
Classifications are based on SILVA database analysis. Group names are shown in bold if they were found in the top 10 list for both coral species.
Fig 5.
Class-level sequence affiliates. Percentages of OTUs are related to the nearest order-level affiliation (labeled if ≥3%) derived from 16S rRNA gene bar-coded pyrosequencing of bacteria associated with the surface mucus layer of M. faveolata (a) and P. astreoides (b) corals from St. Thomas, USVI, the Florida Keys inshore site, and Belize. Taxonomic affiliations were derived from SILVA database hits (>97% identity). Pie charts are shaded by class-level affiliation, indicating Alpha-, Beta-, and Gammaproteobacteria and “Other,” which includes Cyanobacteria, Firmicutes, Bacteroidetes, and Actinobacteria. Sites are arranged from least to greatest distance from the mainland, from left to right, as a proxy for anthropogenic influence.
The abundance of sequences similar (at a >96% identity threshold) to those of coral disease-associated and black band disease-associated bacteria was generally higher in the M. faveolata libraries than in those from P. astreoides; however, the abundance of sequences affiliated with a database of healthy coral-associated bacterial sequences was also higher in M. faveolata libraries (Table 2). Very few sequence similarities were observed in any of the examined microbial databases for the bacteria associated with P. astreoides, likely because Caribbean Porites spp. were not assessed in any of the studies that comprised the meta-analysis from which these databases were constructed (64). However, the analysis did include six studies that examined the bacterial assemblages associated with healthy and diseased members of the Caribbean Montastraea sp. complex (collected in Barbados, Bonaire, Curaçao, Florida, Panama, Puerto Rico, and St. Croix, USVI) (see Table S2 in the supplemental material). We found that a larger number of sequences from M. faveolata coral samples affiliated with diseased coral-associated bacteria (2.5%) within the databases examined. The high abundance of M. faveolata sequences affiliated with the coral disease database was particularly evident in St. Thomas samples (taken 2.5 km from shore), where an average of 41.5% of sequences were affiliated with the disease database, with much lower percentages for Florida (20.5%; 5 km from shore) and Belize (26.4%; 20 km from shore) (Table 2; see Table S2). Despite the low similarity for Florida sample sequences and the disease database, a larger number of black band disease database-specific sequences occurred in the Florida Keys samples (7.7%) than in the samples from the other sites (Table 2).
DISCUSSION
We utilized culture-independent methods to characterize the structural patterns of bacterial assemblages associated with a large number of coral colonies (n = 52) among disparate regions of the Caribbean (>1,100 km apart). We targeted the bacterial assemblages of coral colonies that were both visually healthy and free of interaction with macroalgae or other macroinvertebrates. These results strengthen the body of knowledge which indicates that healthy corals harbor distinct assemblages of microbial organisms (42, 67), although detectable variability occurs within species across geographic regions, as demonstrated recently for other corals (48, 78, 79). We illustrate two significantly different clusters of bacterial ribotypes associated with either M. faveolata or P. astreoides colonies. Thus, although geographic variability does exist, the coral holobiont (coral animal, Symbiodinium sp., and microbial constituents) appears to actively mediate some degree of microbial specificity.
The host species we examined are two of the most common reef-building (Hexacorallia:Scleractinia) corals in the Caribbean Sea but belong to distinct phylogenetic lineages separated by 240 to 288 million years of evolutionary divergence (52, 68). Recently, Sunagawa and colleagues showed that bacterial assemblages were more similar within phylogenetic clades of coral hosts, suggesting that M. faveolata, a member of the short (robust) clade due to size differences in the mitochondrial rRNA genes, and P. astreoides, belonging to the long (complex) clade, should have distinct bacterial assemblages (52, 78). These two coral species also vary in reproductive life history strategy (brooder versus spawner), which may alter their mode of acquiring bacterial symbionts (vertical versus horizontal transmission). Furthermore, M. faveolata is affected by a range of bacterial diseases, including but not limited to white plague, dark spot, black band, yellow blotch/band, and red band, while P. astreoides has been associated with only two diseases: white plague and yellow blotch/band (25). Massive reef-building corals, especially species within the genus Montastraea, are often susceptible to large numbers of diseases, e.g., M. annularis is susceptible to 9 diseases, M. faveolata is susceptible to 6 diseases, and Montastraea franksi is susceptible to 5 diseases (reviewed in reference 43). In contrast, Indo-Pacific corals in the genus Porites are quite robust, detecting invasion by endolithic fungi and responding by walling off the site of fungal penetration with layers of calcium carbonate (44, 63). Furthermore, the external cell layers of another member of this genus, Porites compressa, are completely devoid of adhering microbes, although the mucus layer still maintains a diverse assemblage of microorganisms (35). Finally, Porites astreoides is the first coral species to definitively illustrate vertical transmission of microbial symbionts from the parent colony to its pelagic larvae (73). Recently, Pocillopora damicornis corals were also found to regulate the abundance of bacteria on their surfaces when faced with high levels of organic enrichment and water-associated bacterial loads (23). Thus, members of the family Poritidae (and others) appear to maintain an inherent and discriminating relationship between coral host tissues and associated microbes.
We found the surface mucus layer of P. astreoides corals to be dominated (>75%) by members of the Oceanospirillales (within the Gammaproteobacteria), the majority of which had highest similarity to the genus Endozoicomonas, within the order Hahellaceae. Another member of this order, Hahella chejuensis, exhibits genes responsible for the biosynthesis of a pigment (prodigiosin) which has lytic activity against a red tide dinoflagellate (34). Thus, it may be important to investigate whether Endozoicomonas spp. have the potential to antagonize or potentiate the algal symbionts of corals (i.e., Symbiodinium spp.). Alternatively, bacterial pigments may provide beneficial protection against solar radiation or protozoan grazing (49). Further research is needed to define the distribution and potential symbiotic association of the Endozoicomonas-like bacteria that comprise the majority of P. astreoides surface mucus-associated bacterial taxa.
The concentration of Oceanospirillales-related sequences also increased significantly with distance from shore, from 77% in Florida to 99% in Belize. The presence of Oceanospirillales as a major constituent of P. astreoides microbial communities was also documented by Rohwer and colleagues (67). Studies have also shown that members of the Oceanospirillales not only are dominant members of the parent colony microbiota but also are vertically transmitted within P. astreoides larvae, suggesting a symbiotic role (73). Members of the Oceanospirillales are dominant components of the heterotrophic marine microbial environment, with pigments and a distinct coccoid resting stage (33). Functioning as aerobic heterotrophs, Oceanospirillum spp. can utilize constituents of coral tissues and mucus, such as organic acids, amino acids, ammonium, and dimethylsulfoniopropionate (DMSP) (24, 61). High concentrations of DMSP and dimethyl sulfate (DMS) have been found within animals that harbor symbiotic algae, such as scleractinian corals and giant clams, providing a potential link between DMSP-degrading Oceanospirillales and corals (8, 9, 81). Preliminary evidence also indicates that Porites spp. are capable of hosting DMSP-degrading organisms because the dmdA gene, which is the most highly represented gene for DMSP degradation in the Global Ocean Survey (GOS) database (29, 30), is present in both P. astreoides (85) and pH-stressed P. compressa (62, 80, 82) microbial metagenomes. Whether these organisms provide a benefit through the degradation of DMSP and other organic compounds requires further understanding of the functional role of Oceanospirillum in the host-microbe association. These and previous findings suggest that a relationship may exist between P. astreoides and members of the Oceanospirillales, particularly given the known symbioses between members of the Oceanospirillales and marine bivalves (33) or the Osedax polychaete, a worm that feeds upon whalebones (30).
Bacterial assemblages in the surface mucus layer of M. faveolata corals were significantly more diverse than those in P. astreoides. Relatively large numbers of bacteria associated with M. faveolata were similar to known nitrogen-fixing taxa, including Cyanobacteria (5%), Rhizobiales (11%), and Burkholderiales (14%). Shashar and colleagues were the first to detect coral-associated bacteria with the required nifH gene for fixing of nitrogen in corals (74). Five percent of the coral SML samples from both St. Thomas and the Florida Keys contained sequences that displayed high similarity to Synechococcus sp. and other uncultured cyanobacterial sequences. A study of the bacteria associated with the conspecific coral Montastraea cavernosa also found coccoid cyanobacteria related to Synechococcus sp. and Prochlorococcus sp. within coral tissues (46). These symbionts are not believed to be pathogens, in contrast to other cyanobacterial species related to Oscillatoria sp. or Phormidium sp., which are among the consortia of bacteria associated with black band disease (16; for a review, see reference 7). Nitrogenase activity has been found in M. cavernosa colonies (46), and subsequent studies found that endosymbiotic Symbiodinium spp. were actively using the products of nitrogen fixation (47). In the present study, >5% of the samples from all locations had sequences similar to those of Burkholderiales, within the class Betaproteobacteria, and Rhizobiales, within the class Alphaproteobacteria. M. faveolata samples from Belize were dominated by these two orders, with approximately 44% of sequences aligning with the Burkholderiales and 25% aligning with the Rhizobiales. Both of these groups are known to fix nitrogen in terrestrial and mangrove systems but have yet to be examined in corals (26). Rhizobia spp. have long been known to form nitrogen-fixing symbioses with leguminous plants, and Burkholderia spp. were also recently shown to nodulate legumes, becoming the first known betaproteobacteria to form nitrogen-fixing symbioses (14). Other studies using M. faveolata and Montipora spp. as model organisms have expanded the concept of nitrogen-fixing symbionts within the coral holobiont beyond Cyanobacteria, to a wide range of bacterial taxa within the Alpha- and Gammaproteobacteria, Firmicutes, Spirochetes, and Archaea, including three classes of Euryarchaeota (37, 45, 58). Clearly, there is still much to learn about the diversity of nitrogen-fixing prokaryotes and their role in coral microbial communities, but our data suggest that both alpha- and beta-rhizobia associate with M. faveolata corals and may provide fixed nitrogen.
A large number of sequences were detected with >97% identity to sequences within a database of disease-associated bacteria within the M. faveolata SML (53), particularly in samples from St. Thomas. This may indicate nearshore pollutants at St. Thomas, where coral samples were collected 2.5 km from the mainland, adjacent to the St. Thomas international airport. However, the highest abundance of sequences that affiliated with the Enterobacteriales was found in coral species samples from the Florida Keys (5 to 12 km from shore). A prevalence of Enterobacteriales organisms is often associated with fecal contamination (87) and indicates the presence of other enteric bacteria, such as Serratia marcescens, the etiological agent of white pox disease on Acropora coral species in the Caribbean (60). Furthermore, we also found the highest abundances of sequences related to two other disease-affiliated orders, Rhodobacterales and Rickettsiales, in M. faveolata samples from the Florida Keys. A recent meta-analysis of coral-associated bacterial assemblages showed that Rhodobacter spp. are globally associated with several coral diseases (e.g., black band, white plague, and white band disease) and two destructive conditions (atramentous necrosis and cyanobacterial patches) (53). There is also a suggested link between Rickettsia-like bacteria and marine diseases afflicting invertebrates such as corals and abalone (1, 13). These results support our field observations that the M. faveolata colonies selected for sampling were generally healthy and unaffected by disease but that the St. Thomas and Florida Keys corals likely experienced greater levels of environmental stress and/or anthropogenic microbial influx. These stressors may cause a detrimental shift in microbial assemblages, leading to a less stable, potentially pathogenic state. Consequently, corals in St. Thomas and Florida may be more susceptible to disease than those in Belize.
Coral-associated bacterial diversity in both M. faveolata and P. astreoides surface mucus samples was greatest in St. Thomas, contrary to our original hypothesis that the Florida Keys samples would have the highest abundance of and most diverse bacteria because Florida reefs are exposed to large numbers of snorkelers (2.86 million/year) and divers (0.8 million/year) (56a). Florida contained 8% of the nation's coastline population in 1960 and 16% of this population by 2008, an increase of >10 million coastal residents during this period, second only to California in the United States (86). Higher human population densities inherently lead to increased nutrient enrichment and sedimentation on adjacent reefs. High nutrient loads are believed to fertilize selected opportunistic and potentially pathogenic bacterial taxa (e.g., Vibrio spp.), allowing them to become dominant on otherwise healthy corals (11, 40). It is possible that the documented pattern is an example of an intermediate disturbance model, whereas diversity peaks at an intermediate level (intensity or frequency) of small-scale disturbances (15, 77). In the U.S. Virgin Islands, high levels of runoff and the resulting sedimentation can be attributed to recent increases in housing development and road construction, particularly in St. John and St. Thomas, where steep mountain slopes allow rapid runoff (10). Heavy rainfall can overload existing sewage systems, which results in intermediate and severe pollution of coastal waters. Additional studies have directly linked shoreline development in St. Thomas to increased sedimentation during periods of heavy rainfall (56). These point source disturbances during rainfall events may have a lasting effect on the coral-associated microbial assemblages in St. Thomas, increasing their overall diversity in comparison to that of assemblages in Belize and the Florida Keys.
Initial 16S rRNA gene pyrosequencing results based on RDP classifications suggested less intersite variability among P. astreoides samples than among M. faveolata samples, while the DGGE results suggested the opposite, with a higher site affinity among P. astreoides bacterial assemblages. Differences in the abundance of Hahellaceae, within the Oceanospirillales, are the likely drivers of this variability between P. astreoides samples. Our initial RDP analysis using QIIME could not classify P. astreoides OTUs beyond the level of Gammaproteobacteria, which explains the high level of sample similarity across sites (see the PCoA results in Fig. 4). Utilizing the SILVA database with Blast2Go, we were able to quantify a higher level of taxonomic resolution and document greater variability in coral bacterium species specificity within and between sites (Fig. 1a versus Fig. 4). Based on these results, a higher level of species specificity was exhibited by M. faveolata colonies than by P. astreoides colonies from all three sites, and this was supported by the DGGE results. Both corals still exhibited higher intrasite specificity than intersite specificity, suggesting that both M. faveolata and P. astreoides are susceptible to environmental changes and are potentially less able to mediate specific microbial assemblages than first believed. Variability might also be attributed to amplification bias as a result of bar-coded PCR primers (5) and pyrosequencing errors (reviewed in reference 89). Amplification bias can produce variable pyrosequencing data from the same environmental DNA template, resulting in questionable beta diversity of microbial communities (89). These biases can be alleviated through a number of steps prior to amplification (5) and during subsequent analysis (89). Regardless, alpha diversity metrics should be less problematic and more reproducible (89). In our study, we showed that P. astreoides hosts a much less diverse population of bacteria, which is dominated in all samples (>75%) by one type of Oceanospirillales, suggesting that this coral species may be more robust than M. faveolata in its microbial affiliations. Furthermore, the literature suggests that P. astreoides may have a mechanism for fostering a specific bacterial community within its tissues or mucus (35), which may be aided by the vertical transmission of bacterial symbionts (73). P. astreoides also consistently associates with clade A Symbiodinium spp., whereas M. faveolata is more variable in this association (Table 2). It is known that Symbiodinium spp. release carbon exudates (32) and arabinose (21) into coral mucus. Thus, because coral mucus composition is thought to play an important role in shaping coral microbial assemblages (65), variation in photosynthetic activity and products by different Symbiodinium clades may also contribute to the higher diversity observed within M. faveolata microbial assemblages.
We demonstrate here that two prevalent Caribbean corals, M. faveolata and P. astreoides, harbor distinct assemblages of bacteria in their SML. However, we also document shifts in the bacterial assemblages which relate to specific Caribbean sampling sites. Changing environmental conditions (e.g., temperature, irradiance, nutrients, and sedimentation) are known to influence coral microbial assemblages (6, 48, 54, 59), potentially influencing microbial diversity and disease susceptibility. Identification of the microorganisms that comprise healthy coral assemblages in a wide range of species types and geographic locations will allow us to better determine coral stressors and disease causation. Corals exposed to high levels of anthropogenic and environmental stress, such as those adjacent to St. Thomas and the Florida Keys (inshore reefs), likely experience physiological and biochemical changes that alter their microbial assemblages long before visible disease symptoms occur. The shifts documented here for P. astreoides, from a microbial assemblage composed almost entirely (>99%) of Oceanospirillales in Belize to a 77% prevalence of that order in St. Thomas, and in M. faveolata assemblages, from high levels of potentially beneficial nitrogen fixers (e.g., Rhizobiales and Burkholderiales) in Belize to disease-associated taxa (e.g., Rhodobacteriales and Enterobacteriales) in the Florida Keys, demonstrate how studies of coral-associated microbial communities on healthy reefs can be used to indicate reefs that require immediate remedial action to reduce coral stress (e.g., improving sanitation or reducing sedimentation). By developing a globally effective method of diagnosing coral-microbe dynamics, coral reef biologists and coastal management organizations will be better able to identify coral reefs in the greatest danger. Such information will further support global reef recovery efforts.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Florida Keys National Marine Sanctuary for granting collection and research permits (FKNMS-2008-019) and the Belize Fisheries Department for permits to collect coral mucus samples. The Mote Marine Laboratory, the University of the Virgin Islands MacLean Marine Science Center, the Smithsonian Marine Station at Fort Pierce, and the Smithsonian's Carrie Bow Cay Field Station all provided invaluable logistical support. We particularly thank N. Kirk for his help with Symbiodinium clade identification. N. Fogarty, L. Huebner, M. Newman, V. Paul, R. Ritson-Williams, J. Szczebak, J. Voss, and D. Weese provided critical field and laboratory assistance.
This work was supported by a National Oceanic and Atmospheric Administration National Marine Sanctuary Program Nancy Foster Scholarship (K.M.M.) and by Puerto Rico Sea Grant R-101-1-08 (N.E.C.), NSF grant MCB034827 (A.G.M.), and NSF grant EPSCoR EPS0447675 (Auburn University CECST Program).
Footnotes
Published ahead of print 6 July 2012
This is contribution 91 of the Department of Biological Sciences Marine Biology Program, Auburn University, Auburn, Alabama, USA.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Antonio DB, Andree KB, Moore JD, Friedman CS, Hedrick RP. 2000. Detection of Rickettsiales-like prokaryotes by in situ hybridization in black abalone, Haliotis cracherodii, with withering syndrome. J. Invertebr. Pathol. 75:180–182 [DOI] [PubMed] [Google Scholar]
- 2. Arfi Y, Buee M, Marchand C, Levasseur A, Record E. 2012. Multiple markers pyrosequencing reveals highly diverse and host-specific fungal communities on the mangrove trees Avicennia marina and Rhizophora stylosa. FEMS Microbiol. Ecol. 79:433–444 [DOI] [PubMed] [Google Scholar]
- 3. Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barott KL, et al. 2011. Microbial to reef scale interactions between the reef-building coral Montastraea annularis and benthic algae. Proc. Biol. Soc. 279:1655–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berry D, Mahfoudh KB, Wagner M, Loy A. 2011. Barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl. Environ. Microbiol. 77:7846–7849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bourne DG, Iida Y, Uthicke S, Smith-Keune C. 2008. Changes in coral-associated microbial communities during a bleaching event. ISME J. 2:350–363 [DOI] [PubMed] [Google Scholar]
- 7. Bourne DG, et al. 2009. Microbial disease and the coral holobiont. Trends Microbiol. 17:554–562 [DOI] [PubMed] [Google Scholar]
- 8. Broadbent AD, Jones GB. 2004. DMS and DMSP in mucus ropes, coral mucus, surface films and sediment pore waters from coral reefs in the Great Barrier Reef. Mar. Freshw. Res. 55:849–855 [Google Scholar]
- 9. Broadbent AD, Jones GB, Jones RJ. 2002. DMSP in corals and benthic algae from the Great Barrier Reef. Est. Coast. Shelf Sci. 55:547–555 [Google Scholar]
- 10. Brooks GR, Devine B, Larson RA, Rood BP. 2007. Sedimentary development of Coral Bay, St. John, USVI: a shift from natural to anthropogenic influences. Caribb. J. Sci. 43:226–243 [Google Scholar]
- 11. Bruno J, Petes L, Harvell C, Hettinger A. 2003. Nutrient enrichment can increase the severity of coral diseases. Ecol. Lett. 6:1056–1061 [Google Scholar]
- 12. Caporaso JG, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Casas V, et al. 2004. Widespread association of a Rickettsiales-like bacterium with reef-building corals. Environ. Microbiol. 6:1137–1148 [DOI] [PubMed] [Google Scholar]
- 14. Chen W, Moulin L, Bontemps C. 2003. Legume symbiotic nitrogen fixation by β-Proteobacteria is widespread in nature. J. Bacteriol. 185:7266–7272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Connell JH. 1978. Diversity in tropical rain forests and coral reefs. Science 199:1302–1310 [DOI] [PubMed] [Google Scholar]
- 16. Cooney RP, et al. 2002. Characterization of the bacterial consortium associated with black band disease in coral using molecular microbiological techniques. Environ. Microbiol. 4:401–413 [DOI] [PubMed] [Google Scholar]
- 17. Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. 2005. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438:90–93 [DOI] [PubMed] [Google Scholar]
- 18. Daniels C, et al. 2011. Spatial heterogeneity of bacterial communities in the mucus of Montastraea annularis. Mar. Ecol. Prog. Ser. 426:29–40 [Google Scholar]
- 19. Dinsdale EA, et al. 2008. Microbial ecology of four coral atolls in the Northern Line Islands. PLoS One 3:e1584 doi: 10.1371/journal.pone.0001584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ducklow HW, Mitchell R. 1979. Bacterial populations and adaptations in the mucus layers on living corals. Limnol. Oceanogr. 24:715–725 [Google Scholar]
- 21. Ducklow HW, Mitchell R. 1979. Composition of mucus released by coral reef coelenterates. Limnol. Oceanogr. 24:706–714 [Google Scholar]
- 22. Ferris MJ, Muyzer G, Ward DM. 1996. Denaturing gradient gel electrophoresis profiles of 26 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garren M, Azam F. 2012. Corals shed bacteria as a potential mechanism of resilience to organic matter enrichment. ISME J. 6:1159–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garrity GM, Bell JA, Lilburn T. 2005. Order VIII. Oceanospirillales ord. nov., p 270–292 In Brenner DJ, Krieg NR, Staley JT, Garrity GM. (ed), Bergey's manual of systematic bacteriology, vol 2 Springer, New York, NY [Google Scholar]
- 25. Garzón-Ferreira J, Gil-Agudelo D, Barrios L. 2001. Stony coral diseases observed in southwestern Caribbean reefs. Hydrobiology 460:65–69 [Google Scholar]
- 26. Goffredi SK, et al. 2005. Evolutionary innovation: a bone-eating marine symbiosis. Environ. Microbiol. 7:1369–1378 [DOI] [PubMed] [Google Scholar]
- 27. Guppy R, Bythell JC. 2007. Environmental effects on bacterial diversity in the surface mucus layer of the reef coral Montastraea faveolata. Mar. Ecol. Prog. Ser. 328:133–142 [Google Scholar]
- 28. Hansson L, Agis M, Weinbauer M. 2009. Community composition of bacteria associated with cold-water coral Madrepora oculata: within and between colony variability. Mar. Ecol. Prog. Ser. 397:89–102 [Google Scholar]
- 29. Howard EC, et al. 2006. Bacterial taxa that limit sulfur flux from the ocean. Science 314:649–652 [DOI] [PubMed] [Google Scholar]
- 30. Howard EC, Sun S, Biers EJ, Moran MA. 2008. Abundant and diverse bacteria involved in DMSP degradation in marine surface waters. Environ. Microbiol. 10:2397–2410 [DOI] [PubMed] [Google Scholar]
- 31. Ihaka R, Gentleman R. 1996. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5:299–314 [Google Scholar]
- 32. Ikeda Y, Miyachi S. 1995. Carbon dioxide fixation by photosynthesis and calcification for a solitary coral, Fungia sp. Bull. Inst. Oceanogr. 14:61–67 [Google Scholar]
- 33. Jensen S, Duperron S, Birkeland Hovland N-KM. 2010. Intracellular Oceanospirillales bacteria inhabit gills of Acesta bivalves. FEMS Microbiol. Ecol. 74:523–533 [DOI] [PubMed] [Google Scholar]
- 34. Jeong H, et al. 2005. Genomic blueprint of Hahella chejuensis, a marine microbe producing an algicidal agent. Nucleic Acids Res. 33:7066–7073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Johnston IS, Rohwer F. 2007. Microbial landscapes on the outer tissue surfaces of the reef-building coral Porites compressa. Coral Reefs 26:375–383 [Google Scholar]
- 36. Kauserud H, Kumar S, Brysting AK, Norden J, Carlsen T. 2012. High consistency between replicate 454 pyrosequencing analyses of ectomycorrhizal plant root samples. Mycorrhiza 22:309–315 [DOI] [PubMed] [Google Scholar]
- 37. Kimes NE, Van Nostrand JD, Weil E, Zhou J, Morris PJ. 2010. Microbial functional structure of Montastraea faveolata, an important Caribbean reef-building coral, differs between healthy and yellow-band diseased colonies. Environ. Microbiol. 12:541–556 [DOI] [PubMed] [Google Scholar]
- 38. Klaus JS, Frias-Lopez J, Bonheyo GT, Heikoop JM, Fouke BW. 2005. Bacterial communities inhabiting the healthy tissues of two Caribbean reef corals: interspecific and spatial variation. Coral Reefs 24:129–137 [Google Scholar]
- 39. Klaus JS, Janse I, Heikoop JM, Sanford RA, Fouke BW. 2007. Coral microbial communities, zooxanthellae and mucus along gradients of seawater depth and coastal pollution. Environ. Microbiol. 9:1291–1305 [DOI] [PubMed] [Google Scholar]
- 40. Kline D, Kuntz N, Breitbart M, Knowlton N, Rohwer F. 2006. Role of elevated organic carbon levels and microbial activity in coral mortality. Mar. Ecol. Prog. Ser. 314:119–125 [Google Scholar]
- 41. Koh E. 1997. Do scleractinian corals engage in chemical warfare against microbes? J. Chem. Ecol. 23:379–398 [Google Scholar]
- 42. Kvennefors ECE, Sampayo E, Ridgway T, Barnes AC, Hoegh-Guldberg O. 2010. Bacterial communities of two ubiquitous Great Barrier Reef corals reveal both site- and species-specificity of common bacterial associates. PLos One 5:e10401 doi: 10.1371/journal.pone.0010401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lafferty K, Porter J, Ford S. 2004. Are diseases increasing in the ocean? Annu. Rev. Ecol. Evol. Syst. 35:31–54 [Google Scholar]
- 44. Le Campion-Alsumard T, Golubic S, Hutchlings P. 1995. Microbial endoliths in skeletons of live and dead corals: Porites lobata (Moorea, French Polynesia). Mar. Ecol. Prog. Ser. 117:149–157 [Google Scholar]
- 45. Lema KA, Willis BL, Bourne DG. 2012. Corals form specific associations with diazotrophic bacteria. Appl. Environ. Microbiol. 78:3136–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lesser MP. 2004. Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science 305:997–1000 [DOI] [PubMed] [Google Scholar]
- 47. Lesser M, et al. 2007. Nitrogen fixation by symbiotic cyanobacteria provides a source of nitrogen for the scleractinian coral Montastraea cavernosa. Mar. Ecol. Prog. Ser. 346:143–152 [Google Scholar]
- 48. Littman R, Bourne D, Willis B. 2010. Responses of coral-associated bacterial communities to heat stress differ with Symbiodinium type on the same coral host. Mol. Ecol. 19:1978–1990 [DOI] [PubMed] [Google Scholar]
- 49. Matz C, et al. 2004. Impact of violacein-producing bacteria on survival and feeding of bacterivorous nanoflagellates. Appl. Environ. Microbiol. 70:1593–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McCune B, Grace JB. 2002. Nonmetric-multidimensional scaling, p 125–142 In Analysis of ecological communities. MjM Software Design, Gleneden Beach, OR. [Google Scholar]
- 51. Medina M. 2011. Analyzing coral reefs and their microbial assemblages. Microbe Mag. 6:226–232 [Google Scholar]
- 52. Medina M, Collins AG, Takaoka TL, Kuehl JV, Boore JL. 2006. Naked corals: skeleton loss in Scleractinia. Proc. Nat. Acad. Sci. U. S. A. 103:9096–9100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mouchka ME, Hewson I, Harvell CD. 2010. Coral-associated bacterial assemblages: current knowledge and the potential for climate-driven impacts. Integr. Comp. Biol. 50:662–674 [DOI] [PubMed] [Google Scholar]
- 54. Muller EM, van Woesik R. 2009. Shading reduces coral-disease progression. Coral Reefs 28:757–760 [Google Scholar]
- 55. Muyzer G, de Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nemeth RS, Nowlis JS. 2001. Monitoring the effects of land development on the near-shore reef environment of St. Thomas, U.S.V.I. Bull. Mar. Sci. 69:759–775 [Google Scholar]
- 56a. NOAA 2000. National survey on recreation and the environment. National Oceanic and Atmospheric Administration, Silver Spring, MD: http://coastalsocioeconomics.noaa.gov/core/nsre/welcome.html [Google Scholar]
- 57. Nugues M, Smith GW, Hooidonk RJ, Seabra MI, Bak RPM. 2004. Algal contact as a trigger for coral disease. Ecol. Lett. 7:919–923 [Google Scholar]
- 58. Olson N, Ainsworth T, Gates R. 2009. Diazotrophic bacteria associated with Hawaiian Montipora corals: diversity and abundance in correlation with symbiotic dinoflagellates. J. Exp. Mar. Biol. Ecol. 371:140–146 [Google Scholar]
- 59. Pantos O, et al. 2003. The bacterial ecology of a plague-like disease affecting the Caribbean coral Montastrea annularis. Environ. Microbiol. 5:370–382 [DOI] [PubMed] [Google Scholar]
- 60. Patterson KL, et al. 2002. The etiology of white pox, a lethal disease of the Caribbean elkhorn coral, Acropora palmata. Proc. Natl. Acad. Sci. U. S. A. 99:8725–8730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Raina J, Dinsdale EA, Willis BL, Bourne DG. 2010. Do the organic sulfur compounds DMSP and DMS drive coral microbial associations? Trends Microbiol. 18:101–108 [DOI] [PubMed] [Google Scholar]
- 62. Raina JB, Tapiolas D, Willis BL, Bourne DG. 2009. Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl. Environ. Microbiol. 75:3492–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ravindran J, Raghukumar C, Raghukumar S. 2001. Fungi in Porites lutea: association with healthy and diseased corals. Dis. Aquat. Organ. 47:219–228 [DOI] [PubMed] [Google Scholar]
- 64. Reshef L, Koren O, Loya Y, Zilber-Rosenberg I, Rosenberg E. 2006. The coral probiotic hypothesis. Environ. Microbiol. 8:2068–2073 [DOI] [PubMed] [Google Scholar]
- 65. Ritchie K. 2006. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322:1–14 [Google Scholar]
- 66. Ritchie KB, Smith GW. 2004. Microbial communities of coral surface mucopolysaccharide layers, p 259–264 In Rosenberg E, Loya Y. (ed), Coral health and disease. Springer-Verlag, Berlin, Germany [Google Scholar]
- 67. Rohwer F, Seguritan V, Azam F, Knowlton N. 2002. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243:1–10 [Google Scholar]
- 68. Romano S, Palumbi S. 1996. Evolution of scleractinian corals inferred from molecular systematics. Science 271:640–642 [Google Scholar]
- 69. Rosenberg E, Kellogg CA, Rohwer F. 2007. A sea of microbes: coral microbiology. Oceanography 20:146–154 [Google Scholar]
- 70. Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. 2007. The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5:355–362 [DOI] [PubMed] [Google Scholar]
- 71. Rowan R, Powers DA. 1991. Molecular genetic identification of symbiotic dinoflagellates (zooxanthellae). Mar. Ecol. Prog. Ser. 71:65–73 [Google Scholar]
- 72. Rypien KL, Ward JL, Azam F. 2010. Antagonistic interactions among coral-associated bacteria. Environ. Microbiol. 12:28–39 [DOI] [PubMed] [Google Scholar]
- 73. Sharp KH, Distel D, Paul VJ. 2011. Diversity and dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides. ISME J. 6:790–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Shashar N, Cohen Y, Loya Y, Sar N. 1994. Nitrogen fixation (acetylene reduction) in stony corals: evidence for coral-bacteria interactions. Mar. Ecol. Prog. Ser. 111:259–264 [Google Scholar]
- 75. Skerra A. 1992. Phosphorothioate primers improve the amplification of DNA sequences by DNA polymerases with proofreading activity. Nucleic Acids Res. 20:3551–3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Smith J, et al. 2006. Indirect effects of algae on coral: algae-mediated, microbe-induced coral mortality. Ecol. Lett. 9:835–845 [DOI] [PubMed] [Google Scholar]
- 77. Sousa WP. 1979. Disturbance in marine intertidal boulder fields: the nonequilibrium maintenance of species diversity. Ecology 60:1225–1239 [Google Scholar]
- 78. Sunagawa S, et al. 2009. Bacterial diversity and white plague disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME J. 3:512–521 [DOI] [PubMed] [Google Scholar]
- 79. Sunagawa S, Woodley C, Medina M. 2010. Threatened corals provide underexplored microbial habitats. PLoS One 5:e9554 doi: 10.1371/journal.pone.0009554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Van Alstyne KL, Schupp P, Slattery M. 2006. The distribution of dimethylsulfoniopropionate in tropical Pacific coral reef invertebrates. Coral Reefs 25:321–327 [Google Scholar]
- 81. Vega Thurber RL, et al. 2008. Metagenomic analysis indicates that stressors induce production of herpes-like viruses in the coral Porites compressa. Proc. Natl. Acad. Sci. U. S. A. 105:18413–18418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Vega Thurber RL, et al. 2009. Metagenomic analysis of stressed coral holobionts. Environ. Microbiol. 11:2148–2163 [DOI] [PubMed] [Google Scholar]
- 83. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F. 2007. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 9:2707–2719 [DOI] [PubMed] [Google Scholar]
- 85. Wilson SG, Fischetti TR. 2010. Coastline population trends in the United States: 1960–2008, p 1–28 U.S. Dept. of Commerce, Economics and Statistics Administration, U.S. Census Bureau, Washington, DC [Google Scholar]
- 86. Wu CH, et al. 2010. Characterization of coastal urban watershed bacterial communities leads to alternative community-based indicators. PLoS One 5:e11285 doi: 10.1371/journal.pone.0011285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Xu J, Gordon JI. 2003. Honor thy symbionts. Proc. Natl. Acad. Sci. U. S. A. 100:10452–10459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhou J, et al. 2011. Reproducibility and quantitation of amplicon sequencing-based detection. ISME J. 5:1303–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zilber-Rosenberg I, Rosenberg E. 2008. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol. Rev. 32:723–735 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.