Abstract
Celiac disease (CD) is associated with intestinal dysbiosis, which can theoretically lead to dysfunctions in host-microbe interactions and contribute to the disease. In the present study, possible differences in Bacteroides spp. and their pathogenic features between CD patients and controls were investigated. Bacteroides clones (n = 274) were isolated, identified, and screened for the presence of the virulence genes (bft and mpII) coding for metalloproteases. The proteolytic activity of selected Bacteroides fragilis strains was evaluated by zymography and, after gastrointestinal digestion of gliadin, by high-pressure liquid chromatography/electrospray ionization/tandem mass spectrometry. The effects of B. fragilis strains on Caco-2 cell culture permeability and inflammatory response to digested gliadin were determined. B. fragilis was more frequently identified in CD patients than in healthy controls, in contrast to Bacteroides ovatus. B. fragilis clones carrying virulence genes coding for metalloproteases were more abundant in CD patients than in controls. B. fragilis strains, representing the isolated clones and carrying metalloprotease genes, showed gelatinase activity and exerted the strongest adverse effects on the integrity of the Caco-2 cell monolayer. All B. fragilis strains also showed gliadin-hydrolyzing activity, and some of them generated immunogenic peptides that preserved or increased inflammatory cytokine production (tumor necrosis factor alpha) and showed increased ability to permeate through Caco-2 cell cultures. These findings suggest that increased abundance of B. fragilis strains with metalloprotease activities could play a role in CD pathogenesis, although further in vivo studies are required to support this hypothesis.
INTRODUCTION
Celiac disease (CD) is the most common chronic intestinal inflammatory disorder triggered by ingestion of dietary gluten. This disease is considered as both a food hypersensitivity and an autoimmune disorder that involves genetic and environmental factors (40). The human leukocyte antigen (HLA) class II genes encoding for DQ2 and DQ8 heterodimers are the main hereditary factors predisposing to CD and are present in most CD patients (95%). Nevertheless, while 30 to 35% of the general population are carriers of these genes, only 2 to 5% actually develop CD, indicating that other factors contribute to precipitating the disease (33). The intake of gluten proteins is the critical environmental element responsible for the signs and symptoms of the disease and, in fact, typical cases manifest in early childhood after introduction of gluten into the diet. However, the disease is also being increasingly diagnosed in adulthood (5), suggesting that early exposure to gluten is not the only environmental trigger.
Gluten proteins and their toxic components (gliadins) are partially resistant to proteolytic degradation and can accumulate and interact with the small intestinal mucosa (14). Enzyme deficiency in the small intestinal mucosa of CD patients does not seem to be causally related to the disease (3). However, in CD patients, some peptides, such as the 33-mer of α-gliadin and others containing its main structural epitopes (PFPQPQLPY and PQPQLPYPQ), preferentially drive an adaptive immune response by binding to HLA-DQ2/DQ8 molecules of antigen-presenting cells and activating T-helper 1 (Th1) and Th17 inflammatory responses within the mucosa, with the resulting production of inflammatory cytokines (e.g., gamma interferon [IFN-γ] and interleukin-21 [IL-21]) leading to severe inflammation (24). Other gliadin peptides activate an innate immune response characterized by increased production of IL-15 by epithelial and antigen-presenting cells, which activate the effector function and cytotoxic activity of intraepithelial lymphocytes (15). Gliadin peptides also induce upregulation of the zonulin innate immunity pathway, which leads to increased intestinal permeability and enables paracellular translocation of gliadin and its subsequent interaction with antigen-presenting cells within the intestinal submucosa (11).
In recent years, alterations in the composition of the intestinal microbiota have been associated with CD. The bacterial numbers of the Bacteroides-Prevotella group or the Bacteroides fragilis group in CD patients have been demonstrated to be increased compared to those in healthy controls (8, 26). Bacteroides spp. are generally considered commensals or symbionts inhabiting the human gastrointestinal tract, representing ca. 25% of the total bacterial cells. Nonetheless, members of the normal microbiota can also potentially cause disease in cases of failure of the host defenses and major dysbiosis and can then be considered “pathobionts” (35). In spite of the abundance of Bacteroides spp. in the gut microbiota, their ecological distribution, composition, and impact on health remain unclear (46). Species such as B. fragilis and Bacteroides vulgatus seem to be implicated in the disruption of the integrity of the intestinal epithelial barrier, thereby contributing to the development of inflammation in experimental animal models (38, 42) and, possibly, in patients with inflammatory bowel disease (IBD) (10). The potential pathogenicity of Bacteroides spp. is related to the expression of a variety of virulence factors, including proteolytic and other hydrolytic enzymes (4). Enterotoxigenic B. fragilis (ETBF) strains produce an enterotoxin, termed B. fragilis toxin (BFT), which is a 20-kDa zinc-dependent metalloprotease that has been associated with diarrhea in humans and young animals (37). The B. fragilis toxin gene (bft) is located in a pathogenicity island, present exclusively in ETBF strains and is associated with another gene (mpII) that encodes a second metalloprotease (12).
Here, we hypothesized that changes in the composition of Bacteroides spp. and associated virulence features can turn these commensal bacteria into pathogenic inhabitants of the human intestinal tract that, acting in consortium with gluten peptides, can contribute to CD. To address this question, we determined differences in the diversity of Bacteroides spp. isolated from the feces of patients with active and nonactive CD in comparison with healthy controls and evaluated their virulence features and potential participation in the generation of gliadin peptides with immunotoxic effects on intestinal epithelial cells.
MATERIALS AND METHODS
Subjects and sampling.
Three groups of children were included in the present study: (i) patients with active CD (n = 20; mean age, 3.9 years; range, 1.0 to 8.8 years), who were on a normal gluten-containing diet, showing clinical symptoms of the disease, positive CD serology markers (anti-gliadin antibodies and anti-transglutaminase antibodies), and signs of severe enteropathy, classified as type 3 according to the Marsh classification of CD by duodenal biopsy examination; (ii) patients with nonactive CD (n = 18; mean age, 6.2 years; range, 3.3 to 12.2 years), who were on a gluten-free diet for at least 2 years, showed negative celiac serology markers and normal mucosa or infiltrative lesions classified as type 0-1 according to Marsh classification, and absence of disease symptoms; and (iii) healthy control children (n = 20; mean age, 5.7 years; range, 2.5 to 10.8 years). None of the children included in the study were treated with antibiotics for at least 1 month before the sampling time. The study was conducted in accordance with the ethical standards of the responsible institutional committees on human experimentation and in accordance with the Helsinki Declaration of 1975 as revised in 1983. Children were enrolled in the study after written informed consent was obtained from their parents.
Isolation of Bacteroides spp. from child feces.
Fecal samples were collected in sterile containers, kept under anaerobic conditions (AnaeroGen; Oxoid, Hampshire, United Kingdom), stored at 4°C, and analyzed in less than 12 h to avoid alterations in viability of Bacteroides spp. Samples (2 g [wet weight]) were diluted (1:10 [wt/vol]) in phosphate-buffered saline (PBS; 130 mM sodium chloride, 10 mM sodium phosphate [pH 7.2]) and homogenized in a Lab Blender 400 Stomacher (Seward Medical, London, United Kingdom). Serial dilutions were prepared in PBS, and aliquots were plated on Schaedler agar (Scharlau, Barcelona, Spain) supplemented with kanamycin (100 mg/liter), vancomycin (7.5 mg/liter), and vitamin K (0.5 mg/liter) and then incubated under anaerobic conditions at 37°C for 48 h. In order to analyze the dominant clones in each subject (21), five presumably different individual colonies were isolated from the highest dilution plate from each subject, and their cellular morphology and Gram-staining characteristics were examined.
Strain typing and species identification.
The isolated clones were identified at the species level by partial 16S rRNA gene sequencing using the primer pair Bfra531-f and Bfra766-r (45). The PCR products obtained were purified using GFX PCR DNA and a gel band DNA purification kit (GE Healthcare, Buckinghamshire, United Kingdom) for DNA sequencing. DNA sequencing was carried out by an ABI Prism 3130XL genetic analyzer (Applied Biosystems, California). The closest relatives of the partial 16S rRNA gene sequences were sought in GenBank using the basic local alignment search tool (BLAST) algorithm, and sequences with >97% similarity were considered to belong to the same species. RAPD [random(ly) amplified polymorphic DNA]-PCR was performed to differentiate the isolated clones at the strain level by colony PCR and using the M13 primer as previously described (Table 1) (7).
Table 1.
Primers for PCR and sequencing used in this study
| Primer | Fragment name | Fragment length (bp) | Primer sequence (5′–3′) | Reference |
|---|---|---|---|---|
| M13 | M13 | TTATGAAACGACGGCCAGT | 21 | |
| Bfra531F | Bfra | 289 | ATACGGAGGATCCGAGCGTTA | 20 |
| Bfra766R | CTGTTTGATACCCACACT | |||
| GBF-201 | GAACCTAAAACGGTATATGT | 22 | ||
| GBF-312 | bft-1 | 190 | CCTCTTTGGCGTCGC | |
| GBF-322 | bft-2 | 175 | CGCTCGGGCAACTAT | |
| GBF-334 | bft-3 | 287 | TGTCCCAAGTTCCCCAG | |
| LO1 | mpII | 350 | CCACCGTGCCAATGTCAGATA | 23 |
| RO1 | CTGAAGAACGAGGCGGTATC |
Pathogenicity markers and proteolytic activity of B. fragilis strains.
The presence of bft and mpII genes was screened in all of the isolated B. fragilis clones by PCR (Table 1). One forward primer (GBF-201) and three reverse primers (GBF-312 for bft-1, GBF-322 for bft-2, and GBF-334 for bft-3) were used in the same amplification reaction for the detection of the three isoforms of the bft gene by multiplex PCR (17). B. fragilis clones were also screened for the presence of the mpII gene by PCR (23). PCR products were separated in a 2% agarose gel by electrophoresis and visualized by ethidium bromide staining.
B. fragilis clones were analyzed by RAPD-PCR, and nine different strains were identified (A to I). The proteolytic activity of one representative of each B. fragilis strains was determined in gelatin and gliadin zymograms. For this, strains were grown in brain heart infusion broth (Scharlau, Barcelona, Spain) supplemented with 0.05% (wt/vol) cysteine (Sigma, St. Louis, MO) under anaerobic conditions for 24 h. Bacterial cells were collected by centrifugation (6,000 × g for 15 min), washed, and resuspended in PBS at a final concentration of 108 CFU/ml. Cell suspensions were separated in a discontinuous SDS-PAGE system that consisted of (i) a running gel containing 15% acrylamide (pH 8.8) and either 0.5% gelatin or gliadin and (ii) a stacking gel containing 4% acrylamide (pH 6.8). Gels were run at a constant voltage (120 V) in a MiniProtean 3 cell system (Bio-Rad, Richmond, CA). Gels were washed in 2.5% Triton X-100 at room temperature for 1 h and then incubated in reaction buffer (20 mM Tris-HCl, 150 mM NaCl, 5 mM CaCl2 [pH 7.4]) at 37°C overnight. Hydrolysis bands were visualized as a clear zone after Coomassie brilliant blue R-250 staining (Bio-Rad, Richmond, CA).
In vitro digestion of gliadins.
Gliadins were subjected to a simulated human gastrointestinal digestion as previously described (19). Aliquots (150 mg) of a commercially available extract of gliadin (Sigma) were dissolved in 3 ml of a saline solution (140 mM NaCl, 5 mM KCl [pH 3]) at 60°C for 30 min, with gentle agitation. Gastric digestion with pepsin (800 to 2,500 U/mg in 0.1 M HCl [pH 3] for 1 h), and intestinal digestion with pancreatin (4× USP specification) and bile (Sigma) in 0.1 NaHCO3 at pH 7 for 2 h were conducted at 37°C with agitation. After the gastric digestion, the intestinal digestion was carried out in the upper part of a two-chamber system in six-well plates separated by a 15,000-molecular-weight cutoff dialysis membrane (Spectrum Medical, Gardena, CA). Aliquots of the gastric digested samples were loaded into the upper chambers in the presence or absence of B. fragilis cell suspensions (108 CFU/ml) and incubated for 4 h. Then, saline solution from the basal chamber was recovered for further analysis. The total protein concentrations in both dialysates and retentates were quantified using a Lowry-based commercial kit (Sigma). The stability of bacteria during digestion was confirmed by plate counting under optimal conditions, which remained at 108 CFU/ml.
Reversed-phase HPLC and MS/MS analysis.
Gliadin-derived peptides were analyzed after simulated gastrointestinal digestion, as described elsewhere (20). The separation was conducted in a BioBasic C18 column (5 μm; 4.6 by 250 mm; Thermo, Waltham, MA) using an Agilent high-pressure liquid chromatography (HPLC) system connected in-line to an Esquire-LC electrospray system equipped with a quadrupole ion trap mass spectrometer (Bruker Daltonics, Billerica, MA). The elution phases consisted of 15% (vol/vol) acetonitrile (ACN)–0.1% (vol/vol) trifluoroacetic acid (TFA) (solvent A) and 80% (vol/vol) ACN–0.1% (vol/vol) TFA (solvent B). Aliquots (100 μl) of the dialysates were injected in each analysis. The gradient program started with 95% solvent A and 5% solvent B, and changed linearly to reach 10% solvent A and 90% solvent B within 30 min. UV absorbance was recorded at 214 nm. BioTools version 2.1 (Bruker Daltonics) software was used to process the tandem mass spectrometry (MS/MS) data and to identify peptide sequences by comparison to available gliadin sequences (accession numbers: α/β, AAZ94420; γ, AAQ63856; and ω, AAT74547). Three independent dialysates were analyzed in each case.
Caco-2 cell culture conditions.
The human colon carcinoma Caco-2 cell line was obtained from the American Type Culture Collection (Rockville, MD) at passage 14 and used in experiments at passages 19 to 23. Caco-2 cells were grown in Dulbecco modified Eagle medium (DMEM; AQ Media; Sigma), containing 4.5 g of glucose (Sigma/liter), 25 mM HEPES buffer (Sigma), 0.1% (vol/vol) antibiotic mixture (penicillin, streptomycin, and gentamicin; Sigma), and 10% (vol/vol) fetal bovine serum (Sigma). Cells were grown and maintained at 37°C in 5% CO2 and 95% air, and the culture medium was changed every 2 days (19). Cells at 70% confluence were detached from the flasks by using a trypsin solution (2.5 g/liter; Sigma) and resuspended in DMEM.
Evaluation of intestinal Caco-2 cell monolayer integrity.
Caco-2 cells were seeded at a density of 50,000 cells/cm2 onto polyethylene terephthalate membrane inserts (0.4-μm pore size; Millipore, Billerica, MA) and placed in six-well plates (Costar). In this bicameral system, 1.5 ml of treatment medium was loaded into the apical compartment, and 2 ml of saline solution was loaded into the basal compartment. Cell cultures were used at 7 days after seeding.
To determine the influence of selected Bacteroides strains on the integrity of the intestinal cell monolayer, bacterial cell suspensions (108 CFU/ml) of B. fragilis strains A, B, C, and I grown for 20 h were prepared in DMEM (without antibiotics) and loaded into the upper chamber alone and together with the dialysates of gliadin digested in the presence of these strains. After incubation (4 h), the basal medium was recovered and mixed with 100 μl of 1 M NaOH, and the diffusion of phenol red was determined by measuring the absorbance at 558 nm.
To determine the translocation of gliadin-derived peptides, Caco-2 cells were exposed basolaterally to tumor necrosis factor alpha (TNF-α) (10 ng/ml) for 24 h to simulate inflammatory conditions (31). In vitro digestions of gliadins in the presence of cell suspensions of B. fragilis A, B, C, or I strains were loaded into the upper chamber of the in vitro system. After 4 h of incubation, the basal medium was recovered to determine the concentration of permeated gluten peptides by enzyme-linked immunosorbent assay (ELISA), as described below.
Gluten quantification.
A commercially available quantitative immune-based ELISA kit, designed to detect the toxic fraction of gluten from food samples, was used according to the manufacturer's instructions (GlutenTox; Biomedal, Seville, Spain) (25). The analyses were performed in the fraction that reached the basal compartment after crossing the monolayer of Caco-2 cells subjected to inflammatory conditions and the gliadin samples digested in gastrointestinal conditions and in the presence or absence of selected B. fragilis strains.
Analysis of inflammatory markers.
In the supernatants of Caco-2 cell cultures exposed to the dialysates from digests of gliadins inoculated with the different B. fragilis strains, TNF-α (eBioscience, San Diego, CA), and IL-1β (eBioscience) were determined by ELISA according to the manufacturer's instructions. The sensitivity for these methods is 4 pg/ml.
Statistical analyses.
The Renyi diversity index was used to explore differences in Bacteroides species among the different child groups. This index provides three further diversity index values: species richness (S), the Shannon index (H′), and the Simpson index (1-D) (44).
The chi-square test was used to establish differences in the abundance of Bacteroides spp. and virulence genes. A P value of <0.05 was considered statistically significant. The Bonferroni adjustment test was applied to correct the significance for multiple comparisons among the three child groups studied (active and nonactive CD patients and controls), which has the advantage of reducing type I errors and the disadvantage of increasing type II errors.
For experimental studies with Caco-2 cell cultures, one-way analysis of variance (ANOVA) and the Fisher least significant difference (LSD) post hoc test were applied. Statistical significance was established at P < 0.05, using the SPSS software (v.15; SPSS, Inc., Chicago, IL).
RESULTS
Diversity of Bacteroides spp. in the fecal microbiota of CD patients.
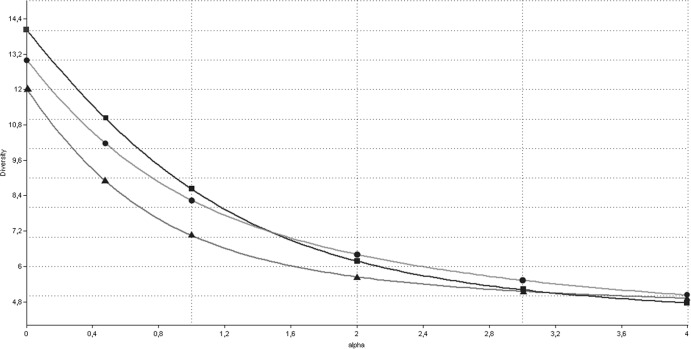
The species richness (S), Shannon (H′), and Simpson (1-D) indexes were very similar between active CD patients (S = 12, H′ = 1.95, and 1-D = 0.82), nonactive CD patients (S = 14, H′ = 2.15, and 1-D = 0.84) and controls (S = 13, H′ = 2.09, and 1-D = 0.84), indicating a similar diversity distribution between the studied child groups. Renyi diversity curves showed that active CD patients had a lower diversity than controls, whereas the diversity curves from nonactive CD patients intersected with the diversity curves from both active CD patients and controls and therefore could not be compared (Fig. 1).
Fig 1.
Renyi index curves of the Bacteroides spp. in active CD patients (▲), nonactive CD patients (▪), and control children (●). The Renyi index estimates total richness for α = 0, the Shannon index for α = 1, and the Simpson index for α = 2.
The total number of clones (n = 274) recovered from feces of healthy subjects and CD patients that were identified as Bacteroides spp. or Parabacteroides spp. is shown in Table 2. B. fragilis was more frequently isolated from CD patients with either active (P = 0.007) or nonactive (P = 0.009) disease than from healthy controls. This species represented up to 12% of the total clones from both groups of CD patients, which differs markedly from the 1% isolated from the controls. Parabacteroides distasonis was more frequently isolated from active CD patients than from nonactive CD patients and controls (P < 0.001). In contrast, Bacteroides ovatus was more frequently detected in controls than in CD patients (P = 0.014), irrespective of the phase of the disease (active or nonactive), and Bacteroides finegoldii was more frequently isolated from controls than from active CD patients (P = 0.014).
Table 2.
Bacteroides and Parabacteroides spp. isolated from fecal samples of active and nonactive CD patients and control children
| Species | Species abundance (%)a |
||
|---|---|---|---|
| Active CD (n = 97) | Nonactive CD (n = 82) | Control (n = 95) | |
| B. acidofaciens | 0A | 2 (2.4)A | 2 (2.1)A |
| B. caccae | 1 (1.0)A | 4 (4.9)A | 6 (6.3)A |
| B. dorei | 4 (4.1)A | 4 (4.9)A | 11 (11.6)A |
| B. finegoldii | 0A | 1 (1.2)AB | 7 (7.4)B |
| B. fragilis | 12 (12.4)A | 10 (12.2)A | 1 (1.1)B |
| B. intestinalis | 1 (1.0)A | 3 (3.7)A | 0A |
| B. massiliensis | 2 (2.1)A | 0A | 0A |
| B. ovatus | 5 (5.2)A | 4 (4.9)A | 16 (16.8)B |
| B. stercoris | 1 (1.0)A | 1 (1.2)A | 2 (2.1)A |
| B. thetaiotaomicron | 1 (1.0)A | 2 (2.4)A | 1 (1.1)A |
| B. uniformis | 22 (22.7)A | 23 (28.0)A | 27 (29.5)A |
| B. vulgatus | 20 (20.6)A | 19 (23.2)A | 13 (13.7)A |
| B. xylanisolvens | 0A | 4 (4.9)A | 2 (2.1)A |
| P. distasonis | 24 (24.7)A | 4 (4.9)B | 6 (6.3)B |
| P. merdae | 4 (4.1)A | 1 (1.2)A | 1 (1.1)A |
That is, the percentage of clones belonging to one specific species related to the total number of clones isolated from each child group (active CD patients, non-active CD patients or controls). Superscript letters (A and B) indicate the statistical differences calculated by using the chi-square test (2 × 2), corrected for a multiple-comparison test (three child groups) by using the Bonferroni adjustment. Significant differences between groups were defined as P < 0.017.
Pathogenicity markers and proteolytic activity of B. fragilis strains.
A total of 23 B. fragilis isolates were identified from the three groups of children and, according to RAPD-PCR fingerprint analyses, these included nine different strains (A to I). The carriage of bft and mpII virulence genes was analyzed in all isolates, and the same pattern was detected for isolates belonging to the same strain as defined by RAPD-PCR. The virulence gene carriage in the different B. fragilis strains is summarized in Table 3. The bft gene was detected in 13 B. fragilis clones from CD patients, 6 of which (identified as strain E or G) were associated with active disease and seven (strain H or I) with nonactive disease. Considering the bft-positive clones, the bft-2 isoform was significantly more prevalent (85%) than the isoforms bft-1 (15%, P = 0.007) and bft-3 (0%, P < 0.001). The bft-2 isoform was detected in all of the bft-positive B. fragilis clones (strains E, G, and H) isolated from active and nonactive CD patients, whereas the bft-1 isoform was only detected in two bft-positive clones (strain I) isolated from nonactive CD patients that also carried the bft-2 isoform. The mpII gene was detected in three of the B. fragilis clones (strains B and D) isolated from active CD patients. Only one B. fragilis clone (strain A) was identified in the healthy controls, and it was bft and mpII negative.
Table 3.
Characterization of virulence-associated genes (bft and mpII) in B. fragilis strains isolated from active CD patients, nonactive CD patients, and control children
| Source and B. fragilis strain (no. of clones) | Presence (+) or absence (−) of virulence genes |
|
|---|---|---|
| bft | mpII | |
| Control children | ||
| Strain A (1) | – | – |
| Active CD patients | ||
| Strain B (2) | – | + |
| Strain C (2) | – | – |
| Strain D (1) | – | + |
| Strain E (2) | + | – |
| Strain F (1) | – | – |
| Strain G (4) | + | – |
| Nonactive CD patients | ||
| Strain F (3) | – | – |
| Strain H (2) | + | – |
| Strain I (5) | + | – |
In zymograms of gelatin, a common clear band of ∼20 kDa was detected for both bft- or mpII-positive B. fragilis strains (B, D, E, G, H, and I) representing all isolated clones; the clearest bands corresponded to B. fragilis strains B and H. In contrast, hydrolysis bands were not detected in gelatin gels corresponding to the B. fragilis strains A, C, F, and J, which were bft and mpII negative (data not shown). In zymograms of gliadins, a proteolytic band of ∼120 kDa was detected in all of the B. fragilis strains tested (Fig. 2). These activities were partially or totally inhibited in the presence of EDTA (data not shown), indicating that metalloproteases were responsible for the gliadin hydrolysis in the gels. According to these results, B. fragilis strains A, B, C, and I, which represent strains of different origins and gene carriages, were selected for further studies on their possible differential pathogenic effects on intestinal epithelial cells and ability to hydrolyze gliadin peptides.
Fig 2.

Gliadin zymograms of the B. fragilis strains (lanes A to I) without EDTA. The identified gliadin-degrading protease pattern is characterized by one clear band (arrow), present in all of the samples with an approximate molecular mass of 120 kDa. Lane M, molecular masses of the protein markers in indicated in kilodaltons.
Effects of B. fragilis strains on monolayer integrity of Caco-2 cells.
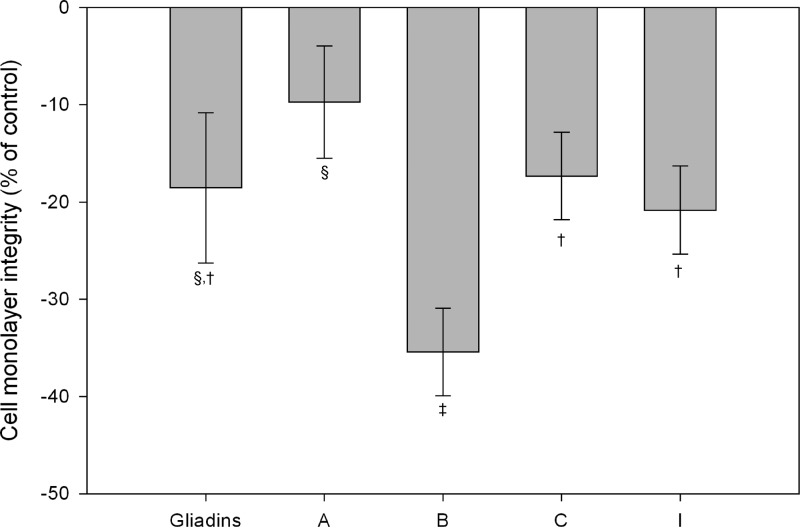
The possible direct adverse effects of the selected B. fragilis strains on cell monolayer integrity were evaluated by a phenol red diffusion assay, whereby diffusion takes place only across the tight junctions (Fig. 3). There was a significant increase (P < 0.05) in the basal content of phenol red in Caco-2 cell cultures incubated with B. fragilis strains B, C, and I compared to control cultures (data not shown), indicating alteration of the integrity of the cell monolayer. B. fragilis strain B (positive for the mpII gene and found in active CD patients) and, secondarily, B. fragilis strain I (positive for bft) caused the most marked impairment of cell monolayer integrity.
Fig 3.
Effect of the dialyzed fraction of gliadins digested in the presence of several B. fragilis strains on sensitized (TNF-α) Caco-2 cells monolayer permeability to phenol red after 4 h of incubation. Values are expressed as means ± the standard deviations (n = 4). Different symbols (†, ‡, and §) indicate statistically significant differences (P < 0.05) among the treatments tested (gliadin and strains A, B, C, and I) by applying ANOVA and the LSD post hoc test. In addition, effects of all treatments were significantly different from those of controls (nonstimulated cells [data not shown]).
Effects of B. fragilis strains in the generation of gliadin-derived peptides during digestion.
The amino acid sequences of gliadin-derived peptides generated during simulated gastrointestinal digestion, in the presence or absence of the selected B. fragilis strains, and that cross the dialysis membrane, are shown in Table 4. The total protein content of the dialysates from in vitro gliadin digestions, not inoculated with B. fragilis strains, was 1.05 ± 0.16 mg, representing up to 7% of the total protein loaded in the upper chamber of the in vitro system. However, inoculation of the selected B. fragilis strains during digestions increased the total dialyzable protein fraction to 2.27 ± 0.38 mg, which constitutes up to 13.3 to 18.7% of the total protein content in the upper chamber, indicating an increase in the degree of gliadin hydrolysis. Peptides generated during in vitro digestions showed high variability in their molecular masses, which ranged from 465.7 to 4,869.8 Da. In samples of gliadins digested in the absence of B. fragilis strains, peptides with amino acid sequences such as α/β-gliadin from amino acid positions 80 to 89 (α/β-Gld[80-89]) and α/β-Gld[80-100], which are inflammatory, and α/β-Gld[124-133], which has an amino acid sequence similar as those that interact with the chemokine receptor CXCR3, were identified (18, 20). In samples inoculated with the different B. fragilis strains different peptides with amino acid sequences of the main epitopes found in the immunodominant 33-mer peptide of α-Gld[56-88] were identified. For example, the peptide sequences for α/β-Gld[62-81], α/β-Gld[82-90], and α/β-Gld[77-85] were identified in gliadin samples digested in the presence of B. fragilis strain A; the sequences for α/β-Gld[84-96] and α/β-Gld[72-87] were identified in gliadin samples digested in the presence of B. fragilis strain I; the sequences for α/β-Gld[56-68], α/β-Gld[77-94], and α/β-Gld[50-86] were identified in gliadin samples digested in the presence of B. fragilis strain C; and the sequences for α/β-Gld[54-94] and α/β-Gld[82-90] were identified in gliadin samples digested in the presence of B. fragilis strain B. The molecular masses of the peptides identified could not be associated with bft or mp II gene carriage, but the shorter peptides were identified in samples inoculated with the strains B. fragilis strains A and I.
Table 4.
Gliadin-derived peptides in dialyzed fractions from different gastrointestinal digestions of gliadins, inoculated or not with B. fragilis strains
| Gliadin/B. fragilisstrain | Peptide | Amino acid sequence | Observed m/z | Calculated m/z | Ion (m/z) selected for MS/MS (charge) |
|---|---|---|---|---|---|
| Gliadins only | α/β-Gld[90-93] | QPQP | 465.7 | 468.3 | 464.7 (1) |
| α/β-Gld[82-89] | PQPQLPYP | 933.4 | 928.5 | 932.4 (1) | |
| α/β-Gld[217-224] | SQVSFQQPQ | 1,033.4 | 1,033.5 | 1,032.4 (1) | |
| α/β-Gld[124-133] | QQQQQQILQQ | 1,270.3 | 1,269.7 | 1,270.0 (1) | |
| α/β-Gld[75-87] | YLQLQPFPQPQLPYPQ | 1,472.7 | 1,471.8 | 1,471.7 (1) | |
| α/β-Gld[75-92] | YLQLQPFPQPQLPYPQPQ | 2,172.8 | 2,172.1 | 1,085.9 (2) | |
| α/β-Gld[80-100] | PFPQPQLPYPQPQPFRPQQPY | 2,541.7 | 2,541.8 | 1,270.4 (2) | |
| Gliadins plus strain: | |||||
| A | γ-Gld[24-46] | QPFSQQPQQIFPQPQQTPHQPQQ | 2,371.5 | 2,370.2 | 2,371.5 (1) |
| α/β-Gld[62-81] | YPQPQPFPSQQPYLQLQPF | 2,402.0 | 2,400.2 | 2,402.0 (1) | |
| α/β-Gld[82-90] | PQPQLPYPQ | 1,067.5 | 1,068.2 | 1,067.5 (1) | |
| α/β-Gld[239-248] | QNPQAQGSFQ | 1,106.5 | 1,105.1 | 1,106.5 (1) | |
| α/β-Gld[77-85] | QLQPFPQPQ | 1,085.0 | 1,083.2 | 1,085.0 (1) | |
| ω-Gld[258-267] | QQPQQPYPQQ | 1,240.9 | 1,241.6 | 1,240.9 (1) | |
| α/β-Gld[97-105] | QQPYPQPQP | 1,082.9 | 1,082.5 | 1,082.9 (1) | |
| B | α/β-Gld[248-279] | FQPQQLPQFEAIRNLALQFLPAMCNVYIPPYC | 3,708.6 | 3,706.8 | 1,854.3 (2) |
| γ-Gld[108-126] | QQSFPQQQPSLIQQSLQQ | 2,242.9 | 2,242.4 | 2,242.9 (1) | |
| α/β-Gld[54-94] | QQPFPPQQPYPQPQPFPSQQPYLQLQPFPQPQLPYPQPQPF | 4,869.8 | 4,868.4 | 2,434.9 (2) | |
| α/β-Gld[254-272] | PQFEAIRNLALQTLPAMCN | 2,131.0 | 2,130.1 | 1,065.5 (2) | |
| α/β-Gld[82-90] | PQPQLPYPQ | 1,067.9 | 1,068.2 | 1,067.9 (1) | |
| C | γ-Gld[82-95] | FPQTQQPQQPFPQS | 1,658.7 | 1,658.8 | 1,659.7 (1) |
| ω-Gld[376-391] | YPQQQPYGSSLTSIGG | 1,683.1 | 1,683.8 | 1,684.1 (1) | |
| γ-Gld[174-190] | QQLQCAAIHSVVHSIIM | 1,877.3 | 1,879.2 | 1,878.3 (1) | |
| α/β-Gld[42-62] | VPLVQQQQFPGQQQPFPPQQP | 2,416.2 | 2,416.2 | 1,208.6 (2) | |
| α/β-Gld[56-68] | PFPPQQPYPQPQP | 1,520.8 | 1,521.7 | 1,521.8 (1) | |
| α/β-Gld[77-94] | QLQPFPQPQLPYPQPQPF | 2,149.3 | 2,150.1 | 2,150.3 (1) | |
| α/β-Gld[50-86] | FPGQQQPFPPQQPYPQPQPFPSQQPYLQLQPFPQPQL | 4,343.3 | 4,343.2 | 1,448.1 (3) | |
| I | α/β-Gld[207-231] | QQQQQQQQPLSQVSFQQPQQQYPSG | 2,944.4 | 2,943.4 | 981.8 (3) |
| ω-Gld[338-355] | PQQPFQQPQQQLSQQPEQ | 2,165.6 | 2,165.3 | 2,166.6 (1) | |
| α/β-Gld[97-113] | QQPYPQPQPQYSQPQQP | 2,041.3 | 2,040.2 | 2,042.3 (1) | |
| α/β-Gld[229-246] | PSGQGFFQPSQQNPQAQG | 1,902.9 | 1,903.9 | 1,903.9 (1) | |
| α/β-Gld[148-167] | QQHNIAQGRSQVLQQSTYQL | 2,329.2 | 2,328.5 | 2,330.2 (1) | |
| ω-Gld[355-371] | QTISQQPQQPFPQQPHQ | 2,018.2 | 2,018.2 | 2,019.2 (1) | |
| α/β-Gld[84-96] | PQLPYPQPQPFRP | 1,565.3 | 1,565.8 | 1,566.3 (1) | |
| α/β-Gld[72-87] | QQPYLQLQPFPQPQLP | 1,924.8 | 1,923.2 | 1,925.8 (1) |
Inflammatory cytokine production by intestinal Caco-2 cells exposed to digested gliadins.
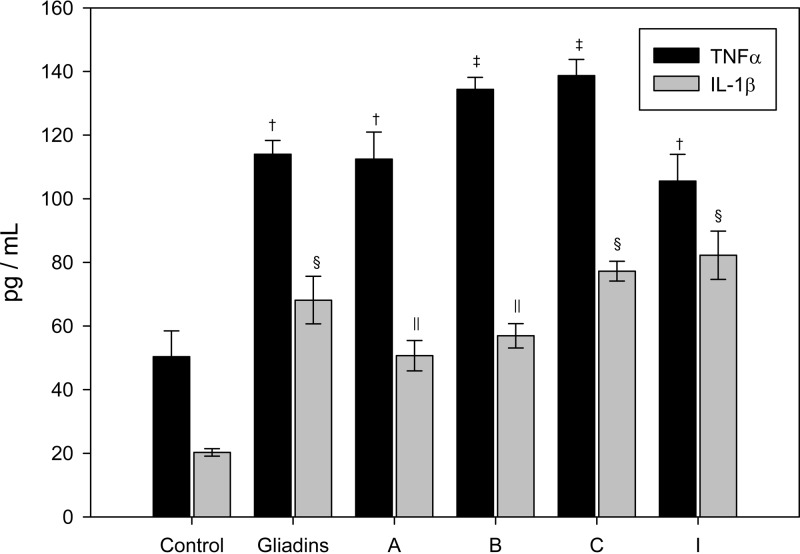
TNF-α and IL-1β production by Caco-2 cell cultures exposed to gliadin digestions, inoculated or not with the different B. fragilis strains selected, are shown in Fig. 4. After gastrointestinal digestion, gliadin-derived peptides significantly induced (P < 0.05) TNF-α and IL-1β production in the presence or absence of all of the B. fragilis strains tested in comparison to the controls. B. fragilis strains B and C induced the highest TNF-α production, whose values were significantly (P < 0.05) higher than those induced by gliadins digested alone, indicating that these B. fragilis strains could increase the gliadin-mediated proinflammatory potential. B. fragilis strains C and I induced the highest production of IL-1β, but the increase was only significant in comparison to the control and not compared to gliadin digested without bacteria.
Fig 4.
TNF-α and IL-1β production by Caco-2 cell cultures exposed to the dialyzed fraction of gliadins digested in the presence of several B. fragilis strains. The results are expressed as means ± the standard deviations (n = 5). Different symbols indicate statistically significant differences (P < 0.05) for TNF-α († and ‡) or for IL-1β (§ and ‖) by applying ANOVA and the LSD post hoc test.
Effect of B. fragilis strains on the permeability of the Caco-2 cell monolayer to gliadins.
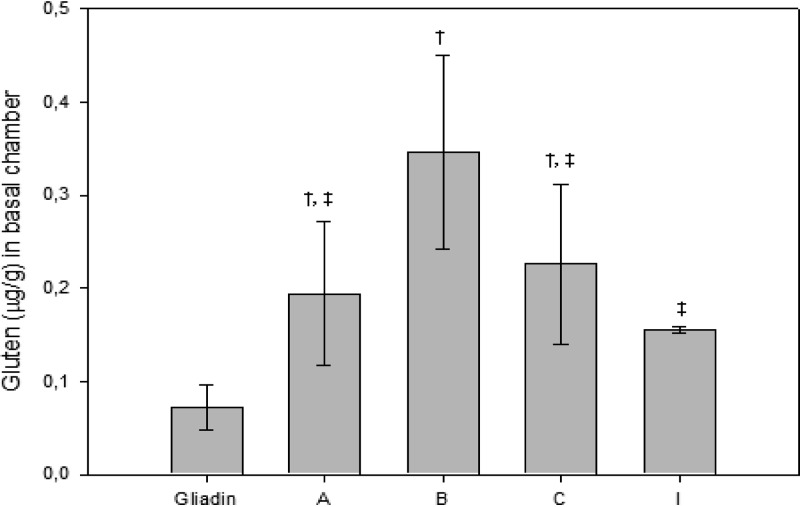
To evaluate whether B. fragilis strains could increase the permeability of Caco-2 cells to gliadin peptides by their direct deleterious effects on intestinal Caco-2 cell monolayer integrity and by increasing the amount of dialyzable and soluble fraction of gliadin through proteolytic degradation, Caco-2 cells were exposed simultaneously to the bacterial cell suspensions and the dialyzable fraction of gliadins digested in the presence of each B. fragilis strain. The toxic fraction of gluten initially loaded in the apical part of the two-chamber system was quantified by ELISA and also in the basal compartment after being incubated with the bacterial strains located in the apical compartment of the two-chamber system. The initial toxic fraction of gluten was significantly (P < 0.05) lower in the samples of gliadins digested alone (104.9 ± 4.4 μg/g) than in samples of gliadins digested in the presence of B. fragilis strains A (124.1 ± 1.1 μg/g), B (126.4 ± 1.9 μg/g), C (124.8 ± 3.3 μg/g), and I (118.4 ± 1.3 μg/g). The toxic fractions of gluten detected in the basolateral compartment of Caco-2 cultures exposed to these gliadin digestions, together with the cell suspensions of B. fragilis strains, were higher (P < 0.05) than in cell cultures exposed to gliadins digested without bacteria (Fig. 5). B. fragilis strain B caused the highest increase in gluten permeability in Caco-2 cells, probably due to its having the most deleterious effect on monolayer integrity related to the presence of the mpII gene and its ability to generate peptides that activated a stronger inflammatory response. This strain also generated the most immunogenic 33-mer (α/β-Gld[54-94], Table 4) identified in α-gliadin.
Fig 5.
Gluten content quantified in the basal chamber of Caco-2 cell cultures exposed to gliadins digested in the presence of cell suspensions of the different B. fragilis strains. Values are expressed as means ± the standard deviations (n = 4). Different symbols († and ‡) indicate statistically significant differences (P < 0.05) among the different strains tested by applying ANOVA and the LSD post hoc test. In addition, effects of all strains (A, B, C, and I) were significantly different from those detected in cells exposed to gliadin alone.
DISCUSSION
This study reports on the composition and potential virulence features of intestinal Bacteroides spp. in CD patients in comparison to healthy controls. Bacteroides spp. are commensal inhabitants of the human gastrointestinal tract, but have also been associated with chronic inflammatory bowel disorders (IBDs), such as ulcerative colitis and Crohn's disease, (2, 41, 43) and with CD (8, 9, 26). In IBD patients, bacteroides and enterobacteria have been considered responsible for >60% of the biofilm mass in the mucosa (41). Our study shows that active CD patients had a higher abundance of B. fragilis and a lower abundance of B. ovatus than controls, and these differences were not restored after long-term adherence to a gluten-free diet, suggesting this could play a primary role in the disease. Nevertheless, differences in the abundance of P. distasonis and B. finegoldii between active CD patients and controls were restored after adherence to the gluten-free diet. Although differences between fecal and duodenal mucosal bacteria may exist, our previous studies reported that B. fragilis numbers of feces and biopsy specimens from CD patients correlated, and their differences in comparison with healthy controls were similar (8), justifying the present study conducted with bacteroides from feces. In accordance with our results, B. fragilis is one of the least common species inhabiting the intestinal tract of healthy subjects, and yet it is most frequently isolated from clinical specimens and is the most virulent species (46). In contrast, B. vulgatus has been generally considered one of the most common intestinal Bacteroides species of healthy subjects (46), although in our study no such association was found. Nevertheless, other human studies have also reported associations between increased abundance of B. vulgatus and/or B. ovatus and chronic IBDs (10, 22), which partly contrast with our findings in CD patients.
It is known that the pathogenic potential of Bacteroides spp. depends on the presence of different virulence factors (e.g., agglutinins, polysaccharide capsules, or lipopolysaccharides) and a variety of proteolytic and hydrolytic enzymes (29, 46). Moreover, the comparative analyses of the whole genomes of several B. fragilis strains are revealing even larger genomic differences among strains (e.g., polysaccharide biosynthesis), which could determine their different virulence as opportunistic pathogens and ability to evade the immune defense mechanisms (27, 28). This evidence stresses the need to characterize the isolates at strain level and determine their specific virulence features to understand their potential pathogenicity in a specific ecosystem. In our study, the presence of genes encoding for metalloproteases was evaluated. B. fragilis clones with genes encoding for metalloproteases were frequently isolated from CD patients, which suggests a role of at least this species and probably these genes in disease pathogenesis according to our preliminary in vitro studies. Enterotoxigenic B. fragilis (ETBF) strains have been related to the inflammatory process in humans (2, 30) or animal models of IBD (30, 32). The enterotoxin produced by ETBF strains is encoded by the bft gene, which can be in three different isoforms; bft-1, bft-2, and bft-3 (37). In the present study, bft-2 was the most common isoform detected in the B. fragilis clones isolated from CD patients, which also seemed to be the most frequently present in clones that colonize the guts of children in comparison to those that colonize adult guts (36). The B. fragilis strains tested exhibited a genetic pattern where only either the bft gene or the mpII gene was present. In accordance with the size of the protease bands detected in our zymograms (Fig. 1), the bft gene encodes a zinc-dependent B. fragilis toxin (BFT), which is translated as a prepro-protein (44.4 kDa) further processed to a biologically active toxin of 168 amino acid residues, with a molecular mass of 20.7 kDa (13). In addition, the mpII gene also encodes a metalloprotease (20 kDa) predicted to be a zinc-dependent protein with 56% similarity to BFT protein (12).
The BFT is a soluble virulence factor secreted to the extracellular medium (13) that increases permeability of the intestinal epithelium by cleavage of the trans-membrane adhesion protein E-cadherin (47). A reduced expression of E-cadherin has been reported in CD patients, (1), although its relation to the microbiota has not yet been directly established. Our study suggests that the carriage of metalloprotease virulence genes by B. fragilis strains is associated with the ability of these strains to increase the permeability of the intestinal epithelium in vitro and that tight junctions are one of the early sites injured in Caco-2 cell cultures. This property was associated with both bft and mpII gene carriage, but the most remarkable effects were detected in the strain carrying the mpII gene. It has been found that both bft-1/bft-2 isoforms and mpII genes are clustered in pathogenicity islands, (12) and suggested that MPII is synthesized as a precursor protein similar to BFT (13). The highly conserved sequence of MPII found in different B. fragilis strains has led to hypotheses of important roles for this protein in ETBF strains and/or in ETBF-induced disease, although, to the best of our knowledge, no biological activity has yet been identified for this protein. B. fragilis strains not endowed with these metalloprotease genes and gelatinase activity also reduced the integrity of the Caco-2 cell monolayer to some extent, suggesting that other factors may also be responsible for this effect.
All of the B. fragilis strains studied exhibited gliadin-hydrolyzing activity. In a previous study, gliadin-hydrolyzing activity from microbial origin was found in biopsy specimens of CD patients, in contrast to controls, suggesting a pathogenic role for this activity, although this has yet to be confirmed (3). For this reason, we also evaluated whether B. fragilis could modify the peptide generated from gliadins and their potential immunotoxicity. Our study shows that the B. fragilis strains studied hydrolyzed gliadins, producing several peptides with the immunogenetic amino acid sequences of the main epitopes of the immunodominant 33-mer of α-Gld[56-88], while the partial digestion of gliadins by the gastrointestinal proteolytic enzymes used in the in vitro system did not produce these particular toxic sequences (19). In order to understand the possible pathological consequences of the hydrolytic activities of B. fragilis strains in this disorder, the inflammatory effects of the peptides generated were evaluated, and we demonstrated that they preserve or even increase their ability to induce inflammatory cytokine production (TNF-α). These increases in TNF-α production by epithelial cells could have adverse consequences on the pathogenesis of CD because this cytokine, in conjunction with IL-1β, increases paracellular permeability, facilitating the translocation of immunogenic peptides derived from gliadin to the lamina propria (39), and also mediates the infiltration of lymphocytes in the intestinal epithelium, thereby promoting tissue inflammation (16).
Although all B. fragilis strains were shown to have gliadin-hydrolyzing activity, the effects of different B. fragilis strains and of the peptides generated were slightly different, probably due to different levels of expression of the possible enzymes responsible or slightly different specificities (6). In addition, the gliadin-hydrolyzing activity of all B. fragilis strains increased the degree of proteolytic degradation and lowered the molecular masses of the peptides generated, increasing the protein content in the bio-accessible fraction that could facilitate interactions with the apical chemokine CXCR3 receptor of enterocytes, triggering inflammatory events (18, 20). In addition, the permeation of immunogenic peptides generated by B. fragilis from α-gliadins across intestinal epithelia was favored, which could promote the interaction of the peptides with the tissue transglutaminase and with antigen-presenting cells that ultimately activate the T cells responsible for the full expression of the disease (34).
Conclusions.
We have demonstrated here that the species B. fragilis is more abundant in the intestinal microbiota of CD patients, whereas B. ovatus is less abundant in comparison to healthy controls. These differences were also detected in CD patients after adherence to a gluten-free diet, suggesting that these alterations are not secondary to the underlying disease. We also demonstrated that B. fragilis clones carrying the bft and mpII metalloprotease genes and activity are abundant in CD patients and cause alterations in epithelial permeability. B. fragilis clones are also endowed with additional metalloproteases with gliadin specificity that generate immunotoxic peptides during in vitro intestinal digestion of gliadin. The generated peptides preserve or even increase their inflammatory properties on intestinal cells and more easily permeate the intestinal epithelial layer, which could favor their interaction with professional immunocompetent cells in the submucosa, although the magnitude of the effects are strain dependent. All in all, our findings indicate that the increased abundance of B. fragilis strains with metalloprotease activities in CD patients could play a pathogenic role, although evidence from in vivo studies is needed to confirm such a hypothesis.
ACKNOWLEDGMENTS
This study was supported by grants AGL2011-25169 and Consolider Fun-C-Food CSD2007-00063 from the Spanish Ministry of Science and Education. Scholarships to E. Sánchez from the Institute Danone and a postdoctoral contract to J. M. Laparra within the program Juan de la Cierva (MICINN, Spain) are acknowledged.
We thank L. Izquierdo for statistical advice.
Footnotes
Published ahead of print 6 July 2012
REFERENCES
- 1. Barshack I, et al. 2001. Immunohistochemical analysis of candidate gene product expression in the duodenal epithelium of children with coeliac sprue. J. Clin. Pathol. 54:684–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Basset C, Holton J, Bazeos A, Vaira D, Bloom S. 2004. Are Helicobacter species and enterotoxigenic Bacteroides fragilis involved in inflammatory bowel disease? Dig. Dis. Sci. 49:1425–1432 [DOI] [PubMed] [Google Scholar]
- 3. Bernardo D, et al. 2009. Is it true that coeliacs do not digest gliadin? Degradation pattern of gliadin in coeliac disease small intestinal mucosa. Gut 58:886–887 [DOI] [PubMed] [Google Scholar]
- 4. Botta GA, Arzese A, Minisini R, Trani G. 1994. Role of structural and extracellular virulence factors in gram-negative anaerobic bacteria. Clin. Infect. Dis. 18(Suppl 4):S260–S264 [DOI] [PubMed] [Google Scholar]
- 5. Catassi C, et al. 2010. Natural history of celiac disease autoimmunity in a USA cohort followed since 1974. Ann. Med. 42:530–538 [DOI] [PubMed] [Google Scholar]
- 6. Cerdeno-Tarraga AM, et al. 2005. Extensive DNA inversions in the Bacteroides fragilis genome control variable gene expression. Science 307:1463–1465 [DOI] [PubMed] [Google Scholar]
- 7. Claros M, et al. 1995. Identification and strain differentiation of “Bacteroides fragilis group” species and Prevotella bivia by PCR fingerprinting. Anaerobe 1:209–217 [DOI] [PubMed] [Google Scholar]
- 8. Collado MC, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. 2009. Specific duodenal and faecal bacterial groups associated with paediatric coeliac disease. J. Clin. Pathol. 62:264–269 [DOI] [PubMed] [Google Scholar]
- 9. De Palma G, Nadal I, Collado MC, Sanz Y. 2009. Effects of a gluten-free diet on gut microbiota and immune function in healthy adult human subjects. Br. J. Nutr. 102:1154–1160 [DOI] [PubMed] [Google Scholar]
- 10. Dicksved J, et al. 2008. Molecular analysis of the gut microbiota of identical twins with Crohn's disease. ISME J. 2:716–727 [DOI] [PubMed] [Google Scholar]
- 11. Fasano A, Catassi C. 2005. Coeliac disease in children. Best Pract. Res. Clin. Gastroenterol. 19:467–478 [DOI] [PubMed] [Google Scholar]
- 12. Franco AA, et al. 1999. Molecular evolution of the pathogenicity island of enterotoxigenic Bacteroides fragilis strains. J. Bacteriol. 181:6623–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Franco AA, et al. 1997. Cloning and characterization of the Bacteroides fragilis metalloprotease toxin gene. Infect. Immun. 65:1007–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hausch F, Shan L, Santiago NA, Gray GM, Khosla C. 2002. Intestinal digestive resistance of immunodominant gliadin peptides. Am. J. Physiol. Gastrointest. Liver Physiol. 283:G996–G1003 [DOI] [PubMed] [Google Scholar]
- 15. Heap GA, van Heel DA. 2009. Genetics and pathogenesis of coeliac disease. Semin. Immunol. 21:346–354 [DOI] [PubMed] [Google Scholar]
- 16. Hoffman RA. 2000. Intraepithelial lymphocytes coinduce nitric oxide synthase in intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 278:G886–G894 [DOI] [PubMed] [Google Scholar]
- 17. Kato N, et al. 2000. A new subtype of the metalloprotease toxin gene and the incidence of the three bft subtypes among Bacteroides fragilis isolates in Japan. FEMS Microbiol. Lett. 182:171–176 [DOI] [PubMed] [Google Scholar]
- 18. Lammers KM, et al. 2008. Gliadin induces an increase in intestinal permeability and zonulin release by binding to the chemokine receptor CXCR3. Gastroenterology 135:194–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Laparra JM, Glahn RP, Miller DD. 2009. Assessing potential effects of inulin and probiotic bacteria on Fe availability from common beans (Phaseolus vulgaris L.) to Caco-2 cells. J. Food Sci. 74:H40–H46 [DOI] [PubMed] [Google Scholar]
- 20. Laparra JM, Sanz Y. 2010. Bifidobacteria inhibit the inflammatory response induced by gliadins in intestinal epithelial cells via modifications of toxic peptide generation during digestion. J. Cell Biochem. 109:801–807 [DOI] [PubMed] [Google Scholar]
- 21. Lidin-Janson G, et al. 1977. Comparison of Escherichia coli from bacteriuric patients with those from feces of healthy schoolchildren. J. Infect. Dis. 136:346–353 [DOI] [PubMed] [Google Scholar]
- 22. Lucke K, Miehlke S, Jacobs E, Schuppler M. 2006. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J. Med. Microbiol. 55:617–624 [DOI] [PubMed] [Google Scholar]
- 23. Moncrief JS, Duncan AJ, Wright RL, Barroso LA, Wilkins TD. 1998. Molecular characterization of the fragilysin pathogenicity islet of enterotoxigenic Bacteroides fragilis. Infect. Immun. 66:1735–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Monteleone I, et al. 2010. Characterization of IL-17A-producing cells in celiac disease mucosa. J. Immunol. 184:2211–2218 [DOI] [PubMed] [Google Scholar]
- 25. Moron B, et al. 2008. Toward the assessment of food toxicity for celiac patients: characterization of monoclonal antibodies to a main immunogenic gluten peptide. PLoS One 3:e2294 doi:10.1371/journal.pone.0002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nadal I, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. 2007. Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J. Med. Microbiol. 56:1669–1674 [DOI] [PubMed] [Google Scholar]
- 27. Patrick S, et al. 2010. Twenty-eight divergent polysaccharide loci specifying within- and amongst-strain capsule diversity in three strains of Bacteroides fragilis. Microbiology 156:3255–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Patrick S, et al. 2011. A unique homologue of the eukaryotic protein-modifier ubiquitin present in the bacterium Bacteroides fragilis, a predominant resident of the human gastrointestinal tract. Microbiology 157:3071–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pumbwe L, Skilbeck CA, Wexler HM. 2006. The Bacteroides fragilis cell envelope: quarterback, linebacker, coach-or all three? Anaerobe 12:211–220 [DOI] [PubMed] [Google Scholar]
- 30. Rabizadeh S, et al. 2007. Enterotoxigenic Bacteroides fragilis: a potential instigator of colitis. Inflamm. Bowel Dis. 13:1475–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Resta-Lenert S, Barrett KE. 2006. Probiotics and commensals reverse TNF-α- and IFN-γ-induced dysfunction in human intestinal epithelial cells. Gastroenterology 130:731–746 [DOI] [PubMed] [Google Scholar]
- 32. Rhee KJ, et al. 2009. Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infect. Immun. 77:1708–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rossi M, Schwartz KB. 2010. Editorial: celiac disease and intestinal bacteria: not only gluten? J. Leukoc. Biol. 87:749–751 [DOI] [PubMed] [Google Scholar]
- 34. Sakly W, Thomas V, Quash G, El AS. 2006. A role for tissue transglutaminase in alpha-gliadin peptide cytotoxicity. Clin. Exp. Immunol. 146:550–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sansonetti PJ. 2011. To be or not to be a pathogen: that is the mucosally relevant question. Mucosal Immunol. 4:8–14 [DOI] [PubMed] [Google Scholar]
- 36. Scotto d'Abusco AS, Del GM, Censini S, Covacci A, Pantosti A. 2000. The alleles of the bft gene are distributed differently among enterotoxigenic Bacteroides fragilis strains from human sources and can be present in double copies. J. Clin. Microbiol. 38:607–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sears CL. 2009. Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin. Microbiol. Rev. 22:349–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shiba T, et al. 2003. The suppressive effect of bifidobacteria on Bacteroides vulgatus, a putative pathogenic microbe in inflammatory bowel disease. Microbiol. Immunol. 47:371–378 [DOI] [PubMed] [Google Scholar]
- 39. Skovbjerg H, Anthonsen D, Knudsen E, Sjostrom H. 2008. Deamidation of gliadin peptides in lamina propria: implications for celiac disease. Dig. Dis. Sci. 53:2917–2924 [DOI] [PubMed] [Google Scholar]
- 40. Sollid LM, Khosla C. 2005. Future therapeutic options for celiac disease. Nat. Clin. Pract. Gastroenterol. Hepatol. 2:140–147 [DOI] [PubMed] [Google Scholar]
- 41. Swidsinski A, Loening-Baucke V, Herber A. 2009. Mucosal flora in Crohn's disease and ulcerative colitis: an overview. J. Physiol. Pharmacol. 60(Suppl 6):61–71 [PubMed] [Google Scholar]
- 42. Sydora BC, et al. 2007. Epithelial barrier disruption allows non-disease-causing bacteria to initiate and sustain IBD in the IL-10 gene-deficient mouse. Inflamm. Bowel Dis. 13:947–954 [DOI] [PubMed] [Google Scholar]
- 43. Takaishi H, et al. 2008. Imbalance in intestinal microflora constitution could be involved in the pathogenesis of inflammatory bowel disease. Int. J. Med. Microbiol. 298:463–472 [DOI] [PubMed] [Google Scholar]
- 44. Tothmeresz B. 1995. Comparison of different methods for diversity ordering. J. Vegetation Sci. 6:283–290 [Google Scholar]
- 45. Vanhoutte T, et al. 2006. Molecular monitoring of the fecal microbiota of healthy human subjects during administration of lactulose and Saccharomyces boulardii. Appl. Environ. Microbiol. 72:5990–5997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wexler HM. 2007. Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20:593–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu S, Lim KC, Huang J, Saidi RF, Sears CL. 1998. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc. Natl. Acad. Sci. U. S. A. 95:14979–14984 [DOI] [PMC free article] [PubMed] [Google Scholar]