Abstract
Salmonella enterica forms aseptate filaments with multiple nucleoids when cultured in hyperosmotic conditions. These osmotic-induced filaments are viable and form single colonies on agar plates even though they contain multiple genomes and have the potential to divide into multiple daughter cells. Introducing filaments that are formed during osmotic stress into culture conditions without additional humectants results in the formation of septa and their division into individual cells, which could present challenges to retrospective analyses of infectious dose and risk assessments. We sought to characterize the underlying mechanisms of osmotic-induced filament formation. The concentration of proteins and chromosomal DNA in filaments and control cells was similar when standardized by biomass. Furthermore, penicillin-binding proteins in the membrane of salmonellae were active in vitro. The activity of penicillin-binding protein 2 was greater in filaments than in control cells, suggesting that it may have a role in osmotic-induced filament formation. Filaments contained more ATP than did control cells in standardized cell suspensions, though the levels of two F0F1-ATP synthase subunits were reduced. Furthermore, filaments could septate and divide within 8 h in 0.2× Luria-Bertani broth at 23°C, while nonfilamentous control cells did not replicate. Based upon the ability of filaments to septate and divide in this diluted broth, a method was developed to enumerate by plate count the number of individual, viable cells within a population of filaments. This method could aid in retrospective analyses of infectious dose of filamented salmonellae.
INTRODUCTION
Numerous conditions cause rod-shaped bacteria to form filamentous cells by interrupting the normal process of division and growth. Repressing the function of any filamentation temperature-sensitive (Fts) protein prevents the division complex from developing but does not block growth and results in filaments with multiple chromosomes (10, 22). FtsZ is a well-studied member of this family of proteins; at nonpermissive temperatures, ftsZ mutants of Escherichia coli are incapable of cellular division (4, 9, 10, 33, 56). Likewise, hydrostatic pressure, the SOS response via SulA, and antimicrobials, such as berberine and cinnamaldehyde, induce filamentation of E. coli by preventing the polymerization of FtsZ (5, 12, 13, 24, 30, 39, 55). β-Lactam antibiotics that bind penicillin-binding protein 3 (PBP3, also known as FtsI) inhibit its enzymatic activity and septum formation but not cellular growth (46, 62).
Bacterial filaments are also generated when DNA replication is inhibited. For example, nalidixic acid binds bacterial DNA gyrase, thereby stalling replication (52). These filaments have a single copy of the bacterial chromosome (27, 52). Additionally, dnaK mutants of E. coli form filaments with lower levels of DNA, protein, and RNA synthesis than the parental strain (23, 44).
Salmonella enterica, Listeria monocytogenes, and E. coli can form filaments in hyperosmotic environments. These osmotic-induced filaments are morphologically similar to fts mutants of E. coli in that they appear aseptate and have numerous chromosomes (1–3, 19, 31, 34, 36, 46, 58). Hyperosmotic conditions can be found in foods and the processing environment. Introducing the filaments into culture media without added humectants encourages the formation of septa and division of the filaments into daughter cells, each with a single chromosome (34). Filaments could have significant implications for public health, including their increased duration in hosts relative to nonfilamented cells (51), and present challenges to accurately monitor S. enterica if filaments form on food items.
Characteristics of osmotic-induced filaments of S. enterica were examined in detail in order to determine the mechanisms that regulate filamentous growth during osmotic stress. DNA and protein levels were similar in filaments and control cells when normalized by biomass. In vitro, the activity of PBP2 was greater in membranes of filaments than in control cells. Also, filaments had significantly more ATP than did control cells despite reduced levels of the β and γ subunits of the F0F1-ATP synthase. Potential mechanisms for the osmotic-induced filamentation of S. enterica are discussed.
MATERIALS AND METHODS
Strains, media, and growth conditions.
All strains of Salmonella used in this study were S. enterica subsp. enterica. LT2 is a laboratory strain of S. enterica serovar Typhimurium (43). S. enterica serovar Typhimurium strain M-09-0001A-1 (M-09) was isolated from an outbreak of salmonellosis involving peanut butter and obtained from the Minnesota Department of Health. S. enterica serovar Enteritidis strain E40 was obtained from the University of Pennsylvania and originally isolated from the ovary of a hen. S. enterica serovar Agona strain S-12 is a clinical isolate obtained from the Wisconsin State Laboratory of Hygiene. S. enterica serovar Tennessee strain 4539H was originally isolated by the United States Food and Drug Administration from an outbreak of salmonellosis involving dry milk. Δdps ΔrecA E. coli is a derivative of strain ATCC 43895 (stxI stxII serotype O157:H7) in which both dps and recA were deleted by homologous recombination (25). Strains were stored at −70°C in nutrient broth (BD Biosciences, Sparks, MD) with 15% (vol/vol) glycerol. Colonies of each strain were maintained on tryptic soy agar (TSA; BD Biosciences) for up to a month at 4°C.
Tryptic soy broth (TSB; BD Biosciences) was inoculated with individual S. enterica colonies and incubated for 20 to 24 h at 37°C with shaking to generate stationary-phase cultures. The optical density at 600 nm (OD600) of cultures in a 1-cm polystyrene cuvette (VWR, Radnor, PA) was measured with a Biomate3 instrument (Thermo Fisher Scientific, Waltham, MA). The OD600 values of stationary-phase cultures were greater than 1.7. Exponential-phase cultures were generated by diluting a stationary-phase culture 1:1,000 in fresh TSB and incubating the culture with shaking for 3 to 4 h at 37°C until an OD600 of 0.4 to 0.6 was achieved.
TSA (BD Biosciences) and TSB were supplemented with 2 to 10% (wt/vol) NaCl (Thermo Fisher Scientific) to induce osmotic stress. These are denoted at TSA-XNaCl and TSB-XNaCl, where X represents the percentage of NaCl supplemented to the media. An AquaLab dew point water activity meter 4TE instrument (Decagon Devices, Inc., Pullman, WA) was used to measure water activity (aw) values at 25°C in three independent experiments. On average, TSA supplemented with 0, 2, 4, 6, 7, 8, or 10% NaCl had aw values of 0.99, 0.98, 0.97, 0.96, 0.95, 0.94, or 0.92, respectively. TSB supplemented with 0, 2, 4, 6, 8, or 10% NaCl had aw values of 0.99, 0.98, 0.97, 0.96, 0.95, or 0.93, respectively.
To generate filaments on agar, 0.1 ml of a stationary- or exponential-phase culture of S. enterica was cultured on TSA or TSA supplemented with NaCl and incubated for 4 days at 30°C. These conditions were found to produce a maximum number of filaments on TSA and 7% NaCl (TSA-7NaCl) (51). Plates were wrapped with parafilm to prevent evaporation. Cells were harvested from the plates in 1× phosphate-buffered saline (PBS) (140 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.47 mM KH2PO4, pH 7.4) and placed on ice to prevent growth or division. To generate filaments in broth, NaCl-supplemented TSB was inoculated with a stationary- or exponential-phase culture of S. enterica at 1:50 in individual wells of a 24-well plate (BD Biosciences) and cultured statically for 4 days at 37°C (51).
Enumeration of Salmonella.
CFU were enumerated by serially diluting cultures 10-fold in 1× PBS and plating 0.1 ml on TSA. Plates were incubated for 20 h at 37°C prior to enumeration. To enumerate CFU in filamented populations, samples were cultured statically in 0.2× Luria-Bertani (0.2LB) broth (BD Biosciences) at a starting OD600 of ∼0.2 at 23°C for up to 24 h to enumerate viable units.
Isolation of genomic DNA.
Genomic DNA was isolated from control or filamented cells harvested from TSA or TSA-7NaCl, respectively. Briefly, cells suspended in 1× Tris-EDTA (TE) (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) with 0.5% (wt/vol) SDS, 167 μg/ml of RNase A, and 50 μg/ml of proteinase K were incubated at 37°C for 1 h. NaCl was added to a final concentration of 1.25 M, and the supernatant was transferred to a new tube following centrifugation in an Eppendorf 5417C centrifuge (18,000 × g, 10 min, 25°C). DNA was extracted with phenol-chloroform-isoamyl alcohol (25:24:1) followed by extraction with chloroform. The DNA was precipitated with 0.6× volumes of isopropanol and washed with 70% (vol/vol) ethanol. DNA was resuspended in 1× TE and the quantity measured with a Nanodrop 1000 (Thermo Fisher Scientific). The number of chromosomes per CFU was calculated using the equation X × (1 mol · bp/660 g) × [1 chromosome/(4.6 × 106 bp)] × [(6.02 × 1023 chromosomes)/1 mol], where X is the concentration of DNA/CFU. The size of the chromosome of S. enterica is approximately 4.6 × 106 bp, and 1 mol of bp is 660 g.
Digestion of genomic DNA with BAL 31.
Genomic DNA was digested with BAL 31 nuclease as previously described (25). Briefly, DNA was digested with 0.2 units of BAL 31 nuclease in 1× BAL 31 nuclease buffer (TaKaRa, Mountain View, CA) for 30 min at 30°C. Afterward, the nuclease was denatured at 70°C and DNA was separated on a 0.8% Tris-acetate-EDTA (TAE) (40 mM Tris, 20 mM acetic acid, 1 mM EDTA, pH 8.5) agarose gel at 9 V/cm. Digested DNA was visualized with ethidium bromide and its migration measured with ImageQuant 5.2 software (GE Healthcare, Piscataway, NJ). DNA isolated from Δdps ΔrecA E. coli O157:H7 that had been cultured for 2 h at 37°C in LB broth, pH 7.0 or 2.0, was used as the negative or positive control, respectively, for the digestion of DNA with BAL 31.
Isolation of WCLs.
Cells harvested from TSA or TSA-7NaCl were suspended in 1× PBS and pelleted via centrifugation using an Eppendorf 5417C centrifuge (18,000 × g, 10 min, 25°C). Pellets were resuspended in 1× lysis buffer (62.5 mM Tris-HCl, 2% [wt/vol] SDS, pH 6.8) and incubated for 10 min at 95°C. Samples were centrifuged again and the supernatants transferred to new tubes. The concentration of protein was quantified with bicinchoninic acid (Pierce, Rockford, IL) as per the manufacturer's instructions. A standard curve was generated with known concentrations of BSA (Pierce). Afterward, samples were diluted to a desired concentration in 1× lysis buffer and 2× sample buffer (62.5 mM Tris-HCl [pH 6.8], 2% [wt/vol] SDS, 20% [vol/vol] glycerol, 0.1 M dithiothreitol [DTT], 0.05% [wt/vol] bromophenol blue) to a final concentration of 1×. Whole-cell lysates (WCLs) were stored at −20°C.
Isolation of membrane proteins.
Membrane proteins were isolated as previously described from S. enterica cultured on TSA or TSA-7NaCl for 4 days at 30°C (15, 50). Briefly, cells harvested in 1× PBS were pelleted via centrifugation using a Sorvall SS34 rotor in a Sorvall RC 5B centrifuge (12,000 × g, 10 min, 4°C) and resuspended in ice-cold 50 mM sodium phosphate buffer, pH 7.0. Cells were sonicated in an ice bath for 20 s every minute for a total of 5 min. Cellular debris was cleared from the supernatant by centrifugation. Membrane proteins were pelleted via centrifugation (100,000 × g, 30 min, 4°C) of the supernatant in a TLA110 rotor in an Optima TLX centrifuge (Beckman Coulter, Brea, CA). Pellets were washed by first resuspending them with a 26-guage needle in fresh 50 mM sodium phosphate buffer, pH 7.0, and then repeating the centrifugation. The membrane pellets were resuspended in 50 mM sodium phosphate buffer, pH 7.0, and the concentration of protein was measured with bicinchoninic acid. Samples were stored at −70°C.
Binding of PBPs to BOCILLIN-FL.
A total of 1.33 μg/μl of membrane proteins was labeled with 25 μM BOCILLIN-FL (Molecular Probes, Eugene, OR) in 50 mM sodium phosphate buffer, pH 7.0, for 15 min at 37°C (18, 63). Afterward, proteins were denatured at 95°C in 1× sample buffer. Equal amounts of protein were separated via SDS-PAGE on a 10% Bis-Tris NuPAGE gel in 1× NuPAGE buffer (Invitrogen, Carlsbad, CA). Bound BOCILLIN-FL was imaged at a resolution of 100 μm with excitation at 473 nm and a long pass blue emission filter (>510 nm) on a Typhoon FLA 9000 scanner (GE Healthcare). The intensity of active PBPs labeled with BOCILLIN-FL was quantified using ImageQuant TL (GE Healthcare). The activity of each PBP was normalized to the total amount of protein detected with Coomassie stain. Values in Table 2 are represented as the normalized fluorescent units of each PBP bound to BOCILLIN-FL relative to the normalized fluorescent units of PBPs 5 and 6 in strain 4539H cultured on TSA. Afterward, proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (0.2-μm pores; Bio-Rad, Hercules, CA) and immunoblotted for total PBP3.
Table 2.
Activity of PBPs in four strains of S. enterica cultured for 4 days on TSA or TSA-7NaCla
| Strain | Activity (relative fluorescent units) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TSA |
TSA-7NaCl |
|||||||||
| PBP1b | PBP2 | PBP3 | PBP4 | PBP5 and PBP6 | PBP1b | PBP2 | PBP3 | PBP4 | PBP5 and PBP6 | |
| E40 | 240 ± 62 | 34 ± 36 | 18 ± 3.0 | 48 ± 27 | 570 ± 500 | 420 ± 59c | 51 ± 7.6 | 59 ± 19c | 95 ± 17 | 930 ± 180 |
| M-09 | 270 ± 110 | 19 ± 13 | 19 ± 7.3 | 61 ± 17 | 860 ± 220 | 500 ± 150 | 64 ± 9.6d | 54 ± 11c | 110 ± 55 | 1,100 ± 200 |
| S-12 | 220 ± 97 | 15 ± 3.0 | 44 ± 52 | 68 ± 34 | 1100 ± 190 | 310 ± 110 | 46 ± 10d | 43 ± 16 | 96 ± 54 | 1,100 ± 418 |
| 4539H | 290 ± 27 | 18 ± 2.8 | 110 ± 40 | 73 ± 11 | 1000 | 380 ± 97 | 100 ± 83c | 90 ± 67 | 110 ± 52 | 1,400 ± 270 |
Activity (in relative fluorescent units) was normalized to the total amount of protein detected with Coomassie stain in each sample and is reported relative to the activity of PBPs 5 and 6 in strain 4539H cultured on TSA, which was given an arbitrary value of 1,000.
Includes PBP1a and PBP1b.
P < 0.05.
P < 0.01.
Immunoblotting.
Equal amounts of proteins were loaded into individual wells of a 4 to 12% Bis-Tris NuPAGE gel and separated by SDS-PAGE in 1× NuPAGE buffer (Invitrogen). Proteins were transferred to PVDF membrane (0.2-μm pores) in transfer buffer (25 mM Tris base, 192 mM glycine, 20% [vol/vol] methanol). Membranes were blocked overnight in Tris-buffered saline Tween 20 (TBST) (150 mM NaCl, 2.7 mM KCl, 25 mM Tris base, 0.05% [vol/vol] Tween 20, pH 7.4) supplemented with 5% (wt/vol) dried skim milk (BD Biosciences) at 4°C. Membranes were incubated with a primary antibody and subsequently incubated with a secondary antibody conjugated to alkaline phosphatase. Bands were visualized with a solution of 0.165 mg/ml of 5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt (Promega, Madison, WI) and 0.33 mg/ml of nitro blue tetrazolium chloride (Promega) in alkaline phosphatase buffer (100 mM Tris-HCl [pH 9.0], 150 mM NaCl, 1 mM MgCl2). Western blots were scanned to digitalize the image and quantified using ImageQuant 5.2 software.
Primary antibodies included rabbit anti-FtsZ polyclonal antibody (Acris Antibodies, Inc., San Diego, CA) at 1:100, mouse anti-DnaK monoclonal antibody (Enzo Life Science, Farmingdale, NY) at 1:2,000, and rabbit anti-PBP3 antisera (kindly provided by David Weiss) at 1:15,000 (58). Secondary antibodies included goat anti-rabbit antibody (Promega) at 1:1,000 and goat anti-mouse antibody (Promega) at 1:6,000. Primary and secondary antibodies were diluted in TBST.
Identification of membrane proteins.
Proteins from bacterial membranes were separated by SDS-PAGE on a 4 to 12% Bis-Tris NuPAGE gel in 1× NuPAGE buffer, stained with 0.1% (wt/vol) Coomassie brilliant blue G-250 (Bio-Rad), and destained with 10% (vol/vol) acetic acid. In two independent experiments, proteins were sequenced from S. Tennessee strain 4539H cultured either on TSA or TSA-7NaCl (26, 28). Briefly, detectable proteins were excised from the gel, destained with 50% (vol/vol) methanol in 100 mM (NH4)HCO3, and dried with acetonitrile. Proteins were reduced with 25 mM DTT in 25 mM (NH4)HCO3 for 20 min at 55°C and alkylated for 20 min at 23°C in the dark with 55 mM iodoacetamide in 25 mM (NH4)HCO3. Samples were washed, dehydrated, and suspended in 25 mM (NH4)HCO3 containing 200 ng of trypsin (Promega) and 0.01% (vol/vol) ProteasMax (Promega). Proteins were digested for 3 h at 42°C, after which the supernatant was transferred to a new tube. ProteasMax was cleaved and trypsin inactivated with 0.3% (vol/vol) trifluoroacetic acid. The proteins were identified via matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry at the University of Wisconsin-Madison Mass Spectrometry facility.
Enumeration of filaments via microscopy.
An Olympus BHS microscope (Center Valley, PA) was used at ×1,000 magnification for all microscopy. Images were captured with an Olympus DP70 digital camera. To enumerate filaments (defined as cells longer than 10 μm), S. enterica cells were fixed in 5% (vol/vol) formalin, fixed to a glass slide (Gorilla Scientific, Silver Spring, MD), stained with 0.3% (wt/vol) crystal violet (BD Biosciences), and viewed with a bright-field objective. On average, more than 40 fields were viewed and 300 to 1,000 cells were enumerated. In the experiments shown in Fig. 2, 1 μg/ml of DAPI (4′,6-diamidino-2-phenylindole) was used to stain filaments on a black polycarbonate filter with pore size of 0.45 μm (Thermo Fisher Scientific). DAPI was visualized with the Olympus UG-1 excitation filter (330 to 385 nm) and L420 emitter (>420 nm). The method of staining did not affect the enumeration of filaments. Bacterial membranes were detected by staining cells with 15 μg/ml of FM4-64 (Molecular Probes) on glass slides coated with poly-l-lysine (Sigma). FM4-64 was visualized with the XF91 filter set (Omega Filters, Brattleboro, VT).
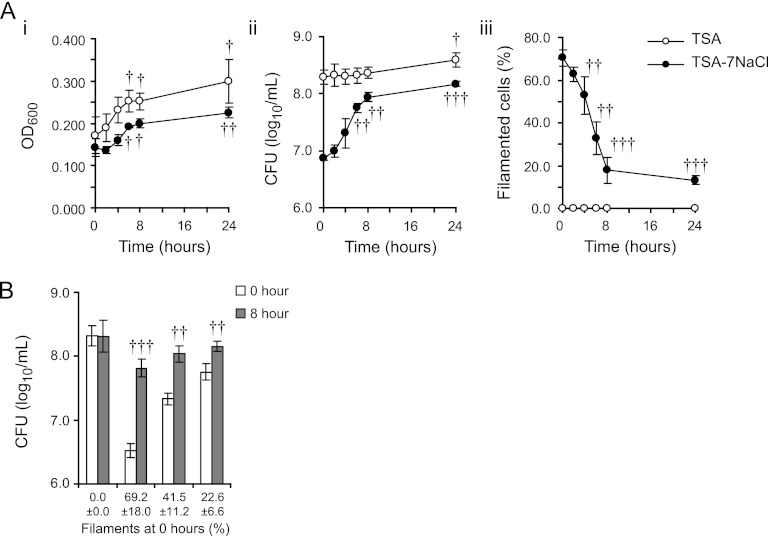
Fig 2.
Filaments of S. Tennessee were viable and septated when osmotic stress was removed. (A) S. Tennessee strain 4539H grown on TSA (open circles) or TSA-7NaCl (closed circles) was inoculated into 0.2LB broth and incubated at 23°C. The OD600 of the cells in suspension (i), log10 CFU/ml (ii), and percent filaments (iii) were calculated over time. (B) Mixed populations of filaments and control cells with various percentages of filaments were incubated at 23°C in 0.2LB broth and the log10 CFU/ml of each was measured at 0 h (white bars) and 8 h (gray bars) of incubation. Filaments were enumerated via fluorescence microscopy with DAPI (1 μg/ml). In both panels A and B, mean values from three independent experiments are reported. Error bars represent the SD of the means. Student's t tests (two-tailed) were performed to identify significant changes of each culture relative to measurements taken at 0 h of incubation (†, P < 0.05; ††, P < 0.01; †††, P < 0.001).
Quantifying amounts of ATP.
Levels of ATP in populations of S. enterica grown for 4 days at 30°C on TSA or TSA-7NaCl were measured using the BacTiter-Glo microbial cell viability assay (Promega). Briefly, harvested cells were suspended in 1× PBS to an OD600 of ∼0.2. Equal volumes of cells and BacTiter-Glo reagent were mixed in individual wells of a black, flat-bottom 96-well plate (Thermo Fisher Scientific) and incubated for 5 min at 23°C. Luminescence was measured with a BioTek Synergy HT multidetection microplate reader (Winooski, VT). Known amounts of ATP (Roche Applied Science, Indianapolis, IN) were used to generate a standard curve.
CTC redox assay.
The BacLight RedoxSensor 5-cyano-2, 3-ditolyl tetrazolium chloride (CTC) vitality kit (Molecular Probes, Eugene, OR) was used to visualize bacterial respiration as described previously (49). Briefly, S. enterica harvested from TSA or TSA-7NaCl was suspended in 1× PBS to an OD600 of ∼0.1 and then diluted 1:100 in 1× PBS. Samples were incubated with 5 mM BacLight RedoxSensor CTC for 30 min at 37°C. Afterward, cells were fixed in 5% (vol/vol) formalin and nucleic acids were counterstained with 1 μM SYTO-24. Cells were wet mounted onto a glass slide and viewed at ×1,000 magnification on an Olympus BHS microscope. SYTO-24 (Molecular Probes) and the red, fluorescent formazan product of reduced CTC were visualized with the XF91 filter set.
Statistics.
All experiments were performed independently at least three times, with the exception of the CTC assay, which was performed twice. Student's t tests were performed to identify significant differences (two-tailed, P < 0.05).
RESULTS
Filamentation was dependent on osmotic stress.
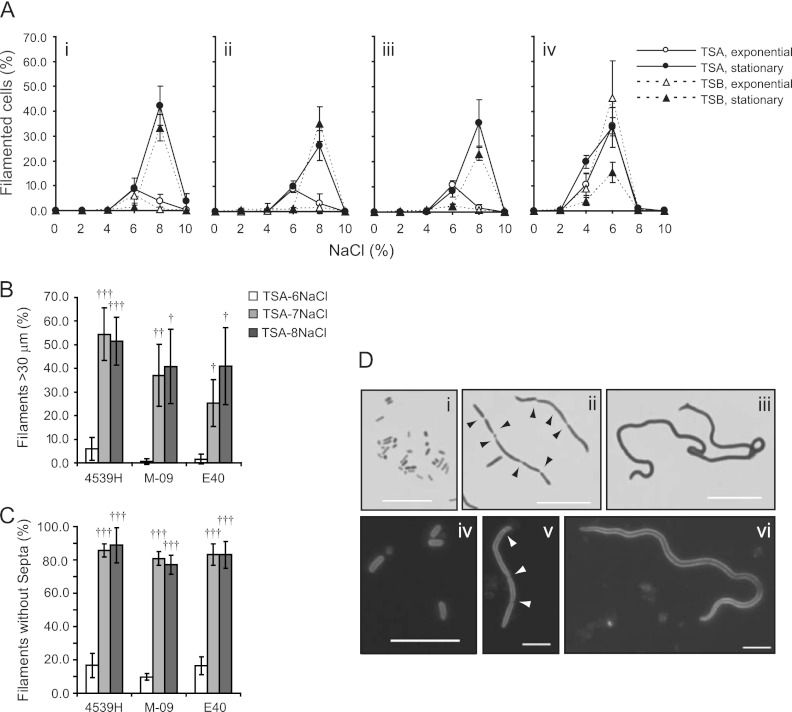
To develop a system for the study of filamentous growth during osmotic stress, four strains of S. enterica (S. Tennessee strain 4539H, S. Enteritidis strain E40, and S. Typhimurium strains M-09 and LT2) were cultured on TSA or in TSB supplemented with various concentrations of NaCl. Strains 4539H, E40, and M-09 formed filaments, which were defined as cells longer than 10 μm, when stationary-phase inocula were cultured on TSA or TSB supplemented with 6 to 10% NaCl (Fig. 1Ai to iii and D). Statistics were performed to identify conditions in which the percentage of filaments was significantly different from the percentage that formed when cells were cultured on TSA or TSB not supplemented with NaCl (see Table S1 in the supplemental material). The maximum frequency of filaments relative to the total number of cells occurred when stationary-phase cells were cultured on TSA-8NaCl or in TSB-8NaCl. For example, 42% and 33% of cells of strain 4539H filamented on TSA-8NaCl and TSB-8NaCl, respectively (Fig. 1Ai). The frequency of filamentation for each strain was less than 5% on TSA-10NaCl or in TSB-10NaCl.
Fig 1.
S. enterica filamented during osmotic stress. (A) Strains 4539H (i), M-09 (ii), E40 (iii), and LT2 (iv) were inoculated from exponential-phase (open symbols) or stationary-phase (closed symbols) cultures onto TSA (circles) or into TSB (triangles) supplemented with 0 to 10% (wt/vol) NaCl. The percentages of filaments (cells greater than 10 μm) generated were calculated after culturing cells for 4 days at 30 (TSA) or 37°C (TSB). In each strain, the maximal percentage of filamented cells was significantly different from the percentage of filaments in media with no added NaCl (Student's t test, two-tailed, P < 0.01). Filaments longer than 30 μm (B) and filaments without septa (C) were quantified for strains 4539H, M-09, and E40 grown on TSA-6NaCl (white bars), TSA-7NaCl (light-gray bars), or TSA-8NaCl (dark-gray bars). Student's t tests (two-tailed) were performed to identify whether filaments on TSA-7NaCl or TSA-8NaCl were significantly different from those generated on TSA-6NaCl (†, P < 0.05; ††, P < 0.01; †††, P < 0.001). Filaments on TSA-7NaCl and TSA-8NaCl were similar. In panels A to C, results are presented as mean values from three independent experiments. Error bars represent the standard deviations (SD) of the means. (D) Representative images of strain 4539H on TSA (i, iv), TSA-6NaCl (ii, v), and TSA-8NaCl (iii, vi) stained with crystal violet (i to iii) or FM4-64 (iv to vi). Arrows point to septa. The bar in panel D represents 10 μm.
Exponential- and stationary-phase inocula of strains 4539H, E40, and M-09 filamented equally well when cultured on TSA-6NaCl or on TSB-6NaCl (Fig. 1Ai to iii). Exponential-phase inocula of strains 4539H, E40, and M-09 did filament when cultured on TSA or in TSB supplemented with 8 or 10% NaCl, albeit less frequently than stationary-phase cells. The percentage of filaments that grew from exponential-phase inocula on TSA or in TSB supplemented with 8 or 10% NaCl was not statistically different from the percentage of filaments generated on TSA or TSB, respectively. The differing ability of exponential- and stationary-phase cells to filament during osmotic stress was not dependent on the concentration of the inoculum. Stationary-phase cultures of strain 4539H diluted prior to inoculation filamented as frequently on TSA-8NaCl and TSB-8NaCl as did undiluted inocula (data not shown).
LT2 encodes a rare start codon for rpoS, which represses the expression of its protein, RpoS (32). LT2 formed filaments as frequently as strains 4539H, E40, and M-09 (Fig. 1Aiv). LT2 cultured on TSA-4NaCl and in TSB-4NaCl filamented, while the other strains that were tested did not. Additionally, LT2 maximally filamented when grown on TSA-6NaCl and in TSB-6NaCl. Unlike E40, M-09, and 4539H, inocula prepared from exponential-phase cells of LT2 filamented more often than inocula prepared from stationary-phase cells in TSB-6NaCl (Fig. 1Aiv).
The frequency of filaments without septa correlated with osmotic stress.
Filaments that formed on TSA-6NaCl or TSA-8NaCl had different morphologies. First, filaments were longer on TSA-8NaCl than TSA-6NaCl (Fig. 1B and Dii and iii). Whereas 6% ± 5% of filaments of strain 4539H grown on TSA-6NaCl were longer than 30 μm, 52% ±10% of filaments that grew on TSA-8NaCl were longer than 30 μm. Second, the percentage of filaments without septa correlated with the concentration of NaCl (Fig. 1C and Dii and iii). A total of 80% of filaments generated on TSA-8NaCl did not have visible septa, while only 15% of filaments on TSA-6NaCl were without septa. This observation was confirmed with an independent staining method, in which FM4-64 was used to stain the bacterial membrane; a greater proportion of filaments generated on TSA-6NaCl had septa than filaments generated on TSA-8NaCl (Fig. 1Div to vi). These findings indicated that the inhibition of bacterial replication or division induced filamentous growth during osmotic stress.
Filaments on TSA-7NaCl were also examined and were morphologically similar to those on TSA-8NaCl (Fig. 1B and C, Table 1). Of these filaments grown on TSA-7NaCl, 80% were aseptate and 40 to 50% were longer than 30 μm. Statistically, the frequency of filamentation on TSA-7NaCl was similar to that on TSA-8NaCl (Student's t test, two-tailed, P > 0.05). In subsequent studies, filaments generated on TSA-7NaCl were studied because they were morphologically similar to filaments generated on TSA-8NaCl and because greater numbers of cells could be collected from TSA-7NaCl than from TSA-8NaCl. Additionally, we chose to study S. Agona strain S-12 rather than LT2. S-12 both grew and filamented better than did LT2 on TSA-7NaCl (Fig. 1Aiv, Table 1).
Table 1.
A comparison of macromolecules isolated from S. enterica grown on TSA or TSA-7NaCl
| Strain | NaCl (%) | % of filamented cells | Amt of protein (μg) |
Amt of chromosomal DNA (μg) |
No. of chromosomes (copies/CFU)a | |||
|---|---|---|---|---|---|---|---|---|
| Isolated from membrane |
In the whole-cell lysate (per OD600) | |||||||
| Per OD600 | Per 108 CFU | Per OD600 | Per 108 CFU | |||||
| E40 | 0 | <0.2 | 16.9 ± 13.3 | 0.7 ± 0.1 | 193 ± 16 | 23.8 ± 5.5 | 1.6 ± 0.2 | 1.9 ± 0.1 |
| 7 | 43.1 ± 9.3b | 14.1 ± 8.4 | 9.1 ± 5.1b | 229 ± 41 | 27.8 ± 6.8 | 10.4 ± 2.9e | 12.1 ± 3.4 | |
| M-09 | 0 | <0.2 | 17.3 ± 2.9 | 0.9 ± 0.4 | 175 ± 8 | 18.8 ± 16.4 | 1.6 ± 0.8 | 1.9 ± 1.1 |
| 7 | 41.3 ± 4.4b | 14.8 ± 2.9 | 12.3 ± 1.2b | 239 ± 41 | 24.0 ± 4.2 | 12.4 ± 1.3b | 14.3 ± 2.0 | |
| S-12 | 0 | <0.1 | 17.1 ± 2.9 | 0.9 ± 0.5 | 174 ± 13 | NDc | ND | ND |
| 7 | 29.7 ± 2.8b | 9.5 ± 1.6d | 4.7 ± 1.3e | 192 ± 9 | ND | ND | ND | |
| 4539H | 0 | <0.2 | 20.9 ± 7.0 | 1.2 ± 0.6 | 188 ± 19 | 11.0 ± 1.0 | 1.1 ± 0.2 | 1.3 ± 0.1 |
| 7 | 63.1 ± 12.4b | 16.1 ± 0.8 | 21.2 ± 5.2e | 272 ± 27d | 18.9 ± 2.7e | 22.3 ± 2.1b | 25.8 ± 2.7 | |
The chromosome of S. enterica is 4.6 × 106 bp.
P < 0.001.
ND, not determined.
P < 0.05.
P < 0.01.
Populations of filaments and control cells had similar amounts of protein and DNA.
E. coli exhibits filamentous growth when DNA (21, 23, 44) replication (21, 23, 44) or protein synthesis is inhibited (23, 27, 44, 52). The concentrations of proteins from membranes or WCLs and of chromosomal DNA in populations of filaments and control cells were measured to address whether deficiencies in the synthesis of protein or DNA contributed to the osmotic-induced filamentation. Control or filamentous populations of strains E40, M-09, S-12, and 4539H were generated on either TSA or TSA-7NaCl, respectively (Table 1). The concentrations of proteins and chromosomal DNA were similar in filamented and control suspensions of cells for each strain when bacterial mass was normalized by OD600. Therefore, filamentation of S. enterica during osmotic stress is not associated with defects in the synthesis of protein or DNA.
It was possible that filamentation was induced by the SOS response. If the SOS response were induced, osmotic stress would be expected to induce DNA damage. Chromosomal DNA isolated from S. enterica grown on TSA or TSA-7NaCl was not sensitive to digestion with BAL 31 and migrated similarly, but DNA from a Δdps ΔrecA mutant of E. coli O157:H7 was sensitive to digestion with BAL 31 after the strain was incubated in LB broth acidified to pH 2.0 (data not shown).
Filaments were multiple viable units.
Filamented populations of strains E40, M-09, and 4539H generated on TSA-7NaCl had 12.1 ± 3.4, 14.3 ± 2.0, and 25.8 ± 2.7 chromosomes/CFU, respectively, while control cells generated on TSA had less than 2 chromosomes/CFU (Table 1). These calculations indicated that filaments were in fact comprised of multiple viable units. Therefore, we predicted a single filamentous cell would produce more than 10 CFU after septation and division.
Each filament of S. enterica forms a single CFU on an agar plate (34). As a result, enumerating CFU of populations of S. enterica containing filaments can underestimate the number of individual, viable bacterial cells. In order to enumerate individual cells within a filament-containing population, a reproducible method was developed for filaments to septate and divide prior to the replication and division of control cells. Suboptimal temperatures and culture medium, which decrease the rate of replication, were employed.
Control and filamented populations of S. Tennessee strain 4539H were generated on TSA or TSA-7NaCl, respectively. Harvested cells were adjusted to an OD600 of 0.2 and incubated in 0.2LB broth at 23°C under static conditions (Fig. 2A). This concentration of LB was chosen because concentrations greater than 0.2× resulted in replication and division of control cells during incubation (data not shown). In the developed assay, the number of CFU of control cells remained unchanged during 8 h of incubation, although the OD600 of the bacterial suspension increased 1.4-fold (Fig. 2Ai and ii). Control cells did appear elongated (4 to 6 μm) after 8 h of incubation in 0.2LB but were not long enough to be considered filaments (Fig. 2Aiii). The elongation of the control cells likely explains the increase in the OD600 of the control cultures. In contrast, the concentration of CFU in the filamented population increased 1.1 log10, while the percentage of filaments in culture decreased from 70.5 ± 3.9% to 17.8 ± 6.1% (Fig. 2Aii and iii). These data indicated that the conditions were favorable for the septation of filaments but not the replication of nonfilamentous control cells during 8 h of incubation. Similar results were obtained for strains E40, M-09, and S-12 (data not shown). The percentages of filaments were similar after 8 and 24 h of incubation. By 24 h, the concentration of control cells had doubled (Fig. 2Aii).
Next, the sensitivity of this method was tested to detect increases in the number of viable cells in filamented populations containing fewer than 70% filaments. Suspensions of control cells and filaments were mixed in various proportions to yield cultures that had different percentages of filaments (Fig. 2B). These percentages correlated inversely with the starting concentration of CFU. Significantly, the concentration of each filament-containing culture increased after 8 h of incubation (P < 0.05) and reached concentrations similar to that of the control culture. For example, the concentration of CFU increased by 0.4 log10/ml when 23% of the initial population was filamented. Consistently, the percentage of filaments decreased by 75% after 8 h of culturing in 0.2LB broth in all strains assayed (Fig. 2Aiii and data not shown).
Osmotic stress did not affect the amount of FtsZ or PBP3.
Proteins of the Fts family are required for the division of enteric bacteria but not cellular growth or replication of DNA (1, 3, 7, 10, 46, 62). The amounts of FtsZ and PBP3 (FtsI) were quantified via immunoblot to address whether the levels of individual Fts proteins differed in osmotically stressed and unstressed cells. The levels of FtsZ and total PBP3 were unaffected (P > 0.05) by the osmolarity of the media after culturing for 4 days (data not shown). Therefore, filamentous growth during osmotic stress is not caused by changes in the expression of FtsZ or PBP3.
Osmotic stress correlated with changes in the activity of PBP2.
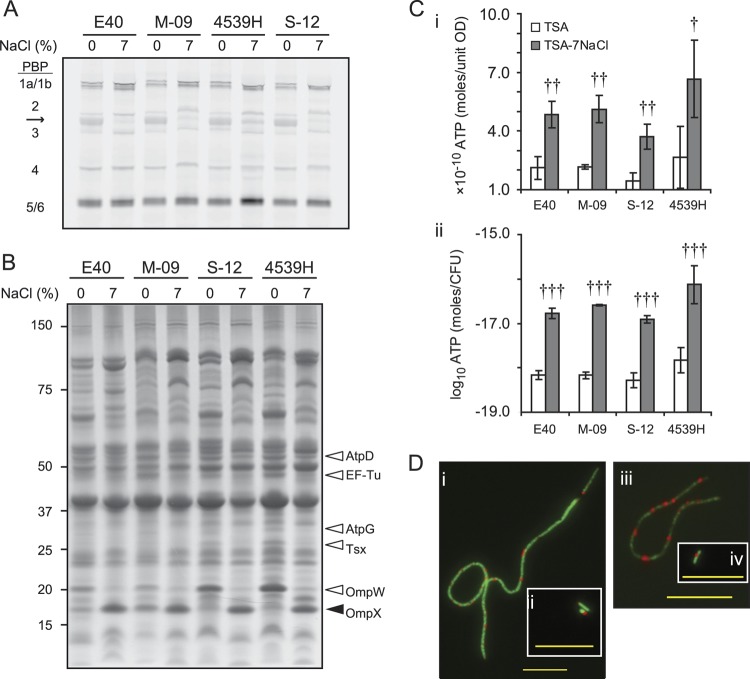
Filaments of S. enterica on TSA-7NaCl and TSA-8NaCl occasionally branched, and their width appeared greater than that of control cells (Fig. 1Diii and vi). PBPs regulate the synthesis of the bacterial cell wall, and unbalanced synthesis can affect cellular morphology (11, 40, 41, 47). PBPs from all four strains of S. enterica cultured on TSA or TSA-7NaCl were active in vitro as determined by their binding to BOCILLIN-FL, a fluorescent derivative of penicillin V (Fig. 3A). To identify which of the fluorescent bands were PBPs, we incubated membrane proteins with increasing concentrations of unlabeled penicillin G and then counterlabeled with BOCILLIN-FL (data not shown). All bands but one (arrow in Fig. 3A) decreased in intensity as the concentration of unlabeled penicillin G increased, confirming their identities as PBPs.
Fig 3.
Growth of S. enterica during osmotic stress correlated with changes in the levels of membrane proteins and ATP. Stationary-phase inocula of strains E40, M-09, S-12, and 4539H were cultured on TSA (white bars) or TSA-7NaCl (gray bars) for 4 days at 30°C to generate populations of control cells and filaments, respectively. (A) PBPs (denoted on the left-hand side of the gel) were bound to BOCILLIN-FL in vitro and detected by SDS-PAGE. The arrow points to a nonspecific, autofluorescent band. (B) Membrane proteins were separated by SDS-PAGE and stained with Coomassie blue G-250. Proteins whose levels decreased (open arrowhead) or increased (closed arrowhead) during osmotic stress in all strains tested were identified and are denoted to the right of the arrowheads. The migration of a protein standard is indicated on the left-hand side of the gel. In panels A and B, a representative image of at least three independent experiments is shown. (C) The amount of ATP in salmonellae grown on TSA (white bars) or TSA-7NaCl (gray bars) was quantified and normalized by OD600 (i) or CFU (ii). Bars represent the means from three independent experiments, and error bars represent the SD of the means. Student's t tests (two-tailed) were performed to identify significant changes in the amount of ATP in populations of control cells and filaments for each strain (†, P < 0.05; ††, P < 0.01; †††, P < 0.001). (D) Representative images from two independent experiments of filaments (i, iii) and control cells (ii, iv) of strains 4539H (i, ii) and M-09 (iii, iv) incubated with CTC. The reduced formazan product of CTC is red, and nucleic acids were stained with SYTO-24 (green). The bar in panel D represents 10 μm.
The activity of each PBP was measured by its binding to BOCILLIN-FL and normalized to the total amount of protein in each sample as determined by Coomassie stain (Table 2, Fig. 3B). Of all the PBPs that were assayed, PBP2 was the only one whose activity was consistently greater in cells cultured on TSA-7NaCl than in cells cultured on TSA (Table 2). The activity of PBP2 increased 3- to 5-fold in filamentous populations of strains M-09, S-12, and 4539H relative to that in control cells of each strain, and these increases were statistically significant (P < 0.05). A 1.5-fold increase in the activity of PBP2 was also observed in filamentous populations of strain E40 relative to that in control cells, but this increase was not significant (P > 0.05). The activities of PBPs 1, 4, and 5/6 were largely similar in filamentous and control populations. The activity of PBP3 increased in filaments of strains E40 and M-09 but remained unchanged in filaments of strains S-12 and 4539H relative to that in control cells. These data suggest that PBP2 is the only candidate of the identified PBPs to contribute to the osmotic-induced filamentation of S. enterica.
The levels of ATP synthase in S. enterica correlated inversely and ATP levels directly with osmotic stress.
To identify other proteins that may contribute to the filamentation of salmonellae during osmotic stress, membrane proteins from four strains of S. enterica were separated via SDS-PAGE and stained with Coomassie G-250 (Fig. 3B). Six proteins correlated either directly or inversely with osmotic stress in all strains tested. These proteins were identified via MALDI-TOF mass spectrometry (Table 3). The levels of two outer membrane proteins (OmpW and Tsx), the translation factor, EF-Tu, and the F0F1-ATP synthase subunits β (AtpD) and γ (AtpG) were less in filamented cells than in control cells. In contrast, the amount of OmpX was greater in filamented cells than in control cells. While five of the proteins were sequenced from samples of strain 4539H that had been grown on TSA or TSA-7NaCl, OmpX could be sequenced only from samples grown on TSA-7NaCl. Another protein not homologous to OmpX—identified as peptidoglycan-associated outer membrane lipoprotein—migrated similarly to OmpX in samples isolated from unstressed cells (Fig. 3B). OmpX was not the major constituent of this band, indicating that the fold difference in the amount of OmpX is likely greater than what was observed (Fig. 3B, Table 3).
Table 3.
Membrane proteins of S. enterica identified via MALDI-TOF mass spectrometry whose levels correlated with osmotic stress
| GI | Name | Description | E value | % of sequence covered | Change of steady-state levels (TSA-7NaCl:TSA) |
|||
|---|---|---|---|---|---|---|---|---|
| E40 | M-09 | S-12 | 4539H | |||||
| 16767149 | AtpD | F0F1-ATP synthase subunit beta | 1.2E−70 | 67 | 0.51 ± 0.20a | 0.36 ± 0.13b | 0.24 ± 0.26b | 0.37 ± 0.24b |
| 16767150 | AtpG | F0F1-ATP synthase subunit gamma | 4.0E−12 | 16 | 0.20 ± 0.09b | 0.14 ± 0.13b | 0.08 ± 0.07a | 0.34 ± 0.24b |
| 16422703 | EF-Tu | Elongation factor | 9.9E−09 | 28 | 0.57 ± 0.25a | 0.47 ± 0.20c | 0.21 ± 0.11b | 0.32 ± 0.30b |
| 16763793 | Tsx | Nucleoside channel, receptor of phage T6 and colicin K | 3.1E−31 | 54 | 0.28 ± 0.20c | 0.15 ± 0.12c | 0.22 ± 0.21c | 0.24 ± 0.21b |
| 16765076 | OmpW | Outer membrane protein | 3.1E−26 | 64 | 0.42 ± 0.23a | 0.26 ± 0.05b | 0.13 ± 0.01b | 0.18 ± 0.08b |
| 16764195 | OmpX | Outer membrane protein | 9.9E−32 | 58 | 3.99 ± 0.86c | 3.34 ± 1.43a | 3.38 ± 0.84c | 2.58 ± 1.37a |
P < 0.05.
P < 0.001.
P < 0.01.
The amount of both AtpD and AtpG in bacterial membranes decreased by 50 to 90% during osmotic stress (Fig. 3B, Table 3). We hypothesized that the decreased levels of these two proteins would negatively affect the amount of ATP in filaments compared to that in control cells and that this difference contributed to filamentation. To test these predictions, the levels of ATP in control and filamented populations of S. enterica were quantified (Fig. 3C). The levels of ATP in E40, M-09, S-12, and 4539H were 3-fold greater in filaments than in control cells when normalized to OD600 (Fig. 3Ci). S. enterica grown on TSA-7NaCl had 1.5 log10/CFU more ATP than cells grown on TSA (Fig. 3Cii). Also, respiration in filaments and control cells was observed microscopically by using CTC as an electron acceptor. When reduced, CTC forms a fluorescent formazan product (Fig. 3D), which was observed in both filaments and control cells. Therefore, electron transport was not inhibited in filaments, nor was the synthesis of ATP.
DISCUSSION
Filamentous growth of rod-shaped bacteria occurs in suboptimal conditions where division, but not growth, is inhibited. Different serovars of S. enterica filament during growth in hyperosmotic, acidic, alkali, and refrigerated environments (19, 31, 34–36). Likewise, both E. coli and L. monocytogenes filament in similar suboptimal conditions (19, 29, 36, 61).
Salmonellae generate filaments at aw values of 0.92 to 0.96 in medium supplemented with glycerol, sucrose, or NaCl (34, 36). Four strains of S. enterica representing different serovars were tested for filamentous growth on agar and in broth medium supplemented with different concentrations of NaCl (Fig. 1A). As the concentrations of NaCl within these media increased, growth decreased, and filaments were observed under conditions permissible for marginal growth. Independent of their concentration during inoculation, strains we examined filamented on TSA and in TSB supplemented with 6 to 8% NaCl (Fig. 1A and D and data not shown). Exponential-phase inocula did not grow or filament when medium was supplemented with 8% or more NaCl, while stationary-phase inocula both grew and filamented under these conditions. Stationary-phase cells did not grow or form filaments in TSB-10NaCl. Therefore, filaments form in hyperosmotic environments that support marginal growth but not septation.
Filamentous growth and RpoS.
It is known that exponential-phase cells are more sensitive to many types of stress than are stationary-phase cells (20). The difference we observed in cells in these two growth phases with regard to their capacity to filament in response to osmotic stress likely reflects the role of RpoS-regulated genes to enable cells to survive osmotic stress (37). Accordingly, LT2, a strain that poorly expresses RpoS (32), formed filaments in the presence of lower concentrations of NaCl than other strains tested (Fig. 1A). Our finding that RpoS is not directly required for filamentation is consistent with the findings of Mattick and colleagues (34).
Enumeration of viable units within filaments.
A multichromosomal filament and a single chromosomal control cell both form a single CFU on an agar plate. Yet filaments generated during osmotic stress are viable and can septate when incubated in medium without added humectants (Fig. 2A) (34). This issue must be considered in retrospective analyses of infectious dose and risk assessment of food-borne pathogens, like Salmonella. Populations of filamented and control cells had equal amounts of protein and chromosomal DNA when normalized by biomass and so cannot be used for the detection and enumeration of filaments (Table 1).
Enumerating viable units of filaments was achieved by incubating a population of filaments in 0.2LB broth for 8 h at 23°C. During incubation, filamentous salmonellae were able to septate and divide, while nonfilamentous salmonellae were not (Fig. 2Aii and iii). This assay was sensitive; when 23% of the cells in a population of S. Tennessee were filamented, CFU increased 0.4 log10/ml (Fig. 2B). The levels of ATP in filaments relative to control cells could in part explain how filaments were able to divide prior to control cells (Fig. 3C). Additionally, control cells had to fully replicate cellular constituents that filaments had already replicated.
DNA, protein, and levels of proteins in filaments.
Filamentous growth occurs by numerous mechanisms, such as when the replication of DNA is blocked with nalidixic acid or protein synthesis is repressed (17, 23, 27, 44, 52). Filaments of S. enterica generated on TSA-7NaCl yielded as much chromosomal DNA and protein as control cells when cell suspensions were normalized by OD600 (Table 1). Many proteins, including DnaK, in control and filamented populations were detected at similar levels (Fig. 3B and data not shown). The activities of numerous PBPs were similar in filamentous and control populations in vitro as assayed by their ability to bind a fluorescent derivative of penicillin G, BOCILLIN-FL (Fig. 3A, Table 2). Collectively, these findings indicate filaments are not likely caused by the repression of DNA or protein synthesis or the misfolding of PBPs.
Filamentous growth during osmotic stress likely does not require an intact SOS response. Chromosomal DNAs isolated from filaments and control cells were equally resistant to the exonuclease, BAL 31, indicating that chromosomal DNA retained its integrity in osmotically stressed S. enterica (data not shown). Also, ΔrecA E. coli serotype O157:H7 is capable of forming filaments on TSA-6NaCl (A. J. Stasic, C. W. Kaspar, and A. C. L. Wong, unpublished data).
Potential role of PBP2 in osmotic-induced filaments.
PBP2 was more active in membranes isolated from strains of S. enterica cultured on TSA-7NaCl than TSA (Fig. 3A). The increase in its activity was statistically significant for three of the four strains assayed (Table 2). Directed by the MreB cytoskeletal complex, PBP2 inserts peptidoglycan into the bacterial cell wall, resulting in elongation (16, 53, 54, 59). Mutations in pbpA, which encodes PBP2, renders E. coli sensitive to osmotic stress (8, 45). Therefore, PBP2 is a candidate for future investigation in the filamentous growth of S. enterica during osmotic stress.
Levels of ATP and subunits of the F0F1-ATP synthase.
S. enterica cultured on TSA-7NaCl contained 3-fold more ATP than salmonellae cultured on TSA when normalized by biomass (Fig. 3Ci). Yet, the amount of the F0F1-ATP synthase subunits β and γ were reduced in filamentous populations relative to control cells (Fig. 3B, Table 3). Both filaments and control cells were capable of oxidative phosphorylation, as measured by the reduction of CTC to a fluorescent formazan product. This assay was qualitative and therefore does not provide information into the rates of oxidative phosphorylation. Furthermore, it does not distinguish between aerobic and anaerobic electron acceptors. During osmotic shock, exponentially growing E. coli accumulates ATP via de novo synthesis, while respiration decreases by 90% (38, 42). Transcription of the genes encoding the F0F1-ATP synthase is negatively regulated when exponentially growing E. coli is osmotically shocked (48, 57).
ATP could contribute to the survival of S. enterica during osmotic stress and regulate the intracellular osmolarity of E. coli during osmotic shock, thereby protecting cells from plasmolysis (42). ATP could also contribute to the outgrowth of filaments (Fig. 2A). The elevated levels of ATP in filaments could indicate a reduction in the activity of ATP-dependent reactions. For example, inhibiting the ATPase activity of either FtsA or FtsE will induce filamentous growth in E. coli (6, 14, 17, 60).
Our findings show that during osmotic stress, S. enterica undergoes filamentous growth that is correlated with an increase in PBP2 and ATP levels and a decrease in levels of the F0F1-ATP synthase in the bacterial membrane. How PBP2 and metabolism contribute to these characteristics of osmotically induced filaments is the subject of ongoing research.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the USDA Agriculture and Food Research Initiative grant 2010-65201-20563 from the National Institute of Food and Agriculture awarded to C.W.K., A.C.L.W., and C.J.C. and the College of Agricultural and Life Sciences at the University of Wisconsin-Madison. B.C. was supported by a summer research fellowship from the Food Research Institute at the University of Wisconsin—Madison.
We thank David Weiss for kindly providing anti-PBP3 antisera, Andrew J. Stasic for sharing his unpublished work on the filamentatous growth of E. coli during osmotic stress, and the University of Wisconsin-Madison Mass Spectrometry facility for the identification of proteins.
Footnotes
Published ahead of print 13 July 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Addinall SG, Bi E, Lutkenhaus J. 1996. FtsZ ring formation in fts mutants. J. Bacteriol. 178:3877–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Addinall SG, Cao C, Lutkenhaus J. 1997. Temperature shift experiments with an ftsZ84(Ts) strain reveal rapid dynamics of FtsZ localization and indicate that the Z ring is required throughout septation and cannot reoccupy division sites once constriction has initiated. J. Bacteriol. 179:4277–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Addinall SG, Lutkenhaus J. 1996. FtsA is localized to the septum in an FtsZ-dependent manner. J. Bacteriol. 178:7167–7172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Begg KJ, Donachie WD. 1985. Cell shape and division in Escherichia coli: experiments with shape and division mutants. J. Bacteriol. 163:615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boberek JM, Stach J, Good L. 2010. Genetic evidence for inhibition of bacterial division protein FtsZ by berberine. PLoS One 5:e13745 doi:10.1371/journal.pone.0013745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bork P, Sander C, Valencia A. 1992. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. U. S. A. 89:7290–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Botta GA, Park JT. 1981. Evidence for involvement of penicillin-binding protein 3 in murein synthesis during septation but not during cell elongation. J. Bacteriol. 145:333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buchnik D, Woldringh CL, Zaritsky A. 1987. Effects of temperature inactivation of penicillin-binding protein 2 on envelope growth in Escherichia coli. Ann. Inst. Pasteur Microbiol. 138:537–547 [DOI] [PubMed] [Google Scholar]
- 9. Burdett ID, Murray RG. 1974. Septum formation in Escherichia coli: characterization of septal structure and the effects of antibiotics on cell division. J. Bacteriol. 119:303–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dai K, Lutkenhaus J. 1991. ftsZ is an essential cell division gene in Escherichia coli. J. Bacteriol. 173:3500–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Pedro MA, Young KD, Holtje JV, Schwarz H. 2003. Branching of Escherichia coli cells arises from multiple sites of inert peptidoglycan. J. Bacteriol. 185:1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Domadia P, Swarup S, Bhunia A, Sivaraman J, Dasgupta D. 2007. Inhibition of bacterial cell division protein FtsZ by cinnamaldehyde. Biochem. Pharmacol. 74:831–840 [DOI] [PubMed] [Google Scholar]
- 13. Domadia PN, Bhunia A, Sivaraman J, Swarup S, Dasgupta D. 2008. Berberine targets assembly of Escherichia coli cell division protein FtsZ. Biochemistry 47:3225–3234 [DOI] [PubMed] [Google Scholar]
- 14. Donachie WD, et al. 1979. Role of the ftsA gene product in control of Escherichia coli cell division. J. Bacteriol. 140:388–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dougherty TJ, Koller AE, Tomasz A. 1980. Penicillin-binding proteins of penicillin-susceptible and intrinsically resistant Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 18:730–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Figge RM, Divakaruni AV, Gober JW. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 51:1321–1332 [DOI] [PubMed] [Google Scholar]
- 17. Fujiwara K, Taguchi H. 2007. Filamentous morphology in GroE-depleted Escherichia coli induced by impaired folding of FtsE. J. Bacteriol. 189:5860–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gee KR, Kang HC, Meier TI, Zhao G, Blaszcak LC. 2001. Fluorescent bocillins: synthesis and application in the detection of penicillin-binding proteins. Electrophoresis 22:960–965 [DOI] [PubMed] [Google Scholar]
- 19. Hazeleger WC, Dalvoorde M, Beumer RR. 2006. Fluorescence microscopy of NaCl-stressed, elongated Salmonella and Listeria cells reveals the presence of septa in filaments. Int. J. Food Microbiol. 112:288–290 [DOI] [PubMed] [Google Scholar]
- 20. Hengge-Aronis R. 2000. The general stress response in Escherichia coli, p In 161–178 Storz G, Hengge-Aronis R. (ed), Bacterial stress responses, 1st ed. ASM Press, Washington, DC [Google Scholar]
- 21. Hirai K, Aoyama H, Irikura T, Iyobe S, Mitsuhashi S. 1986. Differences in susceptibility to quinolones of outer membrane mutants of Salmonella typhimurium and Escherichia coli. Antimicrob. Agents Chemother. 29:535–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hirota Y, Ryter A, Jacob F. 1968. Thermosensitive mutants of E. coli affected in the processes of DNA synthesis and cellular division. Cold Spring Harb. Symp. Quant. Biol. 33:677–693 [DOI] [PubMed] [Google Scholar]
- 23. Itikawa H, Ryu J. 1979. Isolation and characterization of a temperature-sensitive dnaK mutant of Escherichia coli B. J. Bacteriol. 138:339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Janion C. 2008. Inducible SOS response system of DNA repair and mutagenesis in Escherichia coli. Int. J. Biol. Sci. 4:338–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jeong KC, Hung KF, Baumler DJ, Byrd JJ, Kaspar CW. 2008. Acid stress damage of DNA is prevented by Dps binding in Escherichia coli O157:H7. BMC Microbiol. 8:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jimenez CR, Huang L, Qiu Y, Burlingame AL. 2001. In-gel digestion of proteins for MALDI-MS fingerprint mapping. Curr. Protoc. Protein Sci. Chapter 16:Unit 16.4 [DOI] [PubMed] [Google Scholar]
- 27. Kantor GJ, Deering RA. 1968. Effect of nalidixic acid and hydroxyurea on division ability of Escherichia coli fil+ and lon- strains. J. Bacteriol. 95:520–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Katayama H, Nagasu T, Oda Y. 2001. Improvement of in-gel digestion protocol for peptide mass fingerprinting by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 15:1416–1421 [DOI] [PubMed] [Google Scholar]
- 29. Kawarai T, et al. 2009. Biofilm formation by Escherichia coli in hypertonic sucrose media. J. Biosci. Bioeng. 107:630–635 [DOI] [PubMed] [Google Scholar]
- 30. Kawarai T, et al. 2004. SulA-independent filamentation of Escherichia coli during growth after release from high hydrostatic pressure treatment. Appl. Microbiol. Biotechnol. 64:255–262 [DOI] [PubMed] [Google Scholar]
- 31. Kieboom J, et al. 2006. Survival, elongation, and elevated tolerance of Salmonella enterica serovar Enteritidis at reduced water activity. J. Food Prot. 69:2681–2686 [DOI] [PubMed] [Google Scholar]
- 32. Lee IS, Lin J, Hall HK, Bearson B, Foster JW. 1995. The stationary-phase sigma factor sigma S (RpoS) is required for a sustained acid tolerance response in virulent Salmonella typhimurium. Mol. Microbiol. 17:155–167 [DOI] [PubMed] [Google Scholar]
- 33. Lutkenhaus J, Addinall SG. 1997. Bacterial cell division and the Z ring. Annu. Rev. Biochem. 66:93–116 [DOI] [PubMed] [Google Scholar]
- 34. Mattick KL, et al. 2000. Survival and filamentation of Salmonella enterica serovar enteritidis PT4 and Salmonella enterica serovar Typhimurium DT104 at low water activity. Appl. Environ. Microbiol. 66:1274–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mattick KL, Phillips LE, Jorgensen F, Lappin-Scott HM, Humphrey TJ. 2003. Filament formation by Salmonella spp. inoculated into liquid food matrices at refrigeration temperatures, and growth patterns when warmed. J. Food Prot. 66:215–219 [DOI] [PubMed] [Google Scholar]
- 36. Mattick KL, Rowbury RJ, Humphrey TJ. 2003. Morphological changes to Escherichia coli O157:H7, commensal E. coli and Salmonella spp. in response to marginal growth conditions, with special reference to mildly stressing temperatures. Sci. Prog. 86:103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McMeechan A, et al. 2007. Role of the alternative sigma factors sigmaE and sigmaS in survival of Salmonella enterica serovar Typhimurium during starvation, refrigeration and osmotic shock. Microbiology 153(Pt 1): 263–269 [DOI] [PubMed] [Google Scholar]
- 38. Meury J. 1994. Immediate and transient inhibition of the respiration of Escherichia coli under hyperosmotic shock. FEMS Microbiol. Lett. 121:281–286 [DOI] [PubMed] [Google Scholar]
- 39. Mukherjee A, Cao C, Lutkenhaus J. 1998. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 95:2885–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nelson DE, Ghosh AS, Paulson AL, Young KD. 2002. Contribution of membrane-binding and enzymatic domains of penicillin binding protein 5 to maintenance of uniform cellular morphology of Escherichia coli. J. Bacteriol. 184:3630–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nilsen T, Ghosh AS, Goldberg MB, Young KD. 2004. Branching sites and morphological abnormalities behave as ectopic poles in shape-defective Escherichia coli. Mol. Microbiol. 52:1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ohwada T, Sagisaka S. 1987. An immediate and steep increase in ATP concentration in response to reduced turgor pressure in Escherichia coli B. Arch. Biochem. Biophys. 259:157–163 [DOI] [PubMed] [Google Scholar]
- 43. Ozeki H, Howarth S. 1961. Colicine factors as fertility factors in bacteria: Salmonella typhimurium strain LT2. Nature 190:986–988 [DOI] [PubMed] [Google Scholar]
- 44. Paek KH, Walker GC. 1987. Escherichia coli dnaK null mutants are inviable at high temperature. J. Bacteriol. 169:283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Philippe N, Pelosi L, Lenski RE, Schneider D. 2009. Evolution of penicillin-binding protein 2 concentration and cell shape during a long-term experiment with Escherichia coli. J. Bacteriol. 191:909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pogliano J, Pogliano K, Weiss DS, Losick R, Beckwith J. 1997. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc. Natl. Acad. Sci. U. S. A. 94:559–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Popham DL, Young KD. 2003. Role of penicillin-binding proteins in bacterial cell morphogenesis. Curr. Opin. Microbiol. 6:594–599 [DOI] [PubMed] [Google Scholar]
- 48. Shabala L, et al. 2009. Ion transport and osmotic adjustment in Escherichia coli in response to ionic and non-ionic osmotica. Environ. Microbiol. 11:137–148 [DOI] [PubMed] [Google Scholar]
- 49. Sieracki ME, Cucci TL, Nicinski J. 1999. Flow cytometric analysis of 5-cyano-2,3-ditolyl tetrazolium chloride activity of marine bacterioplankton in dilution cultures. Appl. Environ. Microbiol. 65:2409–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Spratt BG. 1977. Properties of the penicillin-binding proteins of Escherichia coli K-12. Eur. J. Biochem. 72:341–352 [DOI] [PubMed] [Google Scholar]
- 51. Stackhouse RR, Faith NG, Kaspar CW, Czuprynski CJ, Wong AC. 2012. Survival and virulence of Salmonella enterica serovar enteritidis filaments induced by reduced water activity. Appl. Environ. Microbiol. 78:2213–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sugino A, Peebles CL, Kreuzer KN, Cozzarelli NR. 1977. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. U. S. A. 74:4767–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vats P, Shih YL, Rothfield L. 2009. Assembly of the MreB-associated cytoskeletal ring of Escherichia coli. Mol. Microbiol. 72:170–182 [DOI] [PubMed] [Google Scholar]
- 54. Vinella D, D'Ari R, Jaffe A, Bouloc P. 1992. Penicillin binding protein 2 is dispensable in Escherichia coli when ppGpp synthesis is induced. EMBO J. 11:1493–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Walker GC, Smith BT, Sutton MD. 2000. The SOS response to DNA damage, p In 131–144 Storz G, Hengge-Aronis R. (ed), Bacterial stress responses, 1st ed. ASM Press, Washington, DC. [Google Scholar]
- 56. Walker JR, Kovarik A, Allen JS, Gustafson RA. 1975. Regulation of bacterial cell division: temperature-sensitive mutants of Escherichia coli that are defective in septum formation. J. Bacteriol. 123:693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Weber A, Jung K. 2002. Profiling early osmostress-dependent gene expression in Escherichia coli using DNA macroarrays. J. Bacteriol. 184:5502–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Weiss DS, et al. 1997. Localization of the Escherichia coli cell division protein Ftsl (PBP3) to the division site and cell pole. Mol. Microbiol. 25:671–681 [DOI] [PubMed] [Google Scholar]
- 59. Wientjes FB, Nanninga N. 1991. On the role of the high molecular weight penicillin-binding proteins in the cell cycle of Escherichia coli. Res. Microbiol. 142:333–344 [DOI] [PubMed] [Google Scholar]
- 60. Yang DC, et al. 2011. An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc. Natl. Acad. Sci. U. S. A. 108:E1052–E1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yoshida S, Udou T, Mizuguchi Y, Tanabe T. 1986. Salt-induced filamentous growth of a Salmonella strain isolated from blood. J. Clin. Microbiol. 23:192–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zak O, Kradolfer F. 1979. Effects of subminimal inhibitory concentrations of antibiotics in experimental infections. Rev. Infect. Dis. 1:862–879 [DOI] [PubMed] [Google Scholar]
- 63. Zhao G, Meier TI, Kahl SD, Gee KR, Blaszczak LC. 1999. BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob. Agents Chemother. 43:1124–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.