Abstract
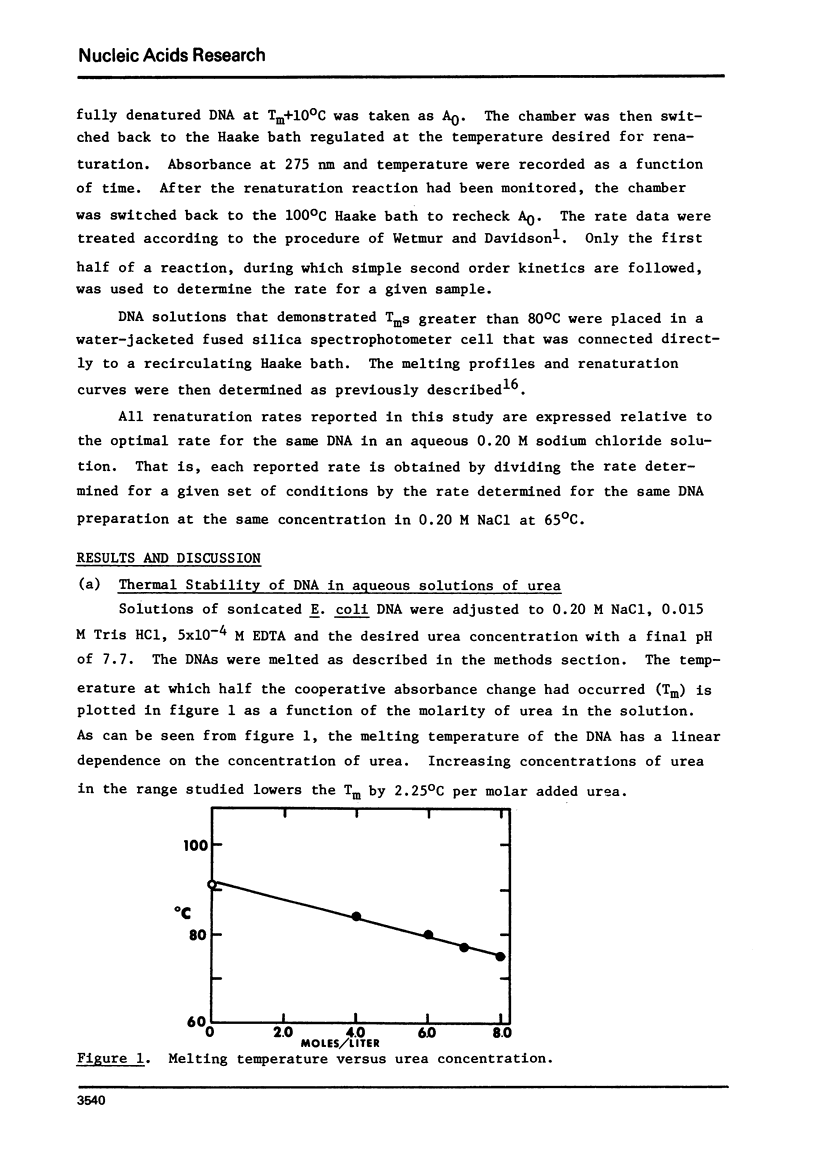
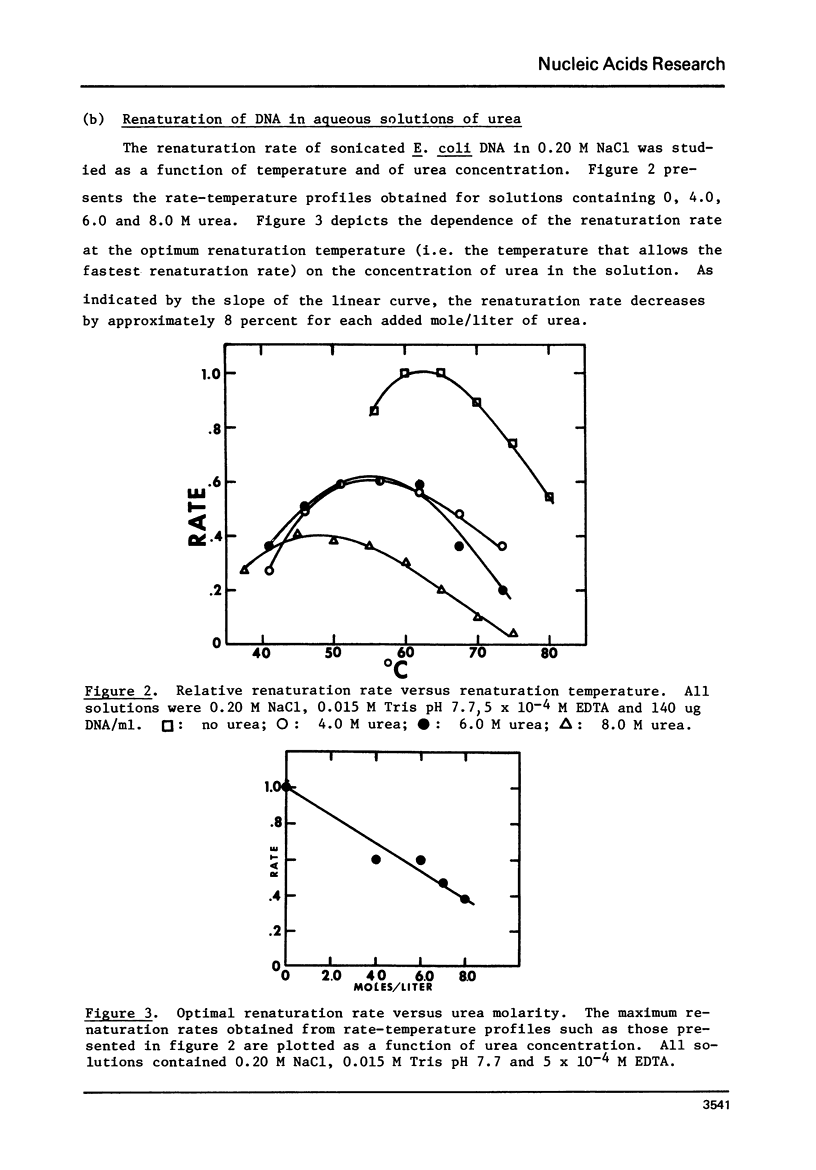
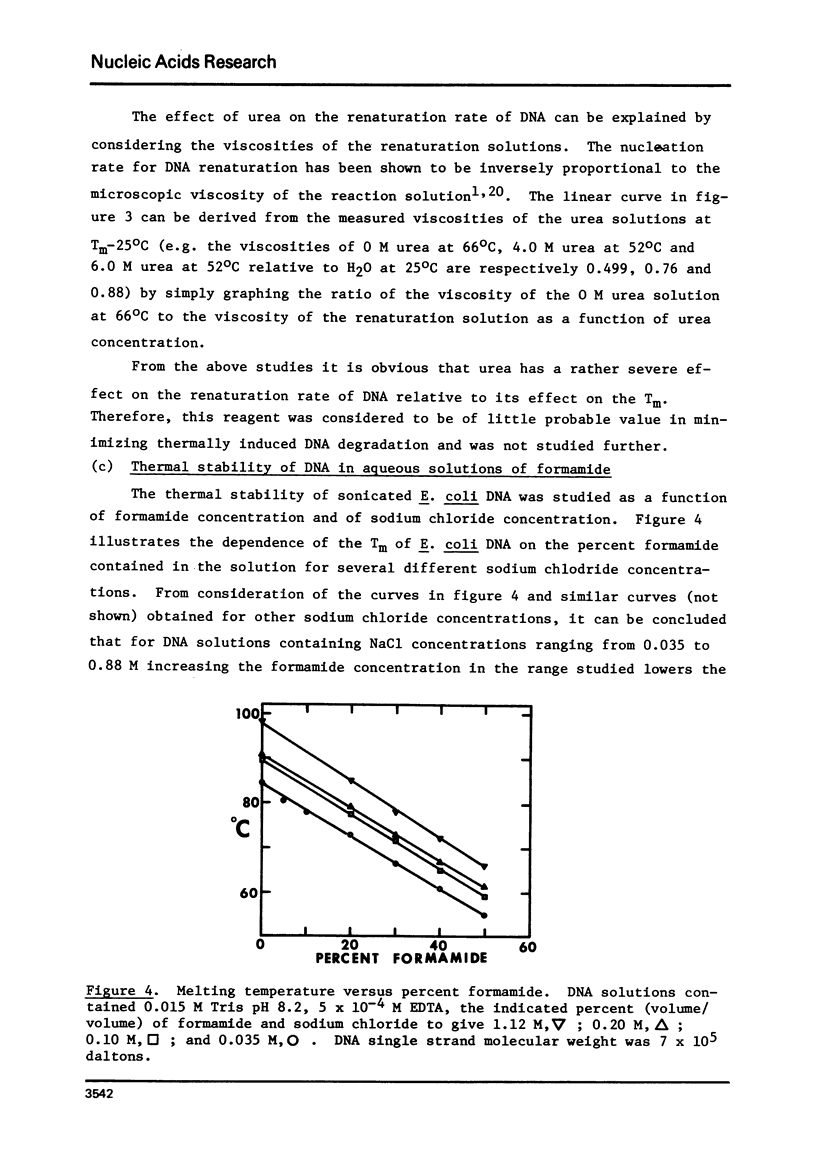
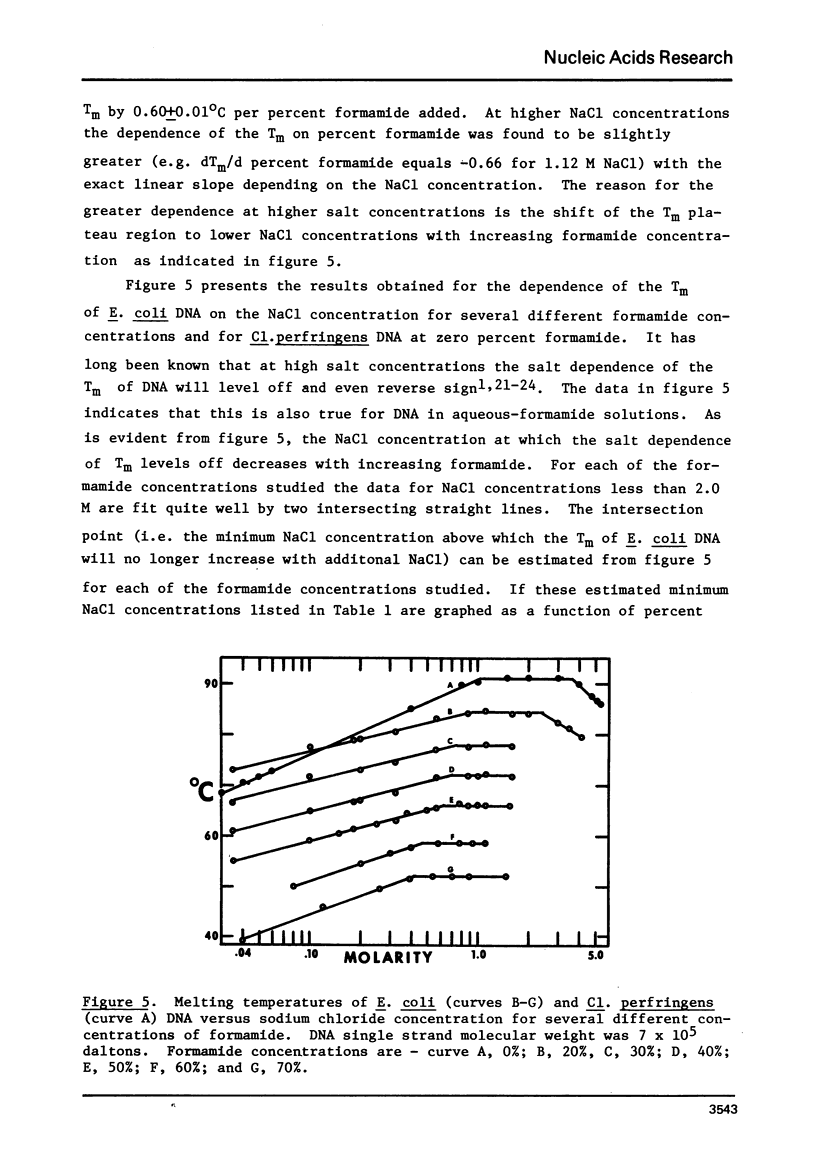
This paper reports the results of a systematic study of the effects of formamide and urea on the thermal stability and renaturation kinetics of DNA. Increasing concentrations of urea in the range 0 to 8 molar lower the Tm by 2.25 degrees C per molar, and decreases the renaturation rate by approximately 8 percent per molar. Increasing concentrations of formamide in the range from 0 to 50 percent lowers the Tm by 0.60 degrees C per percent formamide for sodium chloride concentrations ranging from 0.035M to 0.88M. At higher salt concentrations the dependence of Tm on percent formamide was found to be slightly greater. Increasing formamide concentration decreases the renaturation rate linearly by 1.1% per percent formamide such that the optimal rate in 50% formamide is 0.45 the optimal rate in an identical solution with no formamide. The effects of urea and formamide on the renaturation rates of DNA are explained by consideration of the viscosities of the solutions at the renaturation temperatures.
Full text
PDF





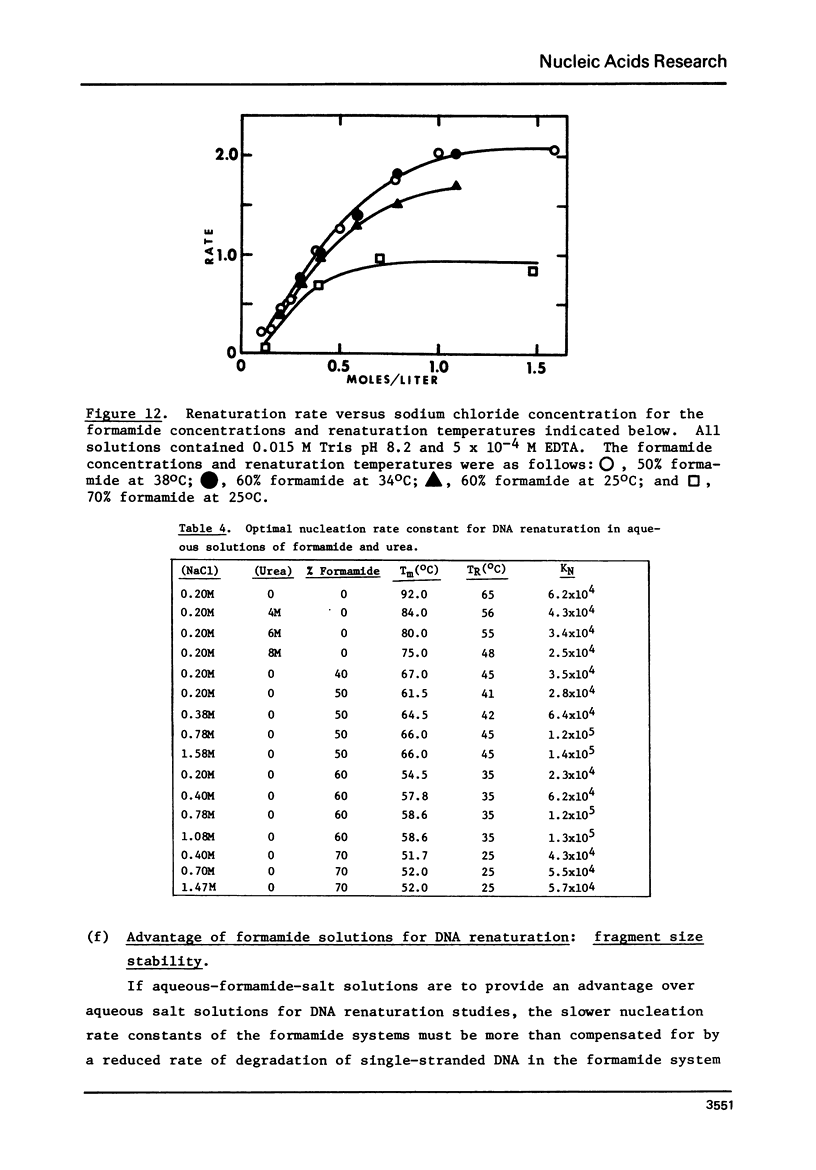

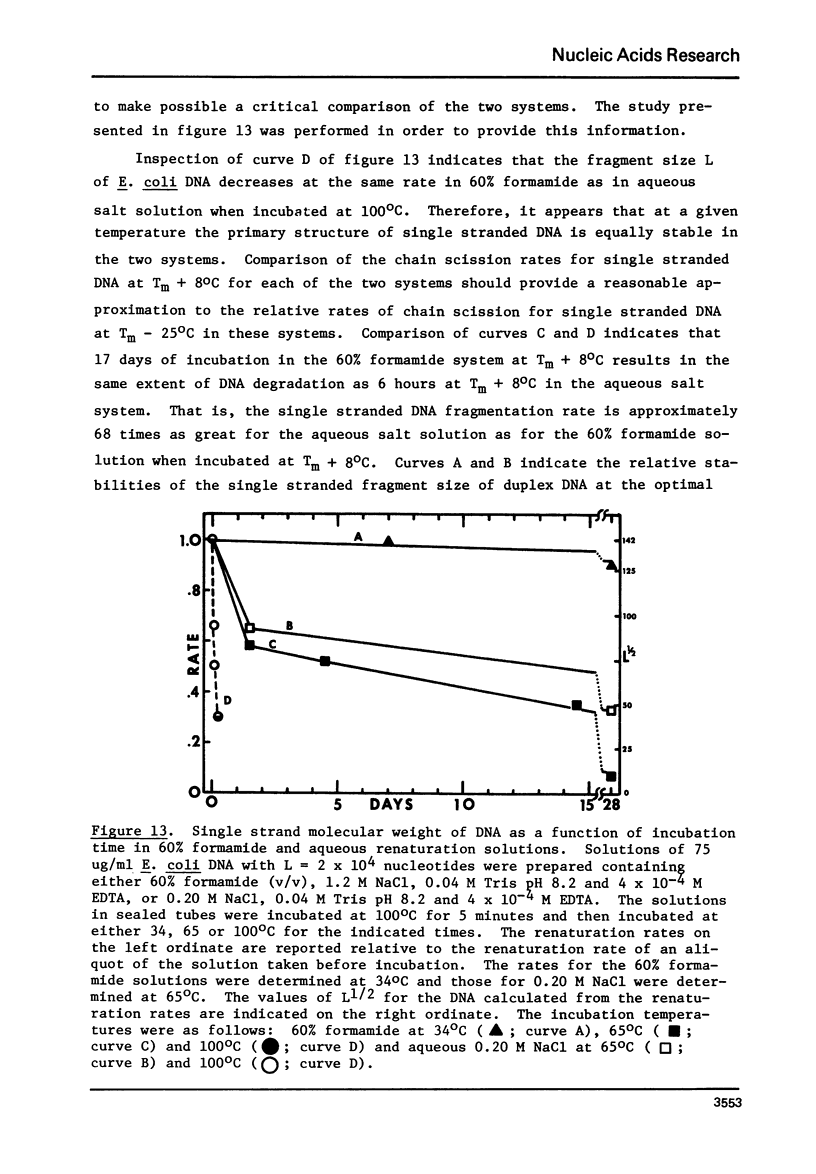


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blüthmann H., Brück D., Hübner L., Schöffski A. Reassociation of nucleic acids in solutions containing formamide. Biochem Biophys Res Commun. 1973 Jan 4;50(1):91–97. doi: 10.1016/0006-291x(73)91068-1. [DOI] [PubMed] [Google Scholar]
- Chang C. T., Hain T. C., Hutton J. R., Wetmur J. G. Effects of microscopic and macroscopic viscosity on the rate of renaturation of DNA. Biopolymers. 1974;13(9):1847–1858. doi: 10.1002/bip.1974.360130915. [DOI] [PubMed] [Google Scholar]
- Elson E. L., Record M. T., Jr The strand-separation transition of T2 bacteriophage DNA. Biopolymers. 1974 Apr;13(4):797–824. doi: 10.1002/bip.1974.360130415. [DOI] [PubMed] [Google Scholar]
- Gillis M., De Ley J., De Cleene M. The determination of molecular weight of bacterial genome DNA from renaturation rates. Eur J Biochem. 1970 Jan;12(1):143–153. doi: 10.1111/j.1432-1033.1970.tb00831.x. [DOI] [PubMed] [Google Scholar]
- Gruenwedel D. W., Hsu C. H., Lu D. S. The effects of aqueous neutral-salt solutions on the melting temperatures of deoxyribonucleic acids. Biopolymers. 1971;10(1):47–68. doi: 10.1002/bip.360100106. [DOI] [PubMed] [Google Scholar]
- Hutton J. R., Wetmur J. G. Activity of endonuclease S1 in denaturing solvents: dimethysulfoxide, dimethylformamide, formamide and formaldehyde. Biochem Biophys Res Commun. 1975 Oct 6;66(3):942–948. doi: 10.1016/0006-291x(75)90731-7. [DOI] [PubMed] [Google Scholar]
- Hutton J. R., Wetmur J. G. Effect of chemical modification on the rate of renaturation of deoxyribonucleic acid. Deaminated and glyoxalated deoxyribonucleic acid. Biochemistry. 1973 Jan 30;12(3):558–563. doi: 10.1021/bi00727a032. [DOI] [PubMed] [Google Scholar]
- Hutton J. R., Wetmur J. G. Renaturation of bacteriophage phiX174 DNA-RNA hybrid: RNA length effect and nucleation rate constant. J Mol Biol. 1973 Jul 15;77(4):495–500. doi: 10.1016/0022-2836(73)90218-0. [DOI] [PubMed] [Google Scholar]
- Kallay M., Record M. T., Jr The strand-separation transition of T7 DNA. Biopolymers. 1974 Apr;13(4):825–841. doi: 10.1002/bip.1974.360130416. [DOI] [PubMed] [Google Scholar]
- Kingsbury D. T. Estimate of the genome size of various microorganisms. J Bacteriol. 1969 Jun;98(3):1400–1401. doi: 10.1128/jb.98.3.1400-1401.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Andersson A. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry. 1972 Sep 12;11(19):3618–3623. doi: 10.1021/bi00769a019. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972 Sep 12;11(19):3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- MANDELL J. D., HERSHEY A. D. A fractionating column for analysis of nucleic acids. Anal Biochem. 1960 Jun;1:66–77. doi: 10.1016/0003-2697(60)90020-8. [DOI] [PubMed] [Google Scholar]
- McConaughy B. L., Laird C. D., McCarthy B. J. Nucleic acid reassociation in formamide. Biochemistry. 1969 Aug;8(8):3289–3295. doi: 10.1021/bi00836a024. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr Electrostatic effects on polynucleotide transitions. I. Behavior at neutral pH. Biopolymers. 1967;5(10):975–992. doi: 10.1002/bip.1967.360051010. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr Electrostatic effects on polynucleotide transitions. II. Behavior of titrated systems. Biopolymers. 1967;5(10):993–1008. doi: 10.1002/bip.1967.360051011. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Woodbury C. P., Lohman T. M. Na+ effects on transition of DNA and polynucleotides of variable linear charge density. Biopolymers. 1976 May;15(5):893–915. doi: 10.1002/bip.1976.360150507. [DOI] [PubMed] [Google Scholar]
- Record M. T. Kinetics of the helix-coil transition in DNA. Biopolymers. 1972;11(7):1435–1484. doi: 10.1002/bip.1972.360110711. [DOI] [PubMed] [Google Scholar]
- Schildkraut C. Dependence of the melting temperature of DNA on salt concentration. Biopolymers. 1965;3(2):195–208. doi: 10.1002/bip.360030207. [DOI] [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- Wetmur J. G. Hybridization and renaturation kinetics of nucleic acids. Annu Rev Biophys Bioeng. 1976;5:337–361. doi: 10.1146/annurev.bb.05.060176.002005. [DOI] [PubMed] [Google Scholar]


