Abstract
The ability to predictably engineer the composition of bowel microbial communities (microbiota) using dietary components is important because of the reported associations of altered microbiota composition with medical conditions. In a synecological study, weanling conventional Sprague-Dawley rats (21 days old) were fed a basal diet (BD) or a diet supplemented with resistant starch (RS) at 5%, 2.5%, or 1.25% for 28 days. Pyrosequencing of 16S rRNA genes and temporal temperature gradient electrophoresis (TTGE) profiles in the colonic digesta showed that rats fed RS had altered microbiota compositions due to blooms of Bacteroidetes and Actinobacteria. The altered microbiota was associated with changes in colonic short-chain fatty acid (SCFA) concentrations, colonic-tissue gene expression (Gsta2 and Ela1), and host physiology (serum metabolite profiles and colonic goblet cell numbers). Comparisons between germ-free and conventional rats showed that transcriptional and serum metabolite differences were mediated by the microbiota and were not the direct result of diet composition. Altered transcriptomic and physiological responses may reflect the young host's attempts to maintain homeostasis as a consequence of exposure to a new collection of bacteria and their associated biochemistry.
INTRODUCTION
Collectively, the complex microbial communities (microbiota) of the large bowel in vertebrates have a diverse and influential metabolic capacity that enables mammalian hosts to extract energy from substrates that they would otherwise lack the ability to utilize (8, 20). Maize-derived resistant starch (RS) is a complex polysaccharide that withstands degradation and absorption in the small bowel and passes to the large bowel, where it is hydrolyzed by members of the microbiota. Maize resistant starch is commonly used as an ingredient in bread marketed as “high in fiber.” Diets rich in fermentable substrates may affect the host by enriching for bacteria that produce short-chain fatty acids (SCFA) as by-products of hydrolysis and fermentation (23, 28). Enrichment of certain microbiota community members might be important for human health, as changes in the composition of large-bowel communities have been reported in relation to allergies, inflammatory bowel diseases, obesity, and metabolic syndrome (9, 12, 36, 38).
While an extensive literature about the manipulation of the composition of the bowel microbiota already exists, experiments using diets supplemented with RS have focused on adult subjects (39). In light of the resilience against long-term diet-induced changes in the microbiota of adult mice (1, 45), it is probably more relevant to study the effect of dietary composition on the development of the bowel microbiota of weanlings. Manipulation of the early microbiota may provide a window of opportunity for long-lasting effects on community composition. For example, reduced diversity in the fecal microbiota composition of infants has been associated with an increased incidence of atopic eczema in later life (42). The bowel ecology of human infants becomes increasingly complex after weaning as new bacterial species become established and the proportions of the different bacterial populations change radically (18, 32). These changes doubtless reflect the changed large-bowel milieu in which milk oligosaccharides are replaced by complex polysaccharides of plant origin as major contributors to bacterial nutrition postweaning (5, 46).
We postulated that RS in the diet of weanlings might enrich for a microbiota with increased fermentative capacity, which in turn could drive host tissue responses. Molecular cross talk between the microbiota, shaped by dietary manipulation, and the host could have impacts on host physiology and predisposition to medical conditions. Therefore, we examined whether feeding RS to conventional weanling rats significantly altered the colonic microbiota composition and whether such changes were associated with altered host physiology in terms of gene expression, colonic mucosal architecture, and serum metabolite profiles.
We report here that the colonic microbiota of rats can be modulated by feeding RS from weaning but, most importantly, that the differentiated microbiota produces distinct mucosal transcriptomes and host physiological changes.
MATERIALS AND METHODS
Animals and diets.
At weaning, 21-day-old conventionally raised male Sprague Dawley rats were individually housed in hanging wire mesh cages. Twenty-one- to 28-day-old germ-free male Sprague-Dawley rats, obtained from Taconic, Germantown, NY, were housed individually in standard rat cages within gnotobiotic isolators. Animal experiments were conducted with the approval of the AgResearch Grasslands Animal Ethics Committee (Palmerston North, New Zealand). The rats were kept under strict 12-h light cycles. Food and water were provided ad libitum and monitored weekly. Germ-free rats were fed irradiated diets and autoclaved water and were weighed weekly. All rats were fed a lactic-casein-based basal diet (BD) or the basal diet supplemented with RS, the compositions of which are described in Table 1. The resistant starch used in this study was Hi-maize 1043 (National Starch and Chemical Company, Bridgewater, NJ), a high-amylose maize RS2-type resistant starch. Diets containing RS were supplemented with 5%, 2.5%, or 1.25% RS by weight. Diets for germ-free rats were sterilized by gamma irradiation at 25 kGy at Schering-Plough Animal Health Ltd. (Upper Hutt, New Zealand). Food intake was measured weekly. Conventional rats were randomly assigned to 1 of 4 groups (BD, RS 5%, RS 2.5%, or RS 1.25%; n = 10 per group), while germ-free rats were placed into 1 of 2 dietary groups (BD or RS 5%; n = 6 per group). After 28 days on the dietary treatment, the rats were euthanized by carbon dioxide overdose, and colon tissue and digesta were collected. Tissue for histology was fixed in formaldehyde. Samples of colon tissue for transcription analysis and digesta for microbiota and SCFA analysis were snap-frozen in liquid nitrogen and stored at −85°C. Weight gain and diet intake results were analyzed using repeated-measures analysis of variance (ANOVA) in R 2.14.1 (R Foundation for Statistical Computing, Vienna, Austria).
Table 1.
Experimental diet compositions
| Component | Amt (g/kg) |
|||
|---|---|---|---|---|
| BD | Resistant starch diet |
|||
| 5% (RS 5) | 2.5% (RS 2.5) | 1.25% (RS 1.25) | ||
| Lactic casein | 120 | 120 | 120 | 120 |
| Vitamin mixa | 50 | 50 | 50 | 50 |
| Mineral mixb | 50 | 50 | 50 | 50 |
| Corn oil | 65 | 65 | 65 | 65 |
| Corn starch | 650 | 600 | 625 | 612.5 |
| Sucrose | 40 | 40 | 40 | 40 |
| Cellulose | 25 | 25 | 25 | 25 |
| RSc | 50 | 25 | 12.5 | |
Vitamin mixture containing the following components (mg/kg diet): retinol acetate, 5.0; dl-α-tocopheryl acetate, 100.0; menadione, 3.0; thiamine hydrochloride, 5.0; riboflavin, 7.0; pyridoxine hydrochloride, 8.0; d-pantothenic acid, 20.0; folic acid, 2.0; nicotinic acid, 20.0; d-biotin, 1.0; myoinositol, 200.0; choline chloride, 1,500.0; ergocalciferol, 25.0 μg/kg diet; cyanocobalamin, 50.0 μg/kg diet.
Mineral mixture contains the following components: Ca, 6.29 g/kg diet; Cl, 7.79 g/kg diet; Mg, 1.06 g/kg diet; P, 4.86 g/kg diet; K, 5.24 g/kg diet; Na, 1.97 g/kg diet; Cr, 1.97 mg/kg diet; Cu, 10.7 mg/kg diet; Fe, 424.0 mg/kg diet; Mn, 78.0 mg/kg diet; Zn, 48.2 mg/kg diet; Co, 29.0 μg/kg diet; I, 151.0 μg/kg diet; Mo, 152.0 μg/kg diet; Se, 151.0 μg/kg diet.
Hi-maize 1043; National Starch and Chemical Company, Bridgewater, NJ.
Histology.
Formalin-fixed transverse sections of the colon from the central position and 1 cm from the start and end positions were stained with hematoxylin and eosin and counterstained with alcian blue. Histological measurements were performed using bright-field microscopy at ×200 magnification and Image-Pro Plus 4.0 (MediaCybernetics, Bethesda, MD). Crypt depths were determined by measuring an average of 80 random fully longitudinally sectioned crypts from the base of the crypt to the flat margin of the colon mucosa in three colon sections per rat. Similarly, goblet cells were counted in an average of 30 random, fully longitudinally sectioned crypts per rat. Histological measurements were analyzed using two-factor ANOVA in R.2.14.1.
SCFA concentrations.
Acids derivatized with N-methyl-N-t-butyldimethylsilytrifluoracetamide were measured in colon digesta using the capillary gas chromatography (GC) method described by Jensen et al. (15), with the following modifications. Colon digesta were homogenized by vortexing with 0.1 g of glass beads (0.3-mm diameter) in 8 ml of homogenization medium (0.9% [wt/vol] NaCl, 0.1% [vol/vol] Tween 20) and 1 ml internal standard (100 mM 2-ethyl butyric acid) per gram of colon digesta. GC separation of SCFA was carried out using a flame ionization detector in a Shimadzu GC-17A chromatograph equipped with an Agilent HP-1 methyl silicone gum column (Agilent Technologies, Santa Clara, CA) with a helium carrier gas at 10 kPa. The column temperature at the time of injection was 70°C, which was then increased to 80°C at 10°C per minute, followed by an increase to 260°C at 20°C per minute. Detector and injector temperatures were both set to 260°C. Short-chain fatty acid concentrations were analyzed by ANOVA in R.2.14.1.
TTGE.
Total RNA from colon digesta was extracted with Qiagen RNA/DNA minikits (Qiagen, Valencia, CA) using the method described by Tannock et al. (33). Temporal temperature gradient electrophoresis (TTGE) profiles were generated from 16S rRNA sequences to calculate profiles of the putatively more metabolically active members of the microbiota (33). Amplification of the V3 region of the 16S rRNA was carried out by reverse transcription (RT)-PCR with bacterial primers HDA1-GC (5′-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAC TCC TAC GGG AGG CAG CAG T-3′) and HDA2 (5′-GTA TTA CCG CGG CTG CTG GCA C-3′) using a previously described program (33). Generation of TTGE profiles from RT-PCR products was carried out using a DCode universal mutation detection system apparatus (Bio-Rad, Hercules, CA) in a 6% polyacrylamide gel with a previously described method (2). Comparison of TTGE profiles using Dice's similarity coefficients and classical multidimensional scaling was achieved using Gelcompar II (Applied Maths, Sint-Martens-Latem, Belgium) and R 2.14.1.
16S rRNA gene sequence analysis.
Bacterial DNA was extracted from colon digesta as described previously (40) and purified using Qiagen RNA/DNA minikits. DNA from colon digesta of individual rats was used as the template to amplify 16S rRNA gene sequences by PCR, using primers with linked adapters for standards for GS FLX Titanium (Roche, Basel, Switzerland) sequencing as follows: Roche adapter A, 10-bp barcode linker primer (CTATGCGCCTTGCCAGCCCGCTCAGNNNNNNNNNNGTATTACCGCGGCTGCTGGCAC), and Roche adapter B, reverse primer (CGTATCGCCTCCCTCGCGCCATCAGGRGTTYGATYMTGGCTCAG). Amplification was carried out using the following cycling protocol: 95°C for 3 min, followed by 30 cycles (95°C for 30 s, 57°C for 1 min, and 72°C for 1 min), and 72°C for 4 min. Amplicons were cleaned using Qiagen PCR cleanup columns and were quantified by Nanodrop (Thermo Fisher). PCR products were sequenced from the linker primer (adapter A) using a GS FLX Titanium sequencer (Roche) at Macrogen, Inc. (Seoul, South Korea). Sequences were processed using QIIME v1.3 (3). Identification of sequences (average length, 522 bp) was performed using the RDP Classifier to assign 16S rRNA gene sequences to the RDP taxonomical hierarchy using an 80% confidence threshold (43). Phylogenetic analysis of sequences was performed by aligning representative sequences clustered at 97% similarity using QIIME. Alpha and beta diversities were calculated using QIIME from the generated phylogenetic tree and table of operational taxonomic units (OTUs) assigned to the different samples. Differences in taxon abundance were analyzed by ANOVA in R 2.14.1.
Transcription analysis of colon tissue.
Total RNA for microarray analysis was prepared from colon tissue using the extraction method described by Knoch et al. (17). RNA quality was assessed using RNA 6000 Nano Labchip kits with an Agilent 2100 Bioanalyzer (Agilent Technologies). Only samples with an RNA integrity number of >8.0 were used for microarray hybridization. RNA from a single rat was cohybridized to individual arrays with reference RNA prepared by pooling equal amounts of RNA extracted from the colon tissues of 10 rats from the BD group. RNAs from individual rats fed a BD or RS 5% diet were analyzed (n = 6). Cy3-labeled cRNA probes, synthesized from sample RNA, and Cy5-labeled probes, synthesized from reference RNA, were prepared with Low RNA Input Linear Amplification kits (Agilent Technologies) and hybridized to 4x44K Agilent Whole Rat Genome Oligo Microarrays (Agilent Technologies; G4131F) using previously described methods (17). The microarrays were scanned with an Agilent DNA Microarray Scanner G2565CA and Agilent Feature Extraction 9.0 Image Analysis software (Agilent Technologies). Differentially expressed genes were determined using R 2.14.1 and Bioconductor (10) with the limma package (31). Intensity ratios for all microarray spots were normalized using a global loess algorithm. Genes with a greater than 1.5-fold change between comparisons and Benjamini and Hochberg false-discovery rate (FDR) adjusted P values of <0.05 were considered to be differentially expressed.
RT-qPCR.
Total RNA was reverse transcribed using Applied Biosystems High Capacity RNA-to-cDNA kits (Applied Biosystems Inc., Foster City, CA). A transcription mixture consisting of 10 μl of 2× RT buffer, 1 μl of 20× RT enzyme mixture, 2 μg of RNA, and H2O up to a total volume of 20 μl was incubated at 37°C for 60 min, followed by 95°C for 5 min. RT-quantitative PCR (qPCR) was performed on a Rotor-Gene 6000 thermocycler (Qiagen) using predesigned and prevalidated Applied Biosystems TaqMan Gene Expression Assays (Applied Biosystems Inc.). Each reaction mixture consisted of 10 μl of 2× TaqMan Gene Expression Master Mix, 1 μl of cDNA template, 1 μl of TaqMan Gene Expression Assay, and 8 μl of nuclease-free water. The reactions were carried out in quadruplicate using the following program: 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. Expression of Gsta2 and Ela1 in individual rats, normalized against expression of the ubiquitin A-52 housekeeping gene (Uba52), was analyzed by two-factor ANOVA in R 2.14.1.
LC-MS metabolomic analysis.
Untargeted analysis of serum metabolite profiles was carried out using a previously described method (25). Blood samples were collected from rats by cardiac puncture without an anticoagulant. The samples were then left at room temperature for 2 h, after which the sera were separated by centrifugation at 1,500 × g for 10 min. Serum samples were prepared for liquid chromatography-mass spectrometry (LC-MS) analysis by mixing 100 ml of serum with 200 ml of acetonitrile. The samples were analyzed in a Thermo LTQ linear ion-trap mass spectrometer (Thermo Electron Corporation, San Jose, CA) using negative electrospray ionization with the ion trap programmed to collect an MS1 spectrum from 100 to 1,000 m/z. The capillary temperature was 275°C, and the source ionization voltage was −4,000 V. Raw LC-MS data were analyzed using the R package xcms (30). Peaks were grouped together across samples using overlapping m/z bins, and peak retention times were corrected using loess normalization. Differentially abundant ions were determined using Welch's two-sample t statistic, and multivariate analysis was performed using principal-component analysis (PCA) in R 2.14.1. Procrustes rotation analysis comparing metabolite profiles with microbiota composition and gene expression was performed using the R 2.14.1 and the ade4 packages (35).
Microarray data accession number.
Microarray data were uploaded to the Gene Expression Omnibus (GEO) database (accession number GSE26108).
RESULTS
Food intake and weight gain.
Food consumption and weight gains by conventional and germ-free rats after 28 days of feeding the BD or RS diets are shown in Table 2. Dietary supplementation with RS did not adversely affect the rats. Animals in all of the dietary groups consumed similar amounts of food and gained similar amounts of weight. However, germ-free rats ate more food but gained less weight than conventional rats fed the same diets.
Table 2.
Body weight gain and food intake in weanling rats after 28 days of feeding
| Rata | Diet | Wt gain (%)b | Total food intake (g)c |
|---|---|---|---|
| CR | BD | 415 ± 16 | 356 ± 11 |
| RS 5% | 392 ± 11 | 360 ± 13 | |
| RS 2.5% | 391 ± 14 | 337 ± 10 | |
| RS 1.25% | 404 ± 11 | 354 ± 7 | |
| GF | BD | 208 ± 8 | 623 ± 13 |
| RS 5% | 215 ± 5 | 677 ± 24 |
CR, conventionally raised; GF, germ free.
Mean body weight gain after 28 days of feeding compared to the start of the study in CR and GF rats fed the BD and RS diets (±SEM); n = 10 (CR) and n = 6 (GF).
Mean total food intake after 28 days of feeding (±SEM). No significant differences in body weight were observed between diets within CR rats or within GF rats.
RS dose response effects on colonic SCFA concentrations.
To investigate the effects of the RS dose on colonic fermentation, conventional rats were fed diets containing RS at a concentration of 5%, 2.5%, 1.25%, or 0% (BD). The colon digesta of rats fed diets supplemented with 5% RS had higher concentrations of acetic, propionic, butyric, lactic, and succinic acid than BD-fed rats (P < 0.05) (Table 3). Moreover, linear regression analysis showed that the concentrations of acetic, propionic, and butyric acids were altered in a dose-dependent manner in relation to the amount of RS in the diet (P < 0.05), indicating a dose-dependent change of microbiota composition and/or metabolism. The linear regression slope of lactic acid concentrations against the RS dose also approached statistical significance (P = 0.07). A minimum dose of 2.5% RS elicited a significant response in acetic and butyric acid concentrations compared to BD-fed rats.
Table 3.
Colonic SCFA and carboxylic acid concentrations in weanling rats
| SCFA | Concna |
|||
|---|---|---|---|---|
| BD | RS |
|||
| 5% | 2.5% | 1.25% | ||
| Acetic | 20.4 ± 3.0 | 32.5 ± 2.3b | 26.8 ± 2.1b | 20.6 ± 1.8 |
| Propionic | 6.9 ± 1.3 | 10.1 ± 1.3b | 9.4 ± 1.1 | 6.0 ± 0.7 |
| Butyric | 5.6 ± 1.9 | 16.3 ± 3.1b | 12.1 ± 2.5b | 6.4 ± 1.2 |
| Lactic | 3.3 ± 0.5 | 6.0 ± 2.2b | 3.6 ± 0.4 | 2.7 ± 0.4 |
| Succinic | 2.3 ± 0.2 | 9.9 ± 5.3b | 4.3 ± 1.8 | 2.8 ± 0.3 |
Mean colonic digesta SCFA concentrations (mmol/g) ± SEM (n = 10) in conventional rats after 28 days of feeding the BD or the RS diet at 5%, 2.5%, or 1.25% RS.
Significantly different from the BD group (P < 0.05).
RS dose-dependent changes in microbiota composition.
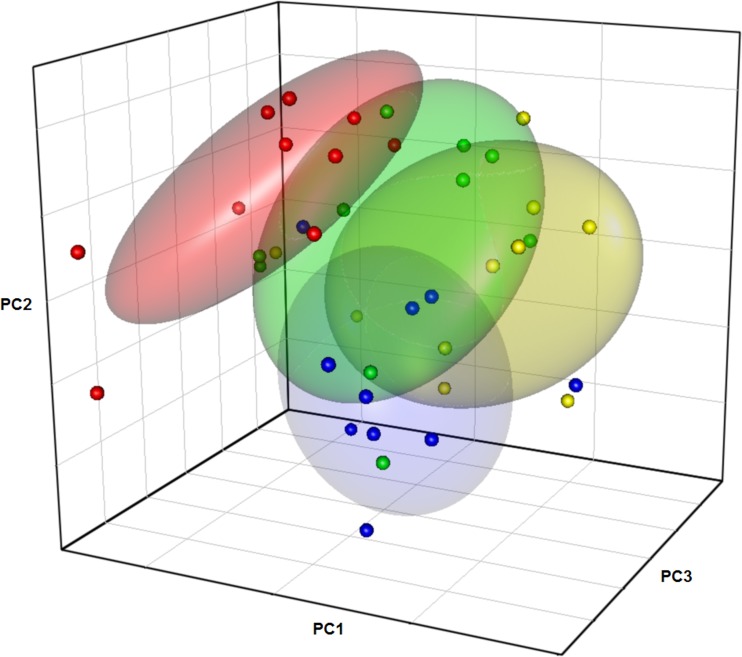
Concomitant with RS dose-dependent changes in SCFA concentrations, colonic RNA-TTGE microbiota profiles were also altered in a dose-dependent manner by feeding RS. Principal-component analysis of RNA-TTGE profile Dice similarity coefficients showed that profiles of rats fed RS at 5% were most dissimilar to rats fed BD (Fig. 1). Rats fed 2.5% and 1.25% RS showed RNA-TTGE profiles that were intermediate between those from rats fed RS 5% and BD. The results aligned well with SCFA concentration results, as RNA-TTGE is more likely to emphasize differences in the metabolically active members of the microbiota than analyses using DNA-TTGE (33).
Fig 1.
Principal-component analysis plot of 16S rRNA TTGE profiles. The points represent profiles of individual rats fed BD (blue), RS 5% (red), RS 2.5% (green), and RS 1.25% (yellow). The ellipses indicate 0.5 confidence interval boundaries.
RS-induced changes in microbiota composition.
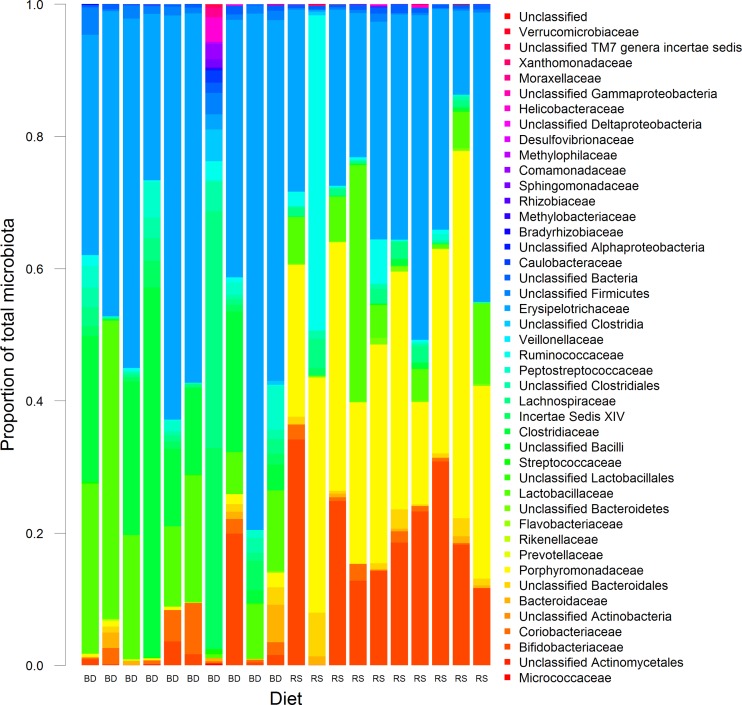
To characterize RS-induced changes in the colonic microbiota composition, DNAs from individual colon digesta samples from rats fed BD and 5% RS were analyzed by high-throughput pyrosequencing of the V1-to-V3 region of the 16S rRNA gene amplified by PCR using barcode-tagged fusion primers, resulting in 42,436 sequences. The minimum, maximum, and average numbers of sequences assigned to each sample were 648, 3,075, and 2,122, respectively, with an average sequence length of 523 bp. Taxonomic assignment of sequences at an 80% confidence threshold confirmed that feeding RS resulted in marked changes in microbiota community composition (Fig. 2 and Table 4). At the phylum level, proportions of Bacteroidetes were dramatically increased by feeding RS (BD = 2.2% ± 3.4% [standard deviation {SD}]; RS = 34.5% ± 12.2%; P < 0.001). Similarly, proportions of Actinobacteria were also significantly enriched in RS-fed rats (BD = 4.9 ± 6.9; RS = 20.0 ± 10.4; P = 0.002). Conversely, feeding RS resulted in a significant decrease in Firmicutes proportions (BD = 91.0 ± 8.4; RS = 44.9 ± 12.8; P < 0.001).
Fig 2.
Abundances of family level taxa as a proportion of total bacterial sequences identified from the colonic digesta of individual conventional rats fed BD or RS 5% for 28 days using high-throughput sequencing of 16S rRNA gene amplicons.
Table 4.
Bacterial-taxon proportions in the colon contents of conventional weanling rats
| Phylum | Proportiona |
Family | Proportion |
Genus | Proportion |
|||
|---|---|---|---|---|---|---|---|---|
| BD | RS | BD | RS | BD | RS | |||
| Actinobacteria | 4.9 | 19.8b | Bifidobacteriaceae | 2.8 | 18.9b | Bifidobacterium | 2.8 | 18.6b |
| Coriobacteriaceae | 2.1 | 0.9 | ||||||
| Bacteroidetes | 2.2 | 34.5b | Bacteroidaceae | 1.0 | 0.4 | Bacteroides | 1.0 | 0.4 |
| Unclassified Bacteroidales | 0.5 | 1.7 | ||||||
| Porphyromonadaceae | 0.6 | 32.0b | ||||||
| Unclassified Bacteroidetes | <0.1 | 0.4b | ||||||
| Firmicutes | 91.0 | 44.9b | Lactobacillaceae | 14.8 | 7.8 | Lactobacillus | 14.8 | 7.7 |
| Clostridiaceae | 15.3 | 0.3b | Anaerobacter | 0.2 | <0.1 | |||
| Clostridium | 13.9 | 0.2b | ||||||
| Clostridiales incertae sedis XIV | 4.5 | 0.2 | Blautia | 4.5 | 0.2 | |||
| Lachnospiraceae | 4.8 | 1.5 | ||||||
| Unclassified Clostridiales | 1.6 | 0.4b | ||||||
| Peptostreptococcaceae | 2.1 | 5.8b | Sporacetigenium | 1.6 | 0.4 | |||
| Ruminococcaceae | 0.6 | 5.8 | Ruminococcus | 0.2 | 5.4 | |||
| Unclassified clostridia | 0.5 | <0.1 | ||||||
| Erysipelotrichaceae | 44.8 | 28.2 | Allobaculum | 34.6 | 16.7 | |||
| Turicibacter | 7.4 | 2.1 | ||||||
| Unclassified Firmicutes | 1.7 | 0.4b | ||||||
| Proteobacteria | 1.3 | 0.1 | Caulobacteraceae | 0.2 | <0.1 | |||
| Comamonadaceae | 0.2 | <0.1 | ||||||
| Helicobacteraceae | 0.4 | 0.1 | Helicobacter | 0.4 | 0.1 | |||
| TM7 | <0.1 | <0.1 | ||||||
| Verrucomicrobia | <0.1 | <0.1 | ||||||
| Unclassified bacteria | 0.6 | 0.7 | ||||||
Mean proportions of bacterial taxa (%) at the phylum, family, and genus levels in colonic digesta of conventionally raised rats fed BD or RS 5% for 28 days (n = 10) determined by high-throughput sequencing of 16S rRNA gene amplicons.
Significantly different from BD-fed rats (P < 0.05).
The RS-induced increase in Bacteroidetes proportions was mainly from a bloom in Porphyromonadaceae proportions (BD = 0.6 ± 0.7; RS = 32.0 ± 10.7; P < 0.001). The increased Actinobacteria proportions in RS-fed rats were primarily the result of an expansion in Bifidobacterium spp. (BD = 2.8 ± 6.0; RS = 18.6 ± 9.9; P < 0.001). In contrast, the decrease in Firmicutes proportions, while less specific, was mostly from a reduction in clostridial proportions (BD = 29.5 ± 26; RS = 8.5 ± 16.6; P = 0.047). Concentrations of propionic and butyric acid were strongly correlated with increasing abundance of Bifidobacterium spp. (Pearson correlation coefficient = 0.85 and 0.76, respectively). Lactic acid concentrations showed a strong correlation with the prevalence of a group consisting of unidentified Bacteroidetes (Pearson correlation coefficient = 0.82). The abundance of the Porphyromonadaceae showed a moderate positive correlation with acetic acid concentrations (Pearson correlation coefficient = 0.61).
Changes in colonic mucosal morphology.
Morphological changes in the colonic mucosal architecture were observed in conventional rats fed RS compared to rats fed BD (Table 5). Feeding RS to conventional rats resulted in a significant increase in colonic crypt length (BD = 207.6 ± 5.1 [standard error of the mean {SEM}] mm; RS = 228.0 ± 6.6 mm) and goblet cell frequency per crypt (BD = 21.1 ± 0.6 [SEM] cells; RS = 24.9 ± 1.4 cells) compared to BD-fed conventional rats. In contrast, feeding RS to germ-free rats did not alter the colonic mucosal structure compared to germ-free rats fed BD.
Table 5.
Colon morphology measurements in weanling rats after 28 days of feeding
| Parametera | Ratb | Valuec |
P value | |
|---|---|---|---|---|
| BD | RS 5% | |||
| Crypt length | CR | 207.6 ± 5.1 | 228.0 ± 6.6d | 0.047 |
| GF | 201.3 ± 4.5 | 205.6 ± 5.5 | 0.644 | |
| Goblet cell no. | CR | 21.1 ± 0.6 | 24.9 ± 1.4d | 0.043 |
| GF | 24.0 ± 0.5 | 25.3 ± 1.4 | 0.347 | |
Mean colon crypt length (mm ± SEM) and mean goblet cell numbers per crypt.
CR, conventionally raised (n = 10); GF, germ free (n = 6).
Rats were fed a BD or RS diet for 28 days.
Significantly different from corresponding BD-fed rats (P < 0.05).
Diet-induced microbiota influences mucosal transcriptomes.
Microarray analyses showed that feeding RS to conventional rats altered the colon tissue expression of 9 genes with existing annotations (fold change [FC] > 1.5; FDR < 0.05) (Table 6). The genes showing the highest fold changes included Gsta2, encoding glutathione S-transferase A2, and Ela1, encoding an elastase protein, which were expressed at 2.6- and 2.5-fold-higher levels, on average, in conventional rats fed RS than in their BD-fed counterparts, respectively (FDR > 0.05). Glutathione S-transferases are involved in the detoxification of exogenous compounds, such as carcinogens and environmental toxins, by conjugation with glutathione. The Ela1 gene encodes a serine protease that is expressed at high levels in the pancreas and at lower levels in the intestine. Neutrophil-derived serine proteases have antimicrobial activities and are important regulators of immunity and inflammation (16). Changes in Gsta2 and Ela1 expression were verified by RT-qPCR, which showed 3.9- and 2-fold-higher expression, on average, in RS-fed conventional rats, respectively (P > 0.05). In contrast, feeding RS to germ-free rats did not affect host gene expression. Therefore, the microbiota, not RS alone, was responsible for the altered mucosal transcriptomes in the large bowel observed between dietary groups.
Table 6.
Fold changes in gene expression in colon tissues of conventional rats fed RS for 28 daysa
| Gene | Description | FCc | FDRd |
|---|---|---|---|
| Gsta2 | Glutathione-S-transferase, alpha type 2 | 2.6 | 0.001 |
| Ela1 | Elastase 1, pancreatic | 2.5 | 0.033 |
| Cdig1l | Cadmium-inducible gene 1L | 1.7 | 0.004 |
| RGD1565715 | Low-density lipoprotein receptor-related protein binding protein (predicted) | −1.7 | 0.032 |
| LOC683313 | Keratin complex 2, basic, gene 6a, transcript variant 2 | 1.7 | 0.048 |
| Wbscr14 | Williams-Beuren syndrome chromosome region 14 homolog | 1.6 | 0.001 |
| LOC680102 | MAbb-21-like 2 (LOC680102) (predicted) | −1.6 | 0.022 |
| Calcb | Calcitonin-related polypeptide, beta | −1.6 | 0.047 |
| Tac1 | Tachykinin 1 | −1.5 | 0.008 |
Changes in differentially expressed genes with known functions between conventionally raised rats fed BD or RS 5% for 28 days (n = 6).
MAb, monoclonal antibody.
FC, fold change in expression between treatments, with a positive value indicating higher expression in RS-fed rats and a negative value indicating higher expression in BD-fed rats.
FDR, false-discovery rate, a multiple-testing adjustment of P values determined by a linear model of microarray analysis (limma).
Alterations in serum metabolite profiles.
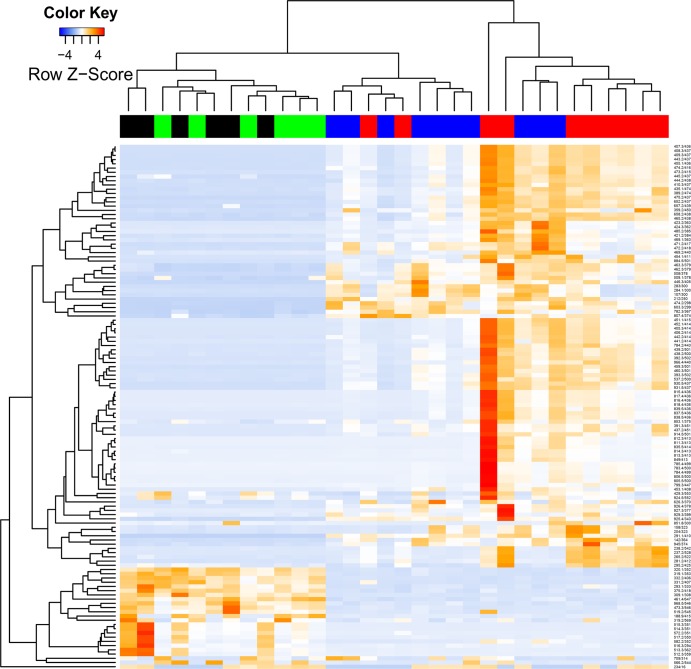
Peak detection and alignment of raw LC-MS data resulted in 1,029 peak groups across all samples, representing the serum metabolite profiles. Hierarchical clustering of metabolite profiles showed a grouping of profiles between conventional rats fed RS and BD (Fig. 3). Metabolite profiles from germ-free rats were clustered separately from profiles from conventional rats, but they showed greater similarity to profiles from BD-fed conventional rats than to those fed RS. However, dietary change in germ-free rats did not result in a clear separation according to diet, unlike that seen in conventional rats.
Fig 3.
Cluster analysis and heat map of serum metabolite ion signal intensities from conventional and germ-free rats fed BD or RS for 28 days, using metabolites that show the top 15% of variation across all samples. The color bar across the top of the heat map indicates the treatment group of the rat: CR-BD (blue), CR-RS (red), GF-BD (black), and GF-RS (green). The right vertical axis indicates metabolite ion identifications (mass/retention time).
Procrustes analysis.
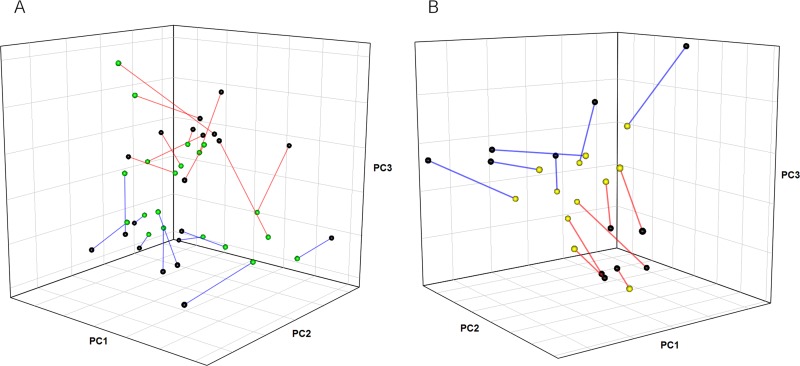
Procrustes analysis is a method for rotating and scaling points from one ordination, such as projections from a PCA plot, to be as close as possible to points from another ordination while maintaining the relative distances between points within each ordination. Procrustes rotation analysis showed that microbiota and metabolite profiles gave similar clustering patterns, indicating a good correlation between the microbiota and serum metabolite composition (Fig. 4A). Similarly, microbiota composition and gene expression profiles also showed strong clustering patterns (Fig. 4B).
Fig 4.
Procrustes rotation analysis of microbiota composition against serum metabolite profiles (A) and microbiota composition against gene expression (B) in conventional rats fed BD or RS for 28 days. The circles represent ordination projections: microbiota (black), serum metabolites (green), and gene expression (yellow). The gene expression and metabolite data used for analyses were those that had the top 1% variation in gene expression levels and the top 15% variation in metabolite intensities across all samples. Each line connects the microbiota projection to the metabolite projection (A) or gene expression projection (B) for each sample, with decreasing length indicating increasing similarity between projections. The overall correlations between rotations were 0.59 (A) and 0.74 (B). The line colors indicate the diet: BD (blue) and RS (red).
DISCUSSION
The results of experiments contained in this report confirmed our hypothesis that RS could be used to modulate the composition of the colonic microbiota when fed to young animals from weaning. Community profiles, measured by TTGE, were altered in a dose-dependent manner by feeding RS, and the change in the microbiota was reflected in the biochemistry of the colon in terms of concentrations of bacterial fermentation products. The minimum dose of RS that produced significant effects on colonic acetic and butyric acid concentrations was 2.5%. Therefore, it seems likely that the addition of relatively small amounts of RS in the diet can profoundly alter microbiota function. Changes in SCFA concentrations and microbiota composition induced by feeding RS were also associated with alterations in colonic gene expression and colonic mucosal architecture. The comparative results from conventional and germ-free rats showed that molecular communication between the microbiota and/or their products and the bowel mucosa of the host occurs; feeding RS to germ-free rats did not elicit significant changes in colon gene expression or phenotype. Broad changes in serum metabolite profiles were also observed in conventional rats fed RS but were not seen in RS-fed germ-free rats. Comparisons of microbiota composition to gene expression and metabolite profiles using Procrustes rotation analysis showed that diet-induced alterations in one data set corresponded to changes in another. These results provide evidence that changes in the microbiota composition have consequences for the host beyond the gastrointestinal tract.
Changes in the fermentation outputs of microbiota were associated with blooms of particular members of the bacterial community. Although average lactic acid concentrations were significantly increased by feeding RS, a strong correlation between lactic acid levels and the prevalence of lactate-producing bacteria, such as Bifidobacterium and Lactobacillus, in individual rats was not observed. Instead, lactic acid concentrations were strongly correlated with a group consisting of unidentified Bacteroidetes. The RS-associated bloom in Bifidobacterium spp. was strongly correlated with increased concentrations of propionic and butyric acid, suggesting conversion of lactate to propionate and butyrate by members of the unidentified Bacteroidetes. Although increases in Bifidobacterium proportions from feeding RS have previously been reported in high-throughput sequencing-based studies, the dramatic blooms in Bacteroidetes and Actinobacteria were greater than those seen in previous reports (19, 39). It appears that modifying the diet at an early age has a greater effect on the microbial community composition than dietary intervention in an older age group. Concomitant with bloom in Bacteroidetes, RS-fed conventional rats also showed a decrease in Firmicutes proportions. Alterations in Bacteroidetes-to-Firmicutes ratios have been associated with obesity (29, 37). Similarly, elevated SCFA concentrations in the large bowel have also been seen in obese subjects (29). In light of these previous studies, the changes we observed in colonic SCFA concentrations and microbiota composition suggest that feeding RS may have important effects on host metabolic processes in the young. The microbiota-mediated effect on host metabolism is further supported by the increased colonic-crypt length seen in RS-fed rats, which is indicative of increased intestinal growth and absorptive capacity (7, 26). The importance of the microbiota to host metabolism was also illustrated by the observation that germ-free rats ate more food but gained less weight than conventional rats fed the same diets. The greater weight gain of conventional relative to germ-free rats has been explained previously in terms of energy harvest by the large-bowel microbiota (37).
Although many studies have explored the health-promoting aspects of consuming RS and other nondigestible carbohydrates (4, 24, 27), the results from this report indicate that the effects on the host are extremely complex. We have shown that RS-induced alterations in microbiota composition resulted in elevated expression of Gsta2, which belongs to one of two supergene families that encode glutathione S-transferases. The enzymes encoded by these genes have an important role in the detoxification of reactive electrophilic compounds derived from allogeneic and autogenic sources (6). Alterations in Gsta2 activity may have implications for health, as genetic polymorphisms of glutathione S-transferase genes are risk factors for colorectal cancer (41). In addition to possible changes in cellular detoxification capacity, alterations in the microbiota structure may also influence apoptosis in the colon, as increased colonic-crypt stem cell apoptosis, associated with increased colonic-crypt length and butyric acid concentrations, has been reported in mice fed RS (21). Microbiota changes induced by feeding RS also increased the expression of a gene encoding a serine protease, Ela1. Serine proteases have an important role in the induction of inflammation at mucosal surfaces (16, 22) and defense against infection (13). Inhibition of serine protease has been shown to reduce inflammation and tissue damage (11, 14, 44). However, serine proteases are also able to promote the generation of immune-suppressing CD4+ FOXP3+ regulatory T cells in vitro (34). The contrasting stimulatory and suppressive effects of serine proteases on inflammation suggest they may play a role in feedback mechanisms that prevent excessive inflammation caused by bacterial stimulation. In addition to changes in gene transcription, RS-induced changes in the microbiota also increased the abundance of mucus-producing goblet cells, another potential defensive response to the altered microbial makeup. Our results strongly suggest that alterations in microbiota composition impart stimuli to the host that result in physiological responses that attempt to maintain homeostasis.
More research is required to reveal the bacterial physiological attributes of the blooms in relation to RS, particularly in light of the broad changes in microbiota composition and host physiology seen by feeding even small doses of RS at an early age. Synecological studies of the bowel ecosystem are clearly enhanced by the use of host transcriptomic and metabolomic data, since they provide measurements that might, with further research, provide biological readouts of large-bowel health. Such readouts could remove the subjectivity from opinions as to what kinds of alterations in bowel microecology are desirable with respect to host well being. In other words, rather than basing interpretations of health status on the prevalence of bacteria commonly thought of as “beneficial” (bifidobacteria and lactobacilli), specific biomarkers that reflect and distinguish between different whole microbiota-host relationships might be discovered.
ACKNOWLEDGMENTS
Wayne Young was the recipient of an Enterprise Ph.D. Scholarship from the Tertiary Education Commission. This research was funded by the New Zealand Ministry of Science and Innovation, the New Zealand Institute of Plant and Food Research Ltd., and AgResearch Ltd.
Footnotes
Published ahead of print 13 July 2012
REFERENCES
- 1. Antonopoulos DA, et al. 2009. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 77:2367–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bibiloni R, et al. 2008. Differential clustering of bowel biopsy-associated bacterial profiles of specimens collected in Mexico and Canada: what do these profiles represent? J. Med. Microbiol. 57:111–117 [DOI] [PubMed] [Google Scholar]
- 3. Caporaso JG, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Conlon MA, et al. 2012. Resistant starches protect against colonic DNA damage and alter microbiota and gene expression in rats fed a western diet. J. Nutr. 142:832–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De Filippo C, et al. 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U. S. A. 107:14691–14696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dourado DF, Fernandes PA, Ramos MJ. 2010. Glutathione transferase classes alpha, pi, and mu: GSH activation mechanism. J. Phys. Chem. B 114:12972–12980 [DOI] [PubMed] [Google Scholar]
- 7. Drozdowski L, Thomson AB. 2006. Intestinal mucosal adaptation. World J. Gastroenterol. 12:4614–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. 2008. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat. Rev. Microbiol. 6:121–131 [DOI] [PubMed] [Google Scholar]
- 9. Frank DN, et al. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U. S. A. 104:13780–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gentleman RC, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80 doi:10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gobbetti T, et al. 2012. Serine protease inhibition reduces post-ischemic granulocyte recruitment in mouse intestine. Am. J. Pathol. 180:141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gore C, et al. 2008. Bifidobacterium pseudocatenulatum is associated with atopic eczema: a nested case-control study investigating the fecal microbiota of infants. J. Allergy Clin. Immunol. 121:135–140 [DOI] [PubMed] [Google Scholar]
- 13. Hirche TO, et al. 2008. Neutrophil elastase mediates innate host protection against Pseudomonas aeruginosa. J. Immunol. 181:4945–4954 [DOI] [PubMed] [Google Scholar]
- 14. Ishizaki M, et al. 2008. Nafamostat mesilate, a potent serine protease inhibitor, inhibits airway eosinophilic inflammation and airway epithelial remodeling in a murine model of allergic asthma. J. Pharmacol. Sci. 108:355–363 [DOI] [PubMed] [Google Scholar]
- 15. Jensen MT, Cox RP, Jensen BB. 1995. Microbial production of skatole in the hind gut of pigs given different diets and its relation to skatole deposition in backfat. Anim. Sci. 61:293–304 [Google Scholar]
- 16. Kessenbrock K, Dau T, Jenne DE. 2011. Tailor-made inflammation: how neutrophil serine proteases modulate the inflammatory response. J. Mol. Med. 89:23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Knoch B, et al. 2010. Dietary arachidonic acid-mediated effects on colon inflammation using transcriptome analysis. Mol. Nutr. Food Res. 54(Suppl. 1): S62–S74 [DOI] [PubMed] [Google Scholar]
- 18. Mah KW, et al. 2007. Effect of a milk formula containing probiotics on the fecal microbiota of Asian infants at risk of atopic diseases. Pediatr. Res. 62:674–679 [DOI] [PubMed] [Google Scholar]
- 19. Martinez I, Kim J, Duffy PR, Schlegel VL, Walter J. 2010. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS One 5:e15046 doi:10.1371/journal.pone.0015046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McNeil NI. 1984. The contribution of the large intestine to energy supplies in man. Am. J. Clin. Nutr. 39:338–342 [DOI] [PubMed] [Google Scholar]
- 21. Mentschel J, Claus R. 2003. Increased butyrate formation in the pig colon by feeding raw potato starch leads to a reduction of colonocyte apoptosis and a shift to the stem cell compartment. Metabolism 52:1400–1405 [DOI] [PubMed] [Google Scholar]
- 22. Miller HR, Pemberton AD. 2002. Tissue-specific expression of mast cell granule serine proteinases and their role in inflammation in the lung and gut. Immunology 105:375–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miller TL, Wolin MJ. 1979. Fermentations by saccharolytic intestinal bacteria. Am. J. Clin. Nutr. 32:164–172 [DOI] [PubMed] [Google Scholar]
- 24. Moreau NM, et al. 2003. Restoration of the integrity of rat caeco-colonic mucosa by resistant starch, but not by fructo-oligosaccharides, in dextran sulfate sodium-induced experimental colitis. Br. J. Nutr. 90:75–85 [DOI] [PubMed] [Google Scholar]
- 25. Otter D, et al. 2011. Identification of urinary biomarkers of colon inflammation in IL10-/- mice using short-column LC-MS metabolomics. J. Biomed. Biotechnol. 2011:974701 doi:10.1155/2011/974701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Riehl TE, Ee X, Stenson WF. 2012. Hyaluronic acid regulates normal intestinal and colonic growth in mice. Am. J. Physiol. Gastrointest. Liver Physiol. doi:10.1152/ajpgi.00034.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rodríguez-Cabezas ME, et al. 2010. The combination of fructooligosaccharides and resistant starch shows prebiotic additive effects in rats. Clin. Nutr. 29:832–839 [DOI] [PubMed] [Google Scholar]
- 28. Roediger WE. 1982. Utilization of nutrients by isolated epithelial cells of the rat colon. Gastroenterology 83:424–429 [PubMed] [Google Scholar]
- 29. Schwiertz A, et al. 2010. Microbiota and SCFA in lean and overweight healthy subjects. Obesity 18:190–195 [DOI] [PubMed] [Google Scholar]
- 30. Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. 2006. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78:779–787 [DOI] [PubMed] [Google Scholar]
- 31. Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:1544–6155 doi:10.2202/1544–6115.1027 [DOI] [PubMed] [Google Scholar]
- 32. Stark PL, Lee A. 1982. The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J. Med. Microbiol. 15:189–203 [DOI] [PubMed] [Google Scholar]
- 33. Tannock GW, et al. 2004. Impact of consumption of oligosaccharide-containing biscuits on the fecal microbiota of humans. Appl. Environ. Microbiol. 70:2129–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tateosian NL, et al. 2011. Neutrophil elastase treated dendritic cells promote the generation of CD4(+)FOXP3(+) regulatory T cells in vitro. Cell Immunol. 269:128–134 [DOI] [PubMed] [Google Scholar]
- 35. Thioulouse J, Dray S. 2007. Interactive multivariate data analysis in R with the ade4 and ade4 TkGUI packages. J. Stat. Softw. 22:1–14 [Google Scholar]
- 36. Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. 2008. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3:213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Turnbaugh PJ, et al. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031 [DOI] [PubMed] [Google Scholar]
- 38. Vijay-Kumar M, et al. 2010. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328:228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Walker AW, et al. 2011. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 5:220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Walter J, et al. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang J, et al. 2011. Genetic polymorphisms of glutathione S-transferase genes and susceptibility to colorectal cancer: a case-control study in an Indian population. Cancer Epidemiol. 35:66–72 [DOI] [PubMed] [Google Scholar]
- 42. Wang M, et al. 2008. Reduced diversity in the early fecal microbiota of infants with atopic eczema. J. Allergy Clin. Immunol. 121:129–134 [DOI] [PubMed] [Google Scholar]
- 43. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yin L, et al. 2010. Differential effects of periopathogens on host protease inhibitors SLPI, elafin, SCCA1, and SCCA2. J. Oral Microbiol. doi:10.3402/jom.v3402i3400.5070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang C, et al. 2012. Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J. doi:10.1038/ismej.2012.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zivkovic AM, German JB, Lebrilla CB, Mills DA. 2011. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl. 1): 4653–4658 [DOI] [PMC free article] [PubMed] [Google Scholar]