Abstract
Salmonella remains the major cause of food-borne diseases worldwide, with chickens known to be the main reservoir for this zoonotic pathogen. Among the many approaches to reducing Salmonella colonization of broilers, bacteriophage offers several advantages. In this study, three bacteriophages (UAB_Phi20, UAB_Phi78, and UAB_Phi87) obtained from our collection that exhibited a broad host range against Salmonella enterica serovar Enteritidis and Salmonella enterica serovar Typhimurium were characterized with respect to morphology, genome size, and restriction patterns. A cocktail composed of the three bacteriophages was more effective in promoting the lysis of S. Enteritidis and S. Typhimurium cultures than any of the three bacteriophages alone. In addition, the cocktail was able to lyse the Salmonella enterica serovars Virchow, Hadar, and Infantis. The effectiveness of the bacteriophage cocktail in reducing the concentration of S. Typhimurium was tested in two animal models using different treatment schedules. In the mouse model, 50% survival was obtained when the cocktail was administered simultaneously with bacterial infection and again at 6, 24, and 30 h postinfection. Likewise, in the White Leghorn chicken specific-pathogen-free (SPF) model, the best results, defined as a reduction of Salmonella concentration in the chicken cecum, were obtained when the bacteriophage cocktail was administered 1 day before or just after bacterial infection and then again on different days postinfection. Our results show that frequent treatment of the chickens with bacteriophage, and especially prior to colonization of the intestinal tract by Salmonella, is required to achieve effective bacterial reduction over time.
INTRODUCTION
In the European Union (EU), salmonellosis is the second most frequently reported zoonotic disease in humans. In fact, in 2009, Salmonella was the most common causative agent, with a total of 1,721 food-borne outbreaks (14). Data from 2006 to 2007 corroborated that Salmonella enterica serovars Enteritidis and Typhimurium persist as the most prevalent Salmonella serovars, although an increase of S. Typhimurium and a decrease of S. Enteritidis were noted (12).
Poultry and their derived products are highly common sources of Salmonella in EU countries. To control Salmonella in animals and foodstuffs, a multifactorial approach through measures applied from farm to fork has been proposed. The first step in this approach is focused on a reduction in the prevalence of Salmonella in farms, which should subsequently diminish its incidence through the food chain. Accordingly, the EU's Salmonella control programs on Gallus gallus have sought to reduce the prevalence of certain Salmonella serovars (especially S. Enteritidis and S. Typhimurium but also S. enterica serovars Hadar, Infantis, and Virchow) at the various levels of poultry production (9). Indeed, the implementation of such measures has led to a significant reduction of these serovars in the EU (14). However, the prevalence of Salmonella in broiler carcasses in the EU is still around 15.6%, with 3.6% ascribed to the Salmonella serovars Enteritidis and Typhimurium (14). In addition, an epidemiological association of Salmonella levels between the flock and carcasses was determined (14). These data are consistent with the fact that the presence of Salmonella on broiler carcasses reflects contamination through the different phases of broiler production. Therefore, other complementary measures must be enacted to reduce Salmonella levels at each step of the food production chain.
Bacteriophage is of particular interest as a biocontrol agent in the prevention of food-borne illnesses due to its target specificity, rapid bacterial killing, and ability to self-replicate. The bacteriophage should be virulent, with a broad host range for the target genus and without bacterial virulence genes. A potential drawback is the survival and persistence of bacteriophage on different surfaces. This is influenced by a variety of physical factors, including those associated with matrix composition, and the particular properties of the bacteriophage itself (13). All these aspects must be investigated and well characterized before an effective biocontrol agent can be established and marketed.
Several studies have addressed the use of bacteriophage to decrease S. enterica concentrations on poultry (2, 4, 6, 7, 15, 24). In the present study, three virulent bacteriophages previously isolated from fecal samples of chickens and swine were selected and characterized, and their efficacy in the control of Salmonella contamination was tested. Special emphasis was placed on the schedule of bacteriophage administration, with the aim of achieving a maximum Salmonella reduction over time.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Salmonella enterica strains were obtained from the Laboratori de Sanitat Animal (Departament d'Agricultura Ramaderia i Pesca [DARP], Generalitat de Catalunya, Spain), the Hospital de la Vall d'Hebron, and the Hospital de la Santa Creu i Sant Pau (Barcelona, Spain). A total of 141 isolates of S. enterica serovar Typhimurium (n = 71) and S. enterica serovar Enteritidis (n = 70) were analyzed by XbaI pulsed-field electrophoresis (PFGE). Among these, 67 different pulsotypes were identified for S. Typhimurium (n = 49) and S. Enteritidis (n = 18). The criterion for discriminating between different pulsotypes was a difference of at least one restriction fragment (20). Based on these results, one representative strain of each pulsotype was chosen for further selection of the bacteriophage (see Table S1 in the supplemental material).
In addition, the virulent strains S. Typhimurium ATCC 14028 (American Type Culture Collection) and S. Enteritidis LK5 (Salmonella Genetic Stock Centre, University of Calgary) were used for in vitro infection assays, which also included the strains S. Virchow 79/S, S. Hadar 8546, and S. Infantis 05544 obtained from the DARP.
The derivative strain ATCC 14028ΩCm (UA1872) was constructed by the insertion of a chloramphenicol resistance cassette in an intergenic sequence, using the red recombinase system (10). This strain was used for in vivo efficacy studies of the bacteriophage.
All bacterial strains were routinely grown in Luria Bertani (LB) broth or on LB agar plates for 18 h at 37°C. Viable counts of ATCC 14028ΩCm in animal samples were routinely determined by plating on xylose lysine deoxycholate (XLD) agar plates, (Pronadisa) supplemented with chloramphenicol (3.4 μg/ml), followed by incubation at 37°C for 24 h.
Bacteriophage.
Fifty-five bacteriophage were obtained from 189 chicken cloacae and pig rectal swabs were collected from farms in different geographical areas of Spain between 2007 and 2009 (unpublished data). From this collection, bacteriophages UAB_Phi20 and UAB_Phi87, isolated from a chicken, and bacteriophage UAB_Phi78, from a pig, were selected for further characterization.
In vitro multiplication of bacteriophage.
Exponential cultures of Salmonella (108 CFU/ml) grown in LB medium at 37°C were infected with bacteriophage at a multiplicity of infection (MOI) of 1 PFU/CFU and incubated at 37°C for 5 h. Afterwards, infected cultures were centrifuged at 8,000 × g for 10 min (Beckman), and the supernatants were filtered through 0.45-μm-pore-size polyvinylidene fluoride (PVDF) membrane filters (Millipore). The bacteriophage titer was determined by plating adequate dilutions onto lawns of the bacterial strain of interest. Plates were incubated at 37°C for 24 h, and bacteriophage plaques were counted. When necessary, phage was concentrated by ultracentrifugation at 51,000 × g for 2 h (Beckman).
For further electron microscopy and DNA studies, the bacteriophage was purified through two successive CsCl gradients as described previously (23). After the second CsCl gradient, the bacteriophage was dialyzed against 10 mM MgSO4 buffer (Dialysis Tubing Cellulose Membrane; Sigma).
Host range of bacteriophage.
The host range of each bacteriophage was determined by spotting 10 μl of the above-described lysates (108 PFU/ml) onto lawns of Salmonella strains (see Table S1 in the supplemental material). The plates were incubated at 37°C for 24 h, and bacterial lysis was recorded.
TEM.
Bacteriophages (1011 PFU/ml) were applied to electron microscopy carbon-coated grids and negatively stained with 2% uranyl acetate. The preparations were allowed to dry and were then examined with a JEOL 1400 transmission electron microscope (TEM) at different magnifications (Servei de Microscopia, UAB). Bacteriophage morphology and dimensions were recorded.
Bacteriophage DNA techniques.
Bacteriophage lysates at 1011 PFU/ml were used to determine the genome sizes of the three bacteriophages by pulsed-field gel electrophoresis (PFGE), as previously described (3).
Bacteriophage DNA was obtained from lysates (1011 PFU/ml) as described previously (5) and digested with the restriction enzymes EcoRI, EcoRV, and HindIII (New England BioLabs), according to the manufacturer's recommendations. Samples were analyzed by electrophoresis in 0.7% agarose gels and visualized by staining with ethidium bromide.
In vitro killing assays.
Cultures of S. Typhimurium ATCC 14028 and S. Enteritidis LK5, grown in LB medium at 37°C until an optical density at 550 nm (OD550) of 0.2, were infected with each bacteriophage at an MOI of 1 PFU/CFU. Likewise, bacterial cultures were infected with a bacteriophage cocktail consisting of a mixture of the three selected bacteriophages (UAB_Phi20, UAB_Phi78, and UAB_Phi87) at a ratio of 1:1:1. Infected cultures were incubated for 8 h at 37°C. The OD550 of each culture was monitored at 30-min intervals. For bacterial enumeration, pellets obtained from 1-ml samples of the cultures centrifuged at 12,000 × g for 1 min (Eppendorf MiniSpin) were resuspended in 1 ml of 0.9% NaCl. Serial dilutions were then prepared for plating onto LB plates. In addition, S. Virchow 79/S, S. Hadar 8546, and S. Infantis 05544 cultures were infected with the bacteriophage cocktail, and the OD550 was monitored as described above.
In vivo assays in animal models.
All procedures involving animals were approved by the Ethical and Animal Welfare Committee from the Universitat Autònoma de Barcelona. Mouse and chicken models were used to assess the efficacy of the phage cocktail. In both models, the animals were orally infected with 0.1 ml of a suspension of S. Typhimurium ATCC 14028ΩCm in LB medium. Each dose of bacteriophage treatment consisted of the oral administration of 0.1 ml of a bacteriophage cocktail at 1011 PFU/ml in LB medium without any antacid.
Necropsies of the animals that had died from the infection and of euthanized animals were performed, and samples of the intestine of mice and of the cecum, liver, and spleen of chickens were taken. To determine the number of Salmonella bacteria, the tissue samples were weighed and homogenized in 0.9% NaCl, and serial dilutions of the homogenates were plated onto XLD plates. When Salmonella bacteria could not be quantified, their presence in tissue samples was detected by an enrichment procedure in which an aliquot (1 ml) from animal homogenized samples was inoculated in buffered peptone water (BPW; Merck) and incubated at 37°C for 18 h. One milliliter of the enrichment culture was then inoculated in 10 ml of Müller-Kauffmann selective broth (Merck) and incubated at 37°C for 24 h, followed by plating of an aliquot of this culture on XLD plates.
For bacteriophage titration, samples of intestine or cecum were weighed and then homogenized in 10 mM MgSO4 buffer. Serial dilutions were plated onto a lawn of S. Typhimurium ATCCΩCm and incubated at 37°C for 24 h. Afterwards, bacteriophage plaques were counted. When direct detection was not possible, a previously reported enrichment procedure (15) was applied.
Assays in mice.
Female BALB/c mice (3 weeks old) obtained from Harlan Iberica, Inc. (Barcelona, Spain), were housed in groups of 6 or 10 animals and then quarantined for 1 week. Food and water were supplied ad libitum. In all trials, one additional animal per assay was added to verify the absence of Salmonella and bacteriophage as described above.
To determine the residence time of the bacteriophage in the intestine of the animals, two groups of 10 mice were treated with the bacteriophage cocktail at 108 and 1010 PFU/animal. Two mice from each group were euthanized after 2 and 4 h of the bacteriophage administration and on days 2, 4, and 10 postadministration, and bacteriophage titration was performed.
Mice infected with S. Typhimurium die from a systemic typhoid fever-like disease similar to that seen in infected humans. Therefore, an effective bacteriophage treatment would delay or prevent the death of the mice. To determine the efficacy of the bacteriophage, three trials were designed. In each trial, two groups of six mice were orally infected with 0.1 ml of S. Typhimurium UA1872 at a concentration 100 times higher than the 50% lethal dose (LD50) of 1.83 × 106 CFU/animal, determined using a procedure previously reported (8). In each trial, group 1 was infected only with Salmonella, whereas group 2 was infected with Salmonella and treated with the bacteriophage cocktail (Table 1 gives details of the trials).
Table 1.
Experimental design of trials in the mouse model
Twelve 21-day-old mice were infected with S. Typhimurium UA1872. Six mice were treated with bacteriophage cocktail, and the other six were the control group.
One dose as a preventive treatment before (−) Salmonella infection.
Three doses as therapeutic treatment.
Four doses to prevent the intestinal tracts of the animals from being fully colonized by the bacterium.
p.i., postinfection.
In all trials, survival of the mice was recorded daily, and the presence of Salmonella in the dead mice was determined as described above. Moreover, the absence of bacteriophage was recorded in all the control animals that died.
Assays in chickens.
White Leghorn chicken (SPF) fertile eggs from Valo Lohmann Tierzucht (Salamanca, Spain) were incubated and hatched in biosafety level three facilities for animals (Centre de Recerca en Sanitat Animal [CReSA], Cerdanyola del Vallès, Spain). The chickens were housed in air-filtered isolation cabinets; food and water were supplied ad libitum. Before each experiment, two chickens were euthanized to confirm that they were free of Salmonella and bacteriophage, following the protocol described above. In addition, the absence of bacteriophage was recorded in the euthanized animals of the control group.
To determine the residence time of bacteriophage in the chicken cecum, 12 3-day-old chickens were orally treated with bacteriophage at 1010 PFU/animal. On days 3, 4, and 8 after phage inoculation, four chickens were euthanized, and cecum samples were obtained for bacteriophage determination following the methodology described above.
The efficacy of bacteriophage administration was studied in three trials for 21 days using two different methods of chicken infection by Salmonella. In each trial, two groups of animals were infected with the bacterium, but only one of them was treated with several doses of bacteriophage, at a concentration of 1010 PFU/animal per dose. The details of the trials are presented in Table 2. Briefly, in trial 1, Salmonella infection was carried out according to the seeder bird method (15), in which only six chickens of each group were orally infected with Salmonella, at a dose of 108 CFU/animal. The infection of the remaining chickens of each group was developed by contact with chickens already infected with the bacterium. We previously determined that all animals were fully colonized with Salmonella after 3 days of infection (data not shown). In trials 2 and 3, each chicken was orally inoculated with Salmonella at 105 CFU/animal. Quantification or detection of Salmonella and bacteriophage in the cecum, liver, and spleen was done following the procedures described above. Bacterial reduction was calculated by subtracting the mean bacterial concentration, expressed in log units, in the cecum of treated chickens from the mean determined in untreated chickens.
Table 2.
Experimental design of trials in the chicken model
| Trial no. | Age of chickens (days) | Total no. of animalsa | Treatment schedule (days)e | Time of euthanasia (days)d |
|---|---|---|---|---|
| 1b | 3 | 60 | 4,5 | 4, 5, 6, 12, 19, 26 |
| 2c | 6 | 46 | −1f, 0, 1, 2, 3, 6, 8, 10, 13, 15 | 1, 2, 6, 8, 10, 13, 15, 17 |
| 3c | 6 | 46 | 0, 1, 2, 3, 6, 8, 10, 13, 15 | 1, 2, 6, 8, 10, 13, 15, 17 |
The chickens were divided into two groups; one was treated with the bacteriophage cocktail, and the other was the control group.
Six 3-day-old chickens from each group were infected with Salmonella, and the rest were infected by contact. We previously determined that all animals were infected by contact after 3 days of infection.
Each chicken was orally inoculated with Salmonella at 105 CFU/animal. Bacteriophage treatment began before the intestinal tract of animals was fully colonized by the bacterium.
In trial 1, on each day of euthanasia, four chickens of the control and treatment groups were sacrificed for further studies. In trials 2 and 3, three chickens of the control and treatment groups were sacrificed for further studies, with the exception of the last day, in which the two remaining animals were sacrificed.
Chickens received two doses daily on the days in boldface.
Bacteriophage cocktail was administered 1 day before (−) Salmonella infection.
Statistical analysis.
The survival rates of the mice were analyzed using a log rank (Mantel-Cox) test. The results of all chicken trials were analyzed with a general lineal model (GLM), using time and group as covariates. When the data did not fulfill a normal distribution (trials 2 and 3), a nonparametric statistic Mann-Whitney-Wilcoxon test was also applied. The significance level was fixed at 5%.
RESULTS
Characterization of bacteriophage.
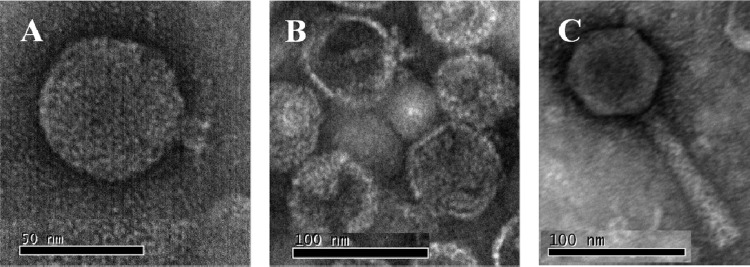
The three bacteriophages used in this study, UAB_Phi20, UAB_Phi78, and UAB_Phi87, were chosen from our collection of Salmonella bacteriophage based on their broad host range (see Table S1 in the supplemental material) and their different restriction patterns with EcoRI, EcoRV, and HindIII (Fig. 1). Together, they produced lytic plaques on lawns of 67 clonally unrelated strains of S. Typhimurium and S. Enteritidis. TEM images revealed that all three belong to the Caudovirales order. UAB_Phi20 has an icosahedral head (60 ± 1.5 nm) and a noncontractile short tail (13 ± 0.7 nm). UAB_Phi78 has the same morphology, with a head of 66 ± 1.7 nm and a tail of 14 ± 0.7 nm. Therefore, both bacteriophages are members of the Podoviridae family (1). UAB_Phi87 has an icosahedral head (68 ± 2.7 nm) but a long contractile tail (114 ± 4.3 nm) and belongs to the Myoviridae family (Fig. 2). The genome size of these bacteriophages as determined by PFGE is 32.3 kb (UAB_Phi20), 31.13 kb (UAB_Phi78), and 84.9 kb (UAB_Phi87).
Fig 1.
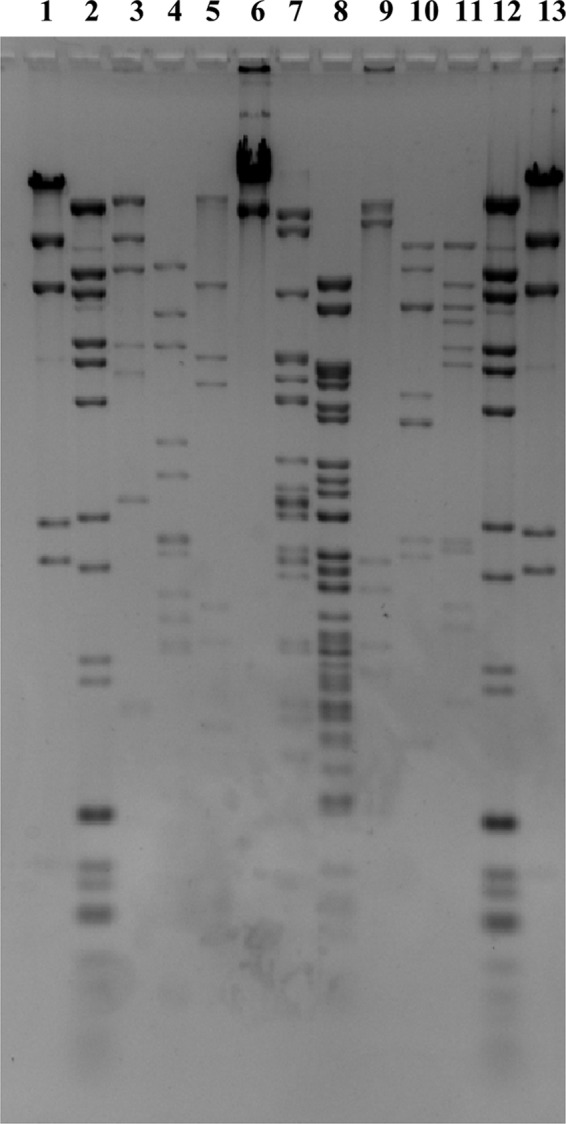
DNA restriction patterns of the bacteriophages UAB_Phi20 (lanes 3 to 5), UAB_Phi87 (lanes 6 to 8), and UAB_Phi78 (lanes 9 to 11) with the restriction enzymes EcoRI (lanes 3, 6, and 9), EcoRV (lanes 4, 7, and 10), and HindIII (lanes 5, 8, and 11). Lanes 1 and 13 correspond to HindIII-restricted bacteriophage lambda DNA markers, and lanes 2 and 12 correspond to BstEII bacteriophage lambda and Xphi174 DNA markers.
Fig 2.
Electron micrographs of bacteriophages UAB_Phi20 (A), UAB_Phi78 (B), and UAB_Phi87 (C).
In vitro infection dynamic of bacteriophage.
The dynamics of infection of each bacteriophage were determined in vitro at an MOI of 1. The greatest reduction in S. Typhimurium by UAB_Phi20, UAB_Phi78, and UAB_Phi87 was 4.4, 3.4, and 5.3 log10 CFU/ml, respectively (Fig. 3A). Higher values were obtained with S. Enteritidis, with a reduction of 5.6, 4, and 5.4 log10 CFU/ml by UAB_Phi20, UAB_Phi78, and UAB_Phi87, respectively (Fig. 3B). However, lysis was more prolonged in cultures of Salmonella infected with a cocktail composed of the three bacteriophages, with a reduction of S. Typhimurium and S. Enteritidis concentrations of 3 log10 and 4 log10, respectively, after 8 h of infection (Fig. 3).
Fig 3.
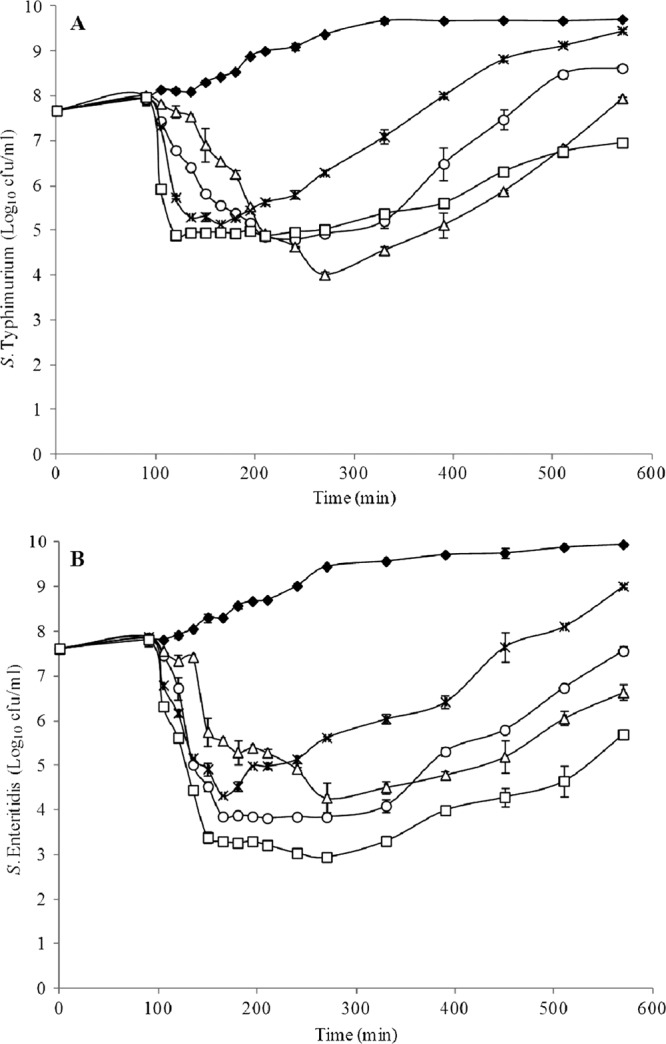
Viable concentration of S. Typhimurium ATCC 14028 (A) and S. Enteritidis LK5 (B) infected with UAB_Phi20 (○), UAB_Phi78 (×), UAB_Phi87 (△), or a bacteriophage cocktail (□). Uninfected (♦) cultures are indicated as well. The data reflect mean values from duplicate experiments, and the error bar represents the standard deviation of these replicates.
In addition, the bacteriophage cocktail was able to promote the lysis of strains of S. enterica serovars Hadar, Infantis, and Virchow (Fig. 4).
Fig 4.
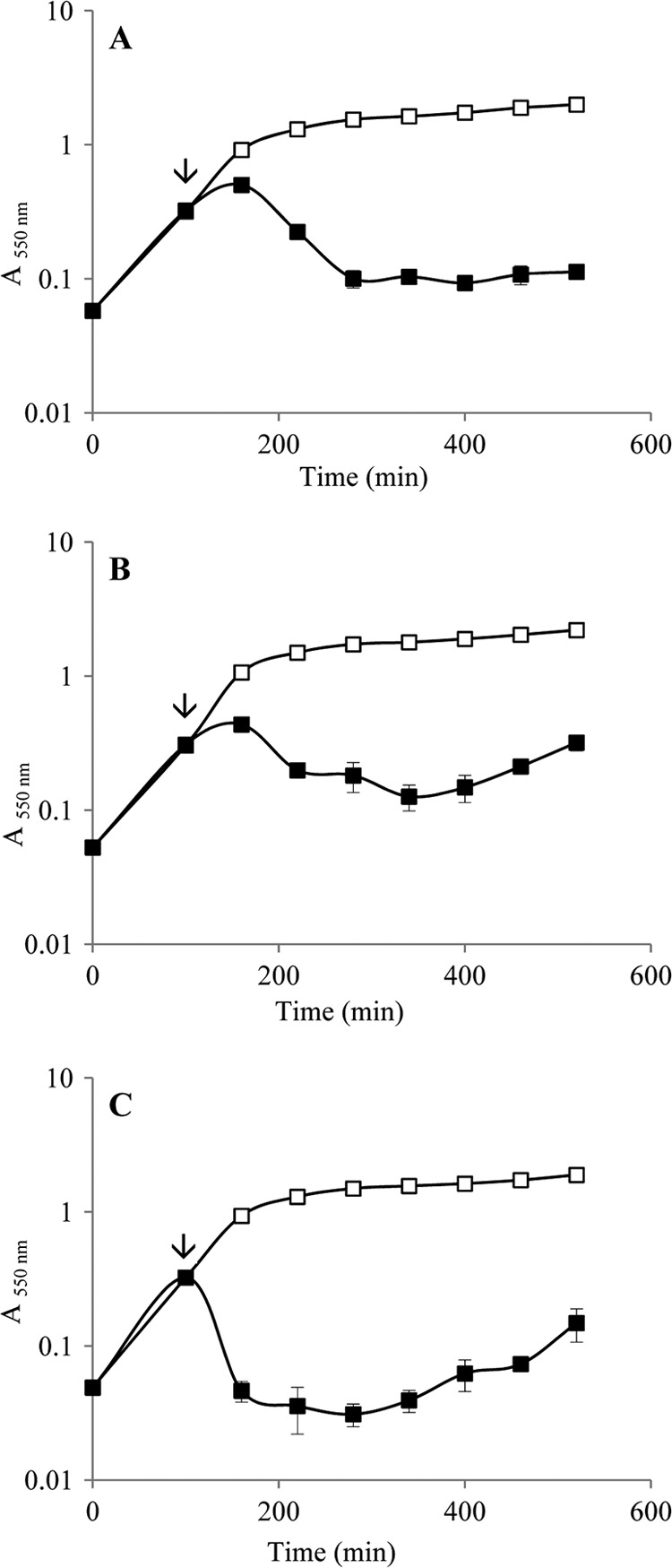
Absorbance of cultures of the S. enterica serovars Virchow (A), Infantis (B), and Hadar (C) infected (■) or not (□) with the bacteriophage cocktail. The time of bacteriophage infection is also shown (↓). The data reflect mean values from triplicate experiments, and the error bar represents the standard deviation of these replicates.
Residence time of bacteriophage in the intestinal tracts of mice and chickens.
The cocktail composed of a 1:1:1 mixture of the three bacteriophages was orally administered to two groups of 10 mice. One group was treated with 108 PFU/animal, and the other was treated with 1010 PFU/animal. The experiments were designed to determine the bacteriophage titer in the intestinal tract of the mice over a period of 10 days. However, the bacteriophage titer could be measured only at 24 h, at which point it was 103 and 104 PFU/g for animals treated with 108 and 1010 PFU/animal, respectively. Bacteriophage could be detected by the enrichment procedure until the 4th and 10th days of treatment, at doses of 108 and 1010 PFU/animal, respectively (data not shown).
In the chicken model, a single dose of 1010 PFU/animal was used because this dose clearly increased the residence time of the bacteriophage in the digestive tract of the mice. In this case, the bacteriophage was detected by the enrichment procedure for only a period of 8 days.
Bacteriophage cocktail efficacy in the mouse model.
Mice administered the bacteriophage cocktail 8 h prior to infection with S. Typhimurium (Table 1, trial 1) survived only slightly longer than mice in the control group (Fig. 5A). A similar effect was seen in mice treated with the cocktail at 4 h, 7 days, and 10 days after S. Typhimurium infection (Trial 2) (Fig. 5B). However, with the administration schedule of trial 3 (Table 1), a survival rate of 50% was obtained (P = 0.068) (Fig. 5C).
Fig 5.
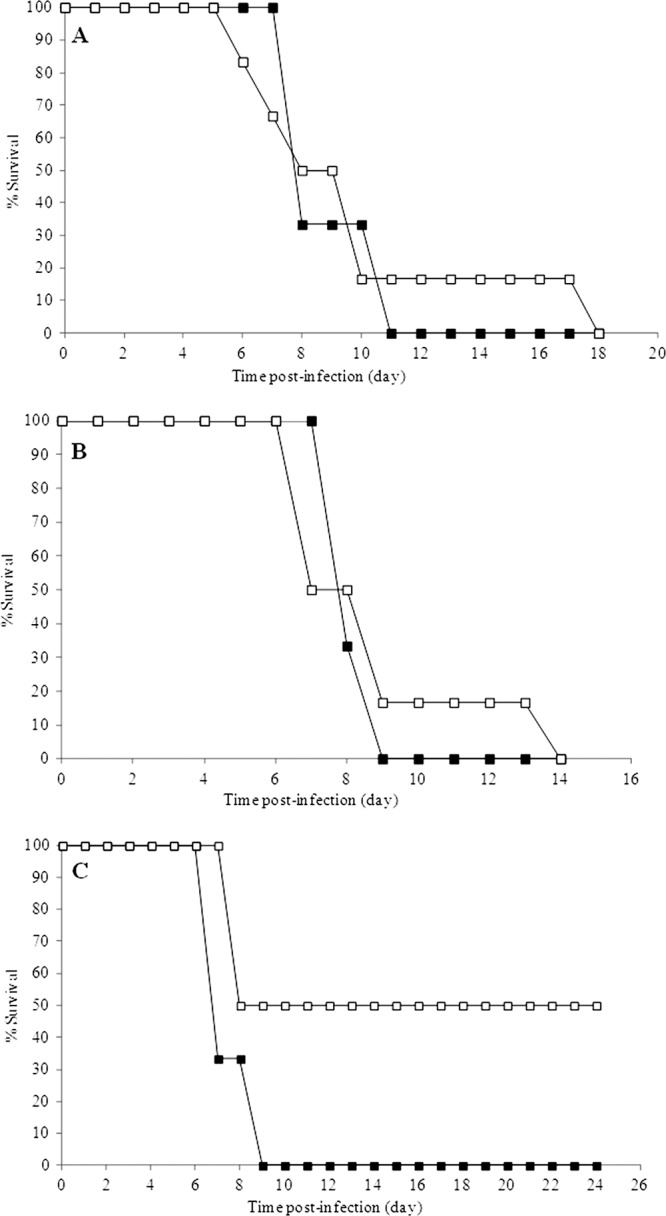
Effect of the bacteriophage cocktail treatment (□) on the survival of mice infected with S. Typhimurium UA1872. (A) Treatment applied at 8 h preinfection. (B) Treatment applied at 4 h postinfection and on days 7 and 10. (C) Treatment administered simultaneously with bacterial inoculations and at 6, 24, and 30 h postinfection. The untreated groups (■) are also shown. Details of each trial are described in Table 1.
Bacteriophage cocktail efficacy in the chicken model.
Animals infected with S. Typhimurium according to the seeder bird infection method (Table 2, trial 1) were treated with the bacteriophage cocktail twice a day on days 4 and 5 postinfection. This treatment resulted in a decrease in the Salmonella concentration in the cecum of infected animals of only 1 log10 compared to the control group during the fifth and sixth days postinfection and a reduction of 0.5 log10 12 days after infection (P = 0.055) (Fig. 6A).
Fig 6.
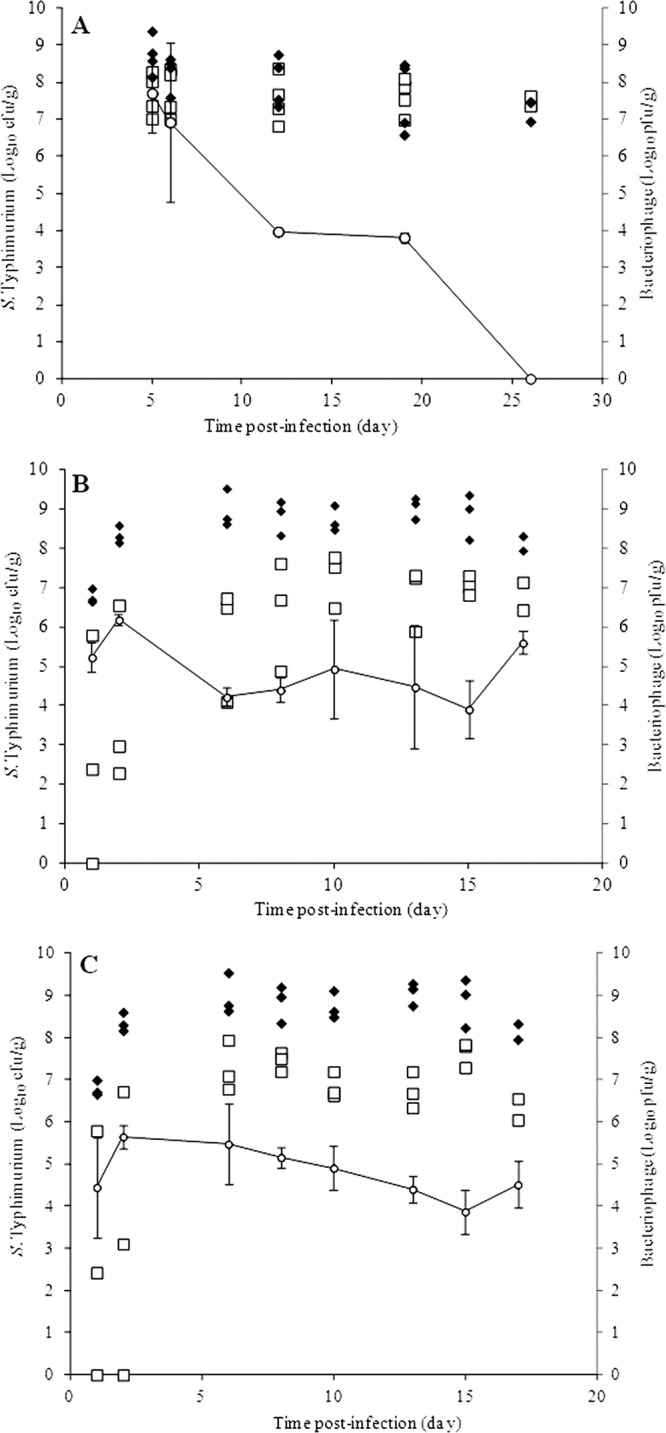
Concentration of S. Typhimurium UA1872 in the cecum of chickens left untreated (♦) or treated (□) with the bacteriophage cocktail, expressed as log10 CFU/g. The titer (○) of bacteriophage in the cecum of treated chickens is also presented as log10 PFU/g. Trials 1, 2, and 3 (Table 2) correspond to panels A, B, and C, respectively.
In trials 2 and 3, all animals were individually infected by oral inoculation of S. Typhimurium UA1872 and then treated with the bacteriophage cocktail, as indicated in Table 2. In trial 2, reductions in the Salmonella concentrations in the chicken cecum of 4.4 log10 and 3.2 log10 were reached by days 2 and 6 postinfection, respectively, and of 2 log10 between day 8 postinfection and the end of the experiment (P < 0.0001) (Fig. 6B). The results of trial 3 were similar, but in this case a decrease of 2 log10 was obtained between day 6 postinfection and the end of the experiment (P < 0.0001) (Fig. 6C).
In all trials, the bacteriophage titer in the chicken cecum was determined over time. As shown in Fig. 6, when only two doses of the cocktail were administered (trial 1), the titer decreased dramatically, reaching <103 PFU/g beginning on day 19 postinfection. In contrast, with the bacteriophage administration schedules of trials 2 and 3, the concentration of bacteriophage in the cecum was in the range of 104 to 105 PFU/g throughout the experiment, albeit with some fluctuations (Fig. 6).
In addition, the presence of S. Typhimurium UA1872 in the liver and spleen of the animals was assayed at each time point of every trial. It is noteworthy that, compared with the control groups, there was a delay in bacterial colonization of these organs in both trial 2 and trial 3 (see Table S2 in the supplemental material).
DISCUSSION
The problems associated with multidrug-resistant bacterial pathogens and the regulations concerning the use of antimicrobials in animal production have led to a resurgence of interest in phage therapy (11). Research into the feasibility of treating food animals has focused on reducing the impact of infections in the animals themselves, as well as decreasing zoonotic pathogens in these reservoirs (18).
In the present study, three virulent Salmonella-specific bacteriophages (UAB_Phi20, UAB_Phi78, and UAB_Phi87) selected from our collection were characterized, and their ability to reduce Salmonella concentrations in mice and chickens was determined. All three bacteriophages belong to the Caudovirales order, and their genomes do not show homology with any known gene involved in virulence, as determined after sequencing of their genomes (data not shown). Moreover, these bacteriophages are highly stable, as confirmed by the observation that their concentrations after a 30-min incubation at pH 2 decreased by a maximum of 3.4 log10 PFU/ml (data not shown), which is better than the values reported by other authors (17, 19). These results suggest that the three bacteriophages are able to resist passage through the low-pH environment of the stomach.
The bacteriophage cocktail produced a faster and longer decrease in the Salmonella concentration than obtained when the bacteriophages were tested individually. In addition, the killing kinetics of the cocktail were better than those of other bacteriophage mixtures, as determined 4 h after bacterial infection (24), and similar to those administered for 8 h (4) although in those studies the MOIs were higher than in our experiments. Moreover, the cocktail was also able to promote the lysis of other S. enterica serovars whose reduction in commercial Gallus gallus production has likewise been targeted by the EU (14).
Based on these results, the efficacy of the bacteriophage cocktail was studied in vivo to determine whether it would significantly reduce the population of Salmonella in the intestinal tract of animals and whether its activity was maintained over a prolonged period of time. These studies were performed in mice and in Gallus gallus, infecting the animals with a derivative of S. Typhimurium ATCC 14028 labeled with a chloramphenicol resistance cassette in an intergenic region.
Mice infected with S. Typhimurium die from a systemic typhoid fever-like disease (21, 25). Therefore, an effect on bacterial infection by the bacteriophage cocktail should increase the survival of the animals. The administration of the bacteriophage cocktail before infection (Fig. 5A, trial 1) or a few hours after infection and then again at 7 and 10 days postinfection (Fig. 5B, trial 2) only delayed the death of the mice. These findings are in accordance with the rapid disappearance of the bacteriophage from the mouse digestive tract. However, when the cocktail was administered simultaneously with S. Typhimurium and readministered at 6, 24, and 30 h postinfection (Fig. 5C, trial 3), a 50% improvement in survival was obtained. Although the latter result is not statistically significant (P = 0.068), it nonetheless suggests the convenience of administering several doses of bacteriophage to animals before their intestinal tract becomes fully colonized by Salmonella.
Results obtained in the chicken model agree with those in mice, which showed that the more affordable mouse model can be used in preliminary studies on the efficacy of bacteriophage in reducing Salmonella. Indeed, as observed in mice, the administration of four doses of the bacteriophage cocktail to chickens whose digestive tracts were already colonized by Salmonella did not result in a significant decrease in the intestinal concentration of this bacterium (Fig. 6A, trial 1). This result is similar to results reported by others (15). In fact, our protocol failed to produce a significant delay in bacterial colonization of the livers and spleens of treated versus untreated animals (data not shown). It should be noted that in this trial the Salmonella concentration in the cecum at the time of bacteriophage administration was ≥108 CFU/g (data not shown) and that in the majority of the animals the bacteriophage titer in the cecum was lower than the bacterial concentration (Fig. 6A). We believe that the lack of effectiveness of the bacteriophage cocktail in this trial was due to the low ratio between bacteriophage and bacteria, which would have seriously hampered an encounter between the two. Consistent with this conclusion was the significant reduction in the S. Typhimurium concentration achieved when the bacteriophage cocktail was first administered to the chickens either before or just after bacterial infection, followed by repeated administration twice a day for 4 days and a daily dose during the next week, as was the case in trials 2 and 3 (Fig. 6B and C). Moreover, both administration schedules resulted in a delay in bacterial colonization of the liver and spleen (see Table S2 in the supplemental material), which corroborates the efficacy of bacteriophage treatment. The slight differences between these two trials could be attributed to the fact that chickens in trial 2 were treated with bacteriophage 1 day before Salmonella infection.
The increase in the S. Typhimurium concentration over time in chickens in trials 2 and 3 may be a consequence of the so-called proliferation threshold, which occurs when a minimum number of bacterial cells are present (22), such that the bacterial density is too low to sustain a growing bacteriophage population. Likely, this effect occurred in most animals of trials 2 and 3, in which the bacterial concentration in the cecum of chickens was <103 CFU/g during the first few days of bacteriophage administration. In addition, it should be noted that we used very young SPF chickens, which facilitated a rapid colonization of their intestines by Salmonella and its translocation to the animals' internal organs. Some authors have shown that certain bacteriophages are able to translocate through the intestinal wall of mice (16); consequently, phage therapy by oral administration could be effective against systemic infection. However, it seems that this might not be relevant in chickens because readministration of the bacteriophage cocktail to the animals was necessary to obtain a significant reduction of Salmonella over time. The bacteriophage effectiveness determined with this treatment schedule was greater than reported in others studies in terms of bacterial cell numbers and the effective duration of treatment (2, 4, 24).
Our model studies in chickens were conducted under very strict conditions, using an infective dose of Salmonella that corresponded to levels of bacterial colonization significantly higher than those typically determined on commercial broiler chickens (26). The fact that our bacteriophage cocktail was highly effective supports its further experimentation with commercial broilers. Nonetheless, according to our findings, frequent treatment of the animals with the bacteriophage cocktail—including prior to bacterial colonization of the intestinal tract—is necessary to achieve a significant reduction of bacterial cell numbers. This is obviously a serious drawback to the implementation of this approach. Therefore, considering the bacteriophage as an inert particle, the current challenge is to increase their removal time in the intestinal tract of the target in order to minimize the number of required doses. This objective is currently the focus of further studies in our laboratory.
Supplementary Material
ACKNOWLEDGMENTS
We are deeply indebted to Montserrat Saco from the Laboratori de Sanitat Animal (Generalitat de Catalunya) and to the Servei de Microbiologia of the Hospitals Vall d'Hebron and Santa Creu i Sant Pau (Barcelona, Spain) for providing us with the Salmonella strains and to Laboratorios Calier S.A. and to Laboratorio de Diagnóstico General S.L. for the fecal samples. Also, we thank R. Dolz and D. Solanes of the Centre de Recerca en Sanitat Animal (CReSA) for their collaboration in poultry experiments. We are grateful to Oliver Valero from the Servei d'Estadística of the Universitat Autònoma de Barcelona for doing the statistical analysis.
This work was supported by grants RTA2006-00065 and SGR1106 from the IRTA (Ministerio de Ciencia e Innovación, Spain) and Generalitat de Catalunya, respectively. C.B. is the recipient of a predoctoral fellowship from the CUR (Departament d'Innovació, Universitats i Empresa, Generalitat de Catalunya and Fons Social Europeu), and D.A.S. received a predoctoral fellowship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil.
Footnotes
Published ahead of print 6 July 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Ackermann HW. 1998. Tailed bacteriophages: the order caudovirales. Adv. Virus Res. 51:135–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andreatti Filho RL, et al. 2007. Ability of bacteriophages isolated from different sources to reduce Salmonella enterica serovar Enteritidis in vitro and in vivo. Poult. Sci. 86:1904–1909 [DOI] [PubMed] [Google Scholar]
- 3. Atterbury RJ, Connerton PL, Dodd CE, Rees CE, Connerton IF. 2003. Isolation and characterization of Campylobacter bacteriophages from retail poultry. Appl. Environ. Microbiol. 69:4511–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Atterbury RJ, et al. 2007. Bacteriophage therapy to reduce Salmonella colonization of broiler chickens. Appl. Environ. Microbiol. 73:4543–4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ausubel FM, et al. 1987. Current protocols in molecular biology, vol 1 Greene Publishing/Wiley-Interscience, New York, NY [Google Scholar]
- 6. Berchieri A, Jr, Lovell MA, Barrow PA. 1991. The activity in the chicken alimentary tract of bacteriophages lytic for Salmonella Typhimurium. Res. Microbiol. 142:541–992 [DOI] [PubMed] [Google Scholar]
- 7. Borie C, et al. 2008. Bacteriophage treatment reduces Salmonella colonization of infected chickens. Avian Dis. 52:64–67 [DOI] [PubMed] [Google Scholar]
- 8. Cárdenas M, et al. 2001. Virulence of Pasteurella multocida recA mutants. Vet. Microbiol. 80:53–61 [DOI] [PubMed] [Google Scholar]
- 9. Commission of the European Communities 2005. Commission regulation (EC) no. 1003/2005 of 30 June 2005 implementing regulation (EC) no. 2160/2003 as regards a Community target for the reduction of the prevalence of certain Salmonella serotypes in breeding flocks of Gallus gallus and amending regulation (EC) no. 2160/2003. OJEU L 170/12. http://faolex.fao.org/docs/pdf/eur52715.pdf
- 10. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deresinski S. 2009. Bacteriophage therapy: exploiting smaller fleas. Clin. Infect. Dis. 48:1096–1101 [DOI] [PubMed] [Google Scholar]
- 12. European Food Safety Authority 2009. The Community summary report on trends and sources of zoonoses and zoonotic agents in the European Union in 2007. EFSA J. 2009:223 doi:10.2903/j.efsa.2009.223r [Google Scholar]
- 13. European Food Safety Authority 2009. Scientific opinion of the Panel on Biological Hazards on a request from European Commission on the use and mode of action of bacteriophages in food production. EFSA J. 2009:1076 doi:10.2903/j.efsa.2009.1076 [Google Scholar]
- 14. European Food Safety Authority, European Centre for Disease Prevention and Control 2011. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2009. EFSA J. 9:2090 doi:10.2903/j.efsa.2011.2090 [Google Scholar]
- 15. Fiorentin L, Vieira ND, Barioni W., Jr 2005. Oral treatment with bacteriophages reduces the concentration of Salmonella Enteritidis PT4 in caecal contents of broilers. Avian Pathol. 34:258–263 [DOI] [PubMed] [Google Scholar]
- 16. Górski A, et al. 2006. Bacteriophage translocation. FEMS Immunol. Med. Microbiol. 46:313–319 [DOI] [PubMed] [Google Scholar]
- 17. Jamalludeen N, She YM, Lingohr EJ, Griffiths M. 2009. Isolation and characterization of virulent bacteriophages against Escherichia coli serogroups O1, O2 and O78. Poult. Sci. 88:1694–1702 [DOI] [PubMed] [Google Scholar]
- 18. Johnson RP, et al. 2008. Bacteriophages for prophylaxis and therapy in cattle, poultry and pigs. Anim. Health Res. Rev. 9:201–215 [DOI] [PubMed] [Google Scholar]
- 19. Kwon HJ, Cho SH, Kim TE. 2008. Characterization of a T7-Like lytic bacteriophage (SG-JL2) of Salmonella enterica serovar Gallinarum biovar Gallinarum. Appl. Environ. Microbiol. 74:6970–6979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liebana E, et al. 2001. Molecular typing of Salmonella serotypes prevalent in animals in England: assessment of methodology. J. Clin. Microbiol. 39:3609–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mastroeni P. 2002. Immunity to systemic Salmonella infections. Curr. Mol. Med. 2:393–406 [DOI] [PubMed] [Google Scholar]
- 22. Payne RJ, Jansen VA. 2001. Understanding bacteriophage therapy as a density-dependent kinetic process. J. Theor. Biol. 208:37–48 [DOI] [PubMed] [Google Scholar]
- 23. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24. Toro H, et al. 2005. Use of bacteriophages in combination with competitive exclusion to reduce Salmonella from infected chickens. Avian Dis. 49:118–124 [DOI] [PubMed] [Google Scholar]
- 25. Tsolis RM, et al. 1999. Of mice, calves, and men. Comparison of the mouse typhoid model with other Salmonella infections. Adv. Exp. Med. Biol. 473:261–274 [PubMed] [Google Scholar]
- 26. Waldroup AL, Rathgeber BM, Forsythe RH. 1992. Effects of six modifications on the incidence and levels of spoilage and pathogenic organisms on commercial processed postchill broilers. J. Appl. Poult. Res. 2:111–116 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.