Abstract
Multicellular communities produced by Bacillus subtilis can adopt sliding or swarming to translocate over surfaces. While sliding is a flagellum-independent motility produced by the expansive forces in a growing colony, swarming requires flagellar functionality and is characterized by the appearance of hyperflagellated swarm cells that associate in bundles or rafts during movement. Previous work has shown that swarming by undomesticated B. subtilis strains requires swrA, a gene that upregulates the expression of flagellar genes and increases swimming motility, and surfactin, a lipopeptide biosurfactant that also facilitates sliding. Through an analysis of swrA+ and swrA mutant laboratory strains with or without a mutation in sfp (a gene involved in surfactin production), we show that both swrA and surfactin upregulate the transcription of the flagellin gene and increase bacterial swimming. Surfactin also allows the nonswarming swrA mutant strain to efficiently colonize moist surfaces by sliding. Finally, we reconfirm the essential role of swrA in swarming and show that surfactin, which increases surface wettability, allows swrA+ strains to produce swarm cells on media at low humidity.
INTRODUCTION
Bacillus subtilis is a flagellated bacterium ubiquitously distributed in the environment and recognized as a potent agent for the biological control of plant diseases. It possesses remarkable metabolic and physiological versatility that allows its growth in diverse settings within the biosphere, including solid substrates in many different environments (9). Depending on nutrient availability and moisture conditions, over a surface, B. subtilis can remain localized, producing biofilms (13) or fruiting bodies (2), or move out to colonize larger areas by swarming (20, 34) or sliding motility (23). Swarming is a surface movement that occurs on moist and nutrient-rich media and is powered by numerous flagella (15, 17, 19). Sliding or spreading is a flagellum-independent motility produced by expansive forces in a growing colony in combination with cell surface properties resulting in reduced friction between the cell monolayer and substrate (17, 23).
Similarly to other Gram-negative and Gram-positive organisms (15, 17, 27), swarming by B. subtilis is accompanied by the production of hyperflagellated swarmer cells predominantly associated in bundles or rafts at the colony edge (20, 34). These cells migrate atop solid surfaces, until they stop and revert to the undifferentiated vegetative state. Swarming behavior has been shown to increase the adaptive resistance of B. subtilis to multiple antibiotics, and it was previously suggested to confer a fitness advantage when the environment contains something harmful (4, 24).
Swarming and sliding motility by B. subtilis were shown to require or be facilitated by the production of the lipopeptide surfactin, a very potent biosurfactant that increases the wettability of surfaces and shows antimicrobial properties (1, 18, 20, 23, 31, 34). Surfactin plays a major role in the B. subtilis-based biocontrol of plant diseases (8), and it is likely that B. subtilis utilizes this compound to better colonize surfaces such as plant roots. However, in contrast to biofilm formation, in which surfactin acts as an autoinducer or a quorum-sensing signal leading to the derepression of genes involved in matrix synthesis (25), nothing is known about the activity of the lipopeptide with respect to B. subtilis motility gene expression.
B. subtilis strains, such as the commonly used laboratory strain 168 and many of its derivatives, do not swarm on common rich media due to a frameshift mutation in the swrA gene (22, 31, 34), also referred to as swrAA (5, 6). However, this gene was recently shown to be dispensable for swarming on synthetic B medium under high-humidity conditions (14). Through the action of the transcription-activating protein DegU (30), SwrA stimulates the transcription of the fla-che operon, which contains the majority of genes for flagellar and chemotaxis proteins and sigD, the σD-encoding gene (21, 32). This increases the synthesis of structural flagellar proteins, including flagellin, and stimulates the expression of swrA itself through a σD-dependent promoter (5). Strains carrying the frameshift mutation in swrA display a reduced number of flagella in liquid media and are unable to produce flagella over rich media containing ≥0.5% agar (6, 22, 34). Very recently, it was shown that a mutation in swrA also causes an inability of laboratory strains to produce robust biofilms (27).
Given that both surfactin and swrA influence the motility and multicellularity of B. subtilis, we wondered what the relative role of these two factors was in swimming behavior and in the development of bacterial communities in motion on a surface. To this aim, an swrA+ laboratory strain able to produce swarm cells, although defective in surfactin production (sfp mutant), and relatives differing in the presence of functional swrA and/or sfp alleles were chosen for the analysis. Here we report that the flagellin expression level is elevated in the presence of surfactin and describe the contribution of swrA and surfactin to B. subtilis sliding and swarming motility.
MATERIALS AND METHODS
Strains and culture conditions.
B. subtilis strains used in this study are listed in Table 1. They differ in the presence of functional swrA and/or sfp alleles. The strains were grown at 37°C on tryptone-NaCl broth medium (Tr) (10 g/liter tryptone, 5 g/liter NaCl). This medium was solidified with different agar (Bacteriological Agar; Oxoid, Basingstoke, United Kingdom) concentrations. Plates were poured when the medium was at ∼50°C and dried for 15 min open-faced in a laminar-flow hood, as described previously by Patrick and Kearns (31). When required, surfactin (Sigma-Aldrich, St. Louis, MO) was added to the liquid media at a final concentration of 20 μM.
Table 1.
B. subtilis strains used in this study
| Strain | Parent strain | Genotype | Derivation and/or reference or source |
|---|---|---|---|
| PB1831 | Laboratory strain | trpC2 pheA1 | JH642a, J. A. Hoch |
| PB5249 | PB1831 | trpC2 pheA1 swrA+ | 34 |
| PB1927 | trpC2 pheA1 sfp+ | OKB105a, 29 | |
| PB5332 | PB1927 | trpC2 pheA1 sfp+swrA+ | PB1927 (tf) PB5249b |
Previous designation of the strain.
Strain constructed by transformation (tf) using donor DNA from PB1927.
Motility assays.
Swimming motility was evaluated by the seeding of stationary-phase cells (∼5.0 × 105 cells) onto the center of motility plates (Tr solidified with 0.2% agar). Plates were incubated at 37°C in a humidified chamber, and the diameters of halos due to bacterial migration were measured 6 and 24 h after inoculation. For each strain, swimming assays were performed in triplicate on four separate days.
Bacterial motility over a surface was analyzed by spotting 2 μl of a culture of each strain grown overnight (∼5.0 × 105 cells) onto the center of Tr plates solidified with agar at various concentrations. Plates were incubated at 37°C in a humidified chamber for 24 h.
Swarming motility was evaluated by searching for the presence of swarm cells and cell rafts in colonies grown on Tr plates for 8 h. Swarm cell production was assessed by measuring the cell length and degree of flagellation, as previously described (10). Briefly, bacteria were stained with a solution containing crystal violet (0.3 g crystal violet, 5 ml isopropanol, and 5 ml ethanol per liter of distilled water), and the cell length was measured with a micrometric-scale graduated ocular by phase-contrast microscopy. The extent of flagellation was evaluated as described below. Raft formation was analyzed by inspecting colonies at a ×400 magnification with an inverted microscope.
Quantification of extracellular flagellin.
The estimation of the amount of extracellular flagellin was considered a measure of the extent of bacterial flagellation (12, 34, 35). Total extracellular flagellin was prepared as previously described (34). Briefly, bacterial cultures grown in Tr broth for 8 h or bacterial suspensions obtained by harvesting cells from solid medium with cold water were normalized with respect to the optical density at 600 nm (OD600), vortexed for 5 min to detach flagella, and centrifuged at 5,000 × g for 15 min at 4°C. Flagellar filaments were collected from supernatants by high-speed centrifugation at 100,000 × g for 1 h and suspended in protein sample buffer containing β-mercaptoethanol. Protein samples were heated at 95°C for 10 min and subjected to SDS-PAGE, and the resulting gels were stained with silver nitrate. The intensity of the flagellin band was measured by densitometric analysis using Image Master ID software (Pharmacia Biotech).
Quantitative real-time RT-PCR.
Cell samples for RNA isolation were obtained from 10-ml Tr cultures incubated at 37°C for 6 h and normalized with respect to the OD600. RNA was extracted with the RNeasy minikit (Qiagen, Valencia, CA), modified as previously described (10). Protein contamination was excluded by measuring the A260/A280 ratio, and the RNA concentration was calculated by measuring the A260. Prior to retrotranscription, total RNA was subjected to PCR to exclude the possibility of DNA contamination. RNA (40 ng) was used for cDNA synthesis using the QuantiTec reverse transcription (RT) kit (Qiagen), as recommended by the manufacturer. Quantitative real-time PCR was carried out with a LightCycler instrument (Roche), using the QuantiFast SYBR green PCR kit (Qiagen). Reactions were performed with primers hagU4 (5′-TCATGCGATCCTTCAACGTG-3′) and hagL4 (5′-GCCATCGAGCAATTTCTTACC-3′), designed within the B. subtilis flagellin gene (hag) to give a 172-bp product. To confirm the specificity of the amplification product, a melting-curve analysis was performed. The quantification of gene expression was determined relative to a standard curve obtained by performing quantitative real-time PCR with known amounts of a hag gene amplicon. This amplicon was generated via conventional PCR using genomic DNA from B. subtilis PB5249 and primers hagU1 (5′-CCACAATATTGCAGCGCTTAA-3′) and hagL1 (5′-TAATAATTGAAGTACGTTTTG-3′). For each strain, quantitative real-time RT-PCR was done in duplicate, and the entire experiment was repeated three times on RNA samples extracted from independent cultures.
Statistical analysis.
Statistical analysis was performed with GraphPad InStat software. A P value of <0.05 was considered significant. Values are expressed as means ± standard deviations.
RESULTS AND DISCUSSION
Swimming motility and flagella.
Our previous data showed that, due to the presence of a functional copy of swrA, strain PB5249 carries a larger number of flagella and is more motile in a liquid environment than its isogenic swrA mutant, PB1831 (6, 34). These two strains carry a mutation in the sfp gene, the product of which is essential for surfactin biosynthesis (28), and are therefore unable to synthesize this biosurfactant.
To evaluate the relative contribution of SwrA and surfactin to B. subtilis swimming motility, PB1831 (swrA sfp mutant), PB5249 (swrA+ sfp mutant), PB1927 (swrA mutant sfp+), and PB5332 (swrA+ sfp+) were centrally seeded onto motility plates (0.2% agar), and the diameters of the growth halos were measured at 6 h postinoculation. All strains moved by swimming, as revealed by microscopic observations of the halos, in which individual cells rapidly alternated between tumbling and smooth swimming. As expected, due to the presence of a larger number of flagella, PB5249 migrated faster than PB1831 (30.4 ± 2.3 mm versus 10.3 ± 1.9 mm) (P < 0.001). The two surfactin-producing strains gave rise to similar swimming halos (40.1 ± 3.2 mm for PB5332 and 42.2 ± 1.6 mm for PB1927) (P = 0.22) that were always wider than those of the sfp mutant strains. The lack of differences in the motilities of the swrA+ and swrA mutant strains in the sfp+ background induced us to evaluate whether surfactin could affect the degree of flagellation in B. subtilis. To explore this hypothesis, all strains were grown in liquid medium, and preparations of cell surface flagellin were analyzed by SDS-PAGE. As shown in Fig. 1A, PB1927 and PB5332 showed 2.2- and 1.4-fold increases, respectively, in the amounts of cell surface flagellin compared to those of their relative sfp mutant strains.
Fig 1.
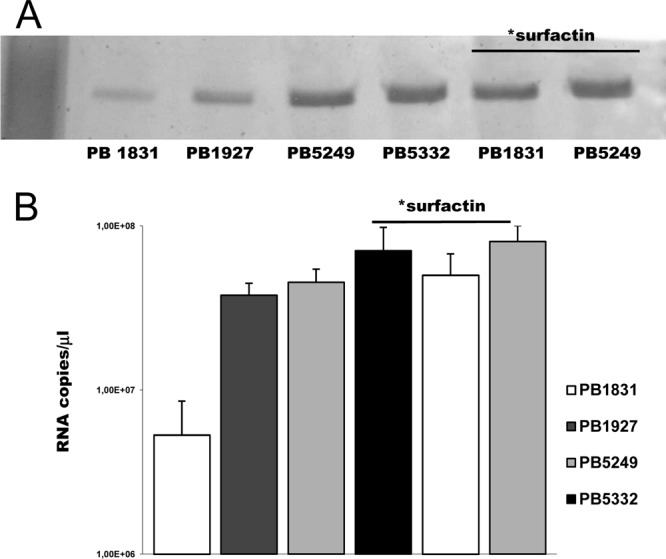
Extracellular flagellin and hag expression of B. subtilis cells grown in liquid cultures. (A) SDS-PAGE analysis of total extracellular flagellin collected from bacteria grown in broth cultures normalized to the same OD600. (B) hag RNA copies/μl determined by quantitative real-time RT-PCR (primer pair hagU4/hagL4) relative to a standard curve obtained with known amounts of a hag gene amplicon. “*surfactin” above the lanes indicates that 20 μM commercial surfactin was added to the culture.
To exclude that the genetic manipulation of PB1927 and PB5332 could have caused the observed increase in the amount of flagellin, sfp mutant strains PB1831 and PB5249 were grown in broth in the presence of commercial surfactin, and the amount of cell surface flagellin was compared to that produced by the same strains in the absence of exogenously added surfactin. As shown in Fig. 1A, growth in the presence of the biosurfactant led to an increase in the amount of extracellular flagellin in both strains (5.6-fold for PB1831 and 1.5-fold for PB5249). Control experiments were performed to exclude that this effect was due to a surfactin-induced detachment of flagella from the cell surface. Flagellin from strain PB5249 was prepared incubating cells in commercial surfactin before shaking for flagellar detachment. Flagellin was then purified and loaded onto SDS-PAGE gels and compared to preparations obtained without the addition of commercial surfactin to the cell suspension. No difference in the amounts of flagellin was evidenced.
To evaluate whether surfactin could have an effect on the expression of hag, encoding flagellin in B. subtilis, quantitative real-time RT-PCR was performed with primers hagU4 and hagL4 on total RNA extracted from the sfp mutant strains, with or without exogenously added surfactin, and the sfp+ strains. The expression level of hag was found to be 8.5-fold increased for PB5249 (4.5 × 107 ± 0.6 × 107 copies/μl) compared to that for PB1831 (5.3 × 106 ± 3.2 × 106 copies/μl) (P < 0.01) (Fig. 1B). This result agrees with previous data obtained by a hag-lacZ transcriptional fusion (34) and with the role of SwrA as positive regulator of σD-directed hag transcription (5). Interestingly, the hag expression level was also found to be increased in the presence of surfactin, either produced or exogenously added. In the swrA mutant background, the expression levels of hag were 7.1- and 9.4-fold lower for PB1831 than for the surfactin producer PB1927 (3.8 × 107 ± 0.6 × 107 copies/μl) and PB1831 grown in the presence of commercial surfactin (5.0 × 107 ± 0.2 × 107 copies/μl), respectively (P < 0.01). An increase in the hag expression level due to surfactin was also observed for the swrA+ strains, with the level of expression being 1.56- and 1.8-fold higher in PB5332 cells (7.0 × 107 ± 0.4 × 107 copies/μl) and PB5249 cells grown with exogenously added surfactin (8.0 × 107 ± 0.2 × 107 copies/μl), respectively, than those in PB5249.
Overall, these results confirm the activity of SwrA in increasing the flagellation of B. subtilis and demonstrate that the hag expression level is elevated in strains that produce surfactin as well as when surfactin is exogenously added. Recently, it was shown that flagellin expression is related to the production of a motility-enabling surfactant in Pseudomonas syringae, thus suggesting an intimate role between surfactant production and flagellar motility (3). Although in-depth studies are required to understand the mechanism of the surfactin effect on flagellar synthesis, a linkage between a biosurfactant molecule and flagella also appears to be valid for B. subtilis.
Surface migration.
To investigate the effect of surfactin and SwrA on surface motility, bacteria were spotted onto the centers of Tr plates with increasing viscosities (0.2%, 0.5%, 0.7%, 1%, 2%, and 3% agar) and allowed to grow for 24 h at 37°C. Among the media used to analyze B. subtilis surface motility, Tr was chosen since it was previously demonstrated to support the swarming of swrA+ strain PB5249 (6, 34). As shown in Fig. 2, all strains efficiently moved over plates with a 0.2% agar viscosity. When the agar concentration was increased to 0.5%, PB5249 and surfactin-producing strains PB1927 and PB5332 still invaded the medium surface, while PB1831 produced a smaller colony. Therefore, in the swrA mutant background, the secretion of surfactin allows B. subtilis to migrate even under conditions in which, without surfactin, it is unable to move and to produce flagella (34). Surfactin also increased surface migration on medium containing 0.7% and 1.0% agar. At 2% and 3% agar, none of the strains migrated efficiently over the surface. Interestingly, on plates containing 3.0% agar, a slight increase in the colony diameter was recorded for PB5332 compared to the colony diameters of the other strains.
Fig 2.
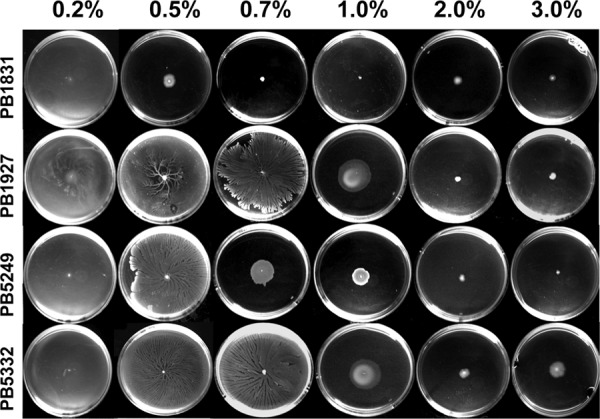
Growth of B. subtilis strains on media with different viscosities. Bacteria were centrally inoculated onto Tr plates solidified with different percentages of agar.
Sliding and swarming motility.
To investigate on the contribution of sliding and swarming to B. subtilis surface migration, we analyzed colonies for the formation of hyperflagellated swarm cells and cell rafting, two peculiar requirements for swarming motility (19). In fact, differently from Proteus mirabilis (16), in which swarm cell differentiation occurs in cyclic rounds, resulting in the appearance of characteristic terraced colonies (15, 26, 33), swarming B. subtilis cells produce a variety of colony patterns (e.g., monolayered, dendritic, or terraced), depending on the environmental conditions (19). Therefore, as also shown previously for other Bacillus species (11), the macroscopic appearance and wideness of colonies do not predict B. subtilis swarming.
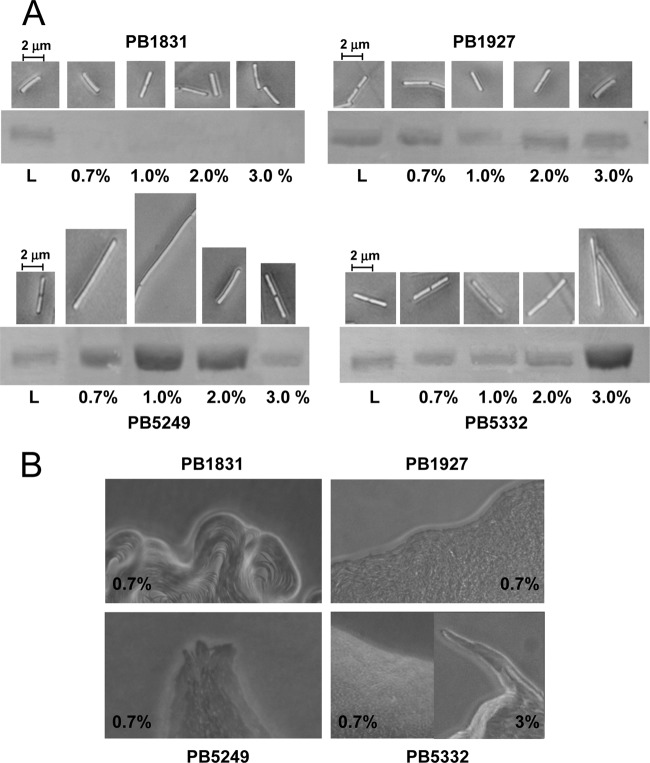
B. subtilis strains were grown on Tr containing increasing agar concentrations (0.7%, 1.0%, 2.0%, and 3.0%) for 6 h. This incubation time was chosen because a 6- to 8-h incubation is ideal to reveal the hyperflagellation of swarming PB5249 cells (6, 34), and an almost 2.5-h-long lag period precedes the initiation of swarming (20). The level of bacterial flagellation was estimated by measuring the amount of extracellular flagellin in preparations of purified flagellar filaments obtained from cell suspensions normalized to the same OD600 (12, 34, 35). Hyperflagellation was defined as a >3-fold increase in the amount of extracellular flagellin of bacteria collected from colonies compared to bacteria grown in liquid cultures. Although cell elongation has recently been debated as an indicator of B. subtilis swarming (19), we also analyzed this feature, since an increase in cell length was shown to occur in PB5249 during swarming (34).
On all solid media, strain PB1831 produced short rods (3.5 ± 1.0 μm) from which flagellin was never detected (Fig. 3A). In this strain, flagellin could be obtained only from swimmer cells collected from broth cultures. In contrast, strain PB1927 exhibited short flagellin-producing rods (3.4 ± 0.7 μm) under all tested culture conditions (Fig. 3A), suggesting that surfactin is also able to stimulate flagellar synthesis on solid media in the swrA mutant strain. The lack of hyperflagellation, cell elongation, and rafting (Fig. 3) in strain PB1927 indicates that, despite surfactin production, functional SwrA is essential for swarming. Due to the release of surfactin, however, this strain can move by sliding on moist surfaces, such as those of media containing ≤1.0% agar. This result agrees with previous work demonstrating that purified surfactin increases the colony expansion of B. subtilis strains deficient in surfactin biosynthesis (20) and favors the sliding motility of strains lacking flagella (23).
Fig 3.
Analysis of swarming features of B. subtilis strains. (A) Cell length (images obtained by phase-contrast microscopy) and surface flagellin (SDS-PAGE of extracellular flagellin) of strains grown on Tr plates solidified with different percentages of agar. L, liquid cultures. (B) Colony edges of B. subtilis strains grown on Tr medium containing 0.7% or 3% agar (magnification, ×200).
When propagated on Tr containing 0.7% to 2.0% agar, strain PB5249 exhibited longer cells (10.5 ± 2.0 μm) and a larger amount of flagellin than bacteria collected from liquid cultures (Fig. 3A). Hyperflagellation and an increase in cell length were also observed for strain PB5332 on Tr containing 3% agar. Under these conditions, bacteria moving in side-by-side cell groups or rafts were observed in a narrow area of the colony rim for both strains (Fig. 3B). Therefore, the presence of a functional copy of swrA makes PB5249 and PB5332 able to swarm. The finding that the agar conditions promoting the swarming of PB5249 and PB5332 cells are different can be explained by the fact that swarming requires a correct fluid environment generated as bacteria extract water from the underlying agar gel (wetness) (7). Swarming is also influenced by the surface wettability and, thus, by the presence of surfactants. When cell wetness is adequate (e.g., 0.7 to 2.0% agar) to support swarming by the swrA+ sfp mutant strain, an increase in wettability due to surfactin inhibits swarming but promotes bacterial spreading. However, when the wetness is reduced (e.g., 3% agar) and the swarmers cannot be produced by the sfp mutant strain, surfactin promotes wettability and allows swarming migration.
Conclusions.
By the analysis of B. subtilis strains able or unable to produce SwrA and surfactin, this work allowed us to highlight an increased flagellar synthesis in the presence of surfactin and to dissect the contribution of SwrA and surfactin to B. subtilis motility over surfaces. In the absence of a functional copy of swrA, which is required for swarming, the biosurfactant enables B. subtilis to colonize by sliding on sufficiently moist surfaces. When functional SwrA is produced, surfactin promotes active spreading at low viscosities and swarming on surfaces at high viscosities. The overall evidence suggests that undomesticated B. subtilis strains, most often provided with surfactin and SwrA, can move fast in liquid environments and use sliding or swarming motility to efficiently colonize different solid matrixes. This property may be of evident adaptive value in the colonization of hydrated soils, fruits, and plant roots by this organism.
ACKNOWLEDGMENTS
We are grateful to Alessandro Galizzi and Daniel Kearns for helpful discussions and suggestions for improving the manuscript.
This work was supported by National Research Project grant 2005058814 from the Ministero dell'Istruzione, dell'Università e della Ricerca.
Footnotes
Published ahead of print 6 July 2012
REFERENCES
- 1. Angelini TE, Roper M, Kolter R, Weitz DA, Brenner MP. 2009. Bacillus subtilis spreads by surfing on waves of surfactant. Proc. Natl. Acad. Sci. U. S. A. 106:18109–18113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc. Nat. Acad. Sci. U. S. A. 98:11621–11626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burch AY, et al. 2012. Pseudomonas syringae coordinates production of a motility-enabling surfactant with flagellar assembly. J. Bacteriol. 194:1287–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Butler MT, Wang Q, Harshey RM. 2010. Cell density and motility protect swarming bacteria against antibiotics. Proc. Natl. Acad. Sci. U. S. A. 107:3776–3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Calvio C, Osera C, Amati G, Galizzi A. 2008. Autoregulation of swrAA and motility in Bacillus subtilis. J. Bacteriol. 190:5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Calvio C, et al. 2005. Swarming differentiation and swimming motility in Bacillus subtilis are controlled by swrA, a newly identified dicistronic operon. J. Bacteriol. 187:5356–5366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen BG, Turner L, Berg HC. 2007. The wetting agent required for swarming in Salmonella enterica serovar Typhimurium is not a surfactant. J. Bacteriol. 189:8750–8753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Compant S, Duffy B, Nowak J, Clément C, Barka EA. 2005. Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71:4951–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Earl AM, Losick R, Kolter R. 2008. Ecology and genomics of Bacillus subtilis. Trends Microbiol. 16:269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ghelardi E, et al. 2002. Requirement of flhA for swarming differentiation, flagellin export, and secretion of virulence-associated proteins in Bacillus thuringiensis. J. Bacteriol. 184:6424–6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ghelardi E, et al. 2007. Swarming behavior of and hemolysin BL secretion by Bacillus cereus. Appl. Environ. Microbiol. 73:4089–4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gygi DM, Bailei G, Allison C, Huges C. 1995. Requirement for FlhA in flagellar assembly and swarm cell differentiation by Proteus mirabilis. Mol. Microbiol. 15:761–769 [DOI] [PubMed] [Google Scholar]
- 13. Hamon MA, Lazazzera BA. 2001. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 42:1199–1209 [DOI] [PubMed] [Google Scholar]
- 14. Hamze K, et al. 2011. Single-cell analysis in situ in a Bacillus subtilis swarming community identifies distinct spatially separated subpopulations differentially expressing hag (flagellin), including specialized swarmers. Microbiology 157:2456–2469 [DOI] [PubMed] [Google Scholar]
- 15. Harshey RM. 2003. Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57:249–273 [DOI] [PubMed] [Google Scholar]
- 16. Hauser G. 1885. Uber faulnissbacterien und deren beziehungen zur septicamie, p 107. FGW Vogel, Leipzig, Germany [Google Scholar]
- 17. Henrichsen J. 1972. Bacterial surface translocation: a survey and a classification. Bacteriol. Rev. 36:478–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. James BL, Kret J, Patrick JE, Kearns DB, Fall R. 2009. Growing Bacillus subtilis tendrils sense and avoid each other. FEMS Microbiol. Lett. 298:12–19 [DOI] [PubMed] [Google Scholar]
- 19. Kearns DB. 2010. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8:634–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kearns DB, Losick R. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 49:581–590 [DOI] [PubMed] [Google Scholar]
- 21. Kearns DB, Losick R. 2005. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 19:3083–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kearns DB, Chu F, Rudner R, Losick R. 2004. Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol. Microbiol. 52:357–369 [DOI] [PubMed] [Google Scholar]
- 23. Kinsinger RF, Shirk MC, Fall R. 2003. Rapid surface motility in Bacillus subtilis is dependent on extracellular surfactin and potassium ion. J. Bacteriol. 185:5627–5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lai S, Trembley J, Déziel E. 2009. Swarming motility: a multicellular behavior conferring antimicrobial resistance. Environ. Microbiol. 11:126–136 [DOI] [PubMed] [Google Scholar]
- 25. López D, Fischbach MA, Chu F, Losick R, Kolter R. 2009. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 106:280–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McCarter LL. 2004. Dual flagellar systems enable motility under different circumstances. J. Mol. Microbiol. Biotechnol. 7:18–29 [DOI] [PubMed] [Google Scholar]
- 27. McLoon AL, Guttenplan SB, Kearns DB, Kolter R, Losick R. 2011. Tracing the domestication of a biofilm-forming bacterium. J. Bacteriol. 193:2027–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakano MM, Corbell N, Besson J, Zuber P. 1992. Isolation and characterization of sfp: a gene that functions in the production of the lipopeptide biosurfactant, surfactin, in Bacillus subtilis. Mol. Gen. Genet. 232:313–321 [DOI] [PubMed] [Google Scholar]
- 29. Nakano MM, Marahiel MA, Zuber P. 1988. Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. J. Bacteriol. 170:5662–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ogura M, Tsukahara K. 9 April 2012, posting date. SwrA regulates assembly of Bacillus subtilis DegU via its interaction with N-terminal domain of DegU. J. Biochem. doi:10.1093/jb/mvs036. [DOI] [PubMed]
- 31. Patrick JE, Kearns DB. 2009. Laboratory strains of Bacillus subtilis do not exhibit swarming motility. J. Bacteriol. 191:7129–7133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Patrick JE, Kearns DB. 2012. Swarming motility and the control of master regulators of flagellar biosynthesis. Mol. Microbiol. 83:14–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rather PN. 2005. Swarmer cell differentiation in Proteus mirabilis. Environ. Microbiol. 7:1065–1073 [DOI] [PubMed] [Google Scholar]
- 34. Senesi S, et al. 2004. Surface-associated flagellum formation and swarming differentiation in Bacillus subtilis are controlled by the ifm locus. J. Bacteriol. 186:1158–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Senesi S, et al. 2002. Swarming motility in Bacillus cereus and characterization of a fliY mutant impaired in swarm cell differentiation. Microbiology 148:1785–1794 [DOI] [PubMed] [Google Scholar]