Abstract
Soilless medium-based horticulture systems are highly prevalent due to their capacity to optimize growth of high-cash crops. However, these systems are highly dynamic and more sensitive to physiochemical and pH perturbations than traditional soil-based systems, especially during nitrification associated with ammonia-based fertilization. The objective of this study was to assess the impact of nitrification-generated acidification on ammonia oxidation rates and nitrifying bacterial community dynamics in soilless growth media. To achieve this goal, perlite soilless growth medium from a commercial bell pepper greenhouse was incubated with ammonium in bench-scale microcosm experiments. Initial quantitative real-time PCR analysis indicated that betaproteobacterial ammonia oxidizers were significantly more abundant than ammonia-oxidizing archaea, and therefore, research focused on this group. Ammonia oxidation rates were highest between 0 and 9 days, when pH values dropped from 7.4 to 4.9. Pyrosequencing of betaproteobacterial ammonia-oxidizing amoA gene fragments indicated that r-strategist-like Nitrosomonas was the dominant ammonia-oxidizing bacterial genus during this period, seemingly due to the high ammonium concentration and optimal growth conditions in the soilless media. Reduction of pH to levels below 4.8 resulted in a significant decrease in both ammonia oxidation rates and the diversity of ammonia-oxidizing bacteria, with increased relative abundance of the r-strategist-like Nitrosospira. Nitrite oxidizers (Nitrospira and Nitrobacter) were on the whole more abundant and less sensitive to acidification than ammonia oxidizers. This study demonstrates that nitrification and nitrifying bacterial community dynamics in high-N-load intensive soilless growth media may be significantly different from those in in-terra agricultural systems.
INTRODUCTION
Greenhouse horticulture has expanded considerably over the past 4 decades due to increased global demand for high-value foods and ornamentals and, specifically, out-of-season high-quality produce (23, 33, 46). In many cases, greenhouse cultivation has moved from soil-based systems to commercial-scale soilless medium-based systems that enable increased crop productivity and efficiency with significant reduction of soil-associated plant pathogens (45). Although medium-free hydroponic systems are sometimes used, most contemporary soilless culture is based on growth in artificial substrates such as sand, stone wool, polyurethane, vermiculite, perlite, and scoria (tuff) (42). These media provide plants with physical support and a steady reservoir of nutrients, water, and oxygen and simplify the capacity to monitor and regulate key agronomic parameters such as electrical conductivity, pH, and temperature (43). Although soilless media used in greenhouse horticulture initially contain very low levels of microbial communities, they are rapidly colonized and develop complex communities of heterotrophic (8, 31, 34) and nitrifying (24) bacteria within a few weeks after planting, and therefore, artificial microbial inoculants are generally not introduced into these systems. The sources of the initial microbial inocula in these systems are believed to be the seedlings, the irrigation water, and aerosols (9).
Fertigation (the application of fertilizers, soil amendments, or other water-soluble products through irrigation systems) is frequently applied in soilless greenhouse cultivation systems because it enables optimization of nutrient (most notably nitrogen, phosphorus, and potassium) supply. Although urea is the cheapest and most concentrated nitrogen source fertilizer, it requires hydrolysis to ammonium before it can be utilized by plants; therefore, soilless culture systems more frequently apply ammonium and nitrate fertilizers (42). The ratio between ammonium and nitrate has great agricultural significance and affects both plants and soil/growth media. Different plant species require different ammonium/nitrate (RN) ratios for optimal growth, and the correct ratio to be applied also varies with temperature, growth stage, rhizosphere pH, and soil properties (42).
Ammonium-based nutrition is generally more beneficial to plant growth than nitrate-based nutrition. In well-buffered systems, moderate reductions in pH due to nitrification and root excretion of protons increase uptake of micronutrients, which prevent plant growth disorders induced by micronutrient deficiencies (42). However, application of ammonium-based fertilizer in low-buffered soils and soilless media may result in rapid nitrification-associated soil acidification, which can significantly modify the soil microbiome and have a deleterious impact on crop growth. The depth and intensity of acidification are affected by a multitude of parameters including soil/medium type, level of organic matter, rate and type of N fertilizer (36), the abundance and activity of nitrifying prokaryotes, and more specifically ammonia-oxidizing prokaryotes (AOP), which lower the medium's pH (1).
The immense significance of nitrification to both soil ecology and plant nutrition has generated a multitude of studies focused on elucidating the diversity, distribution, and activity of AOP in soils in general (2, 5, 20, 21, 27, 28, 41) and specifically in agricultural soils (17–19, 32, 47, 48). In many cases, the functional gene amoA, which encodes the subunit containing the putative ammonia oxygenase (AMO) enzyme active site, is used as a phylogenetic marker for characterization of ammonia-oxidizing bacteria and archaea in environmental samples (14, 37). Several recent studies have assessed how soil pH impacts AOP distribution and abundance, including a recent study that tested the specialization of terrestrial archaeal ammonia oxidizers over a pH gradient (16). Nicol et al. (2008) found that both ammonia-oxidizing bacteria (AOB) and ammonia-oxidizing archaea (AOA) were characterized by distinct lineages in acid versus neutral soils but that archaeal amoA genes were by far the dominant group in acidic soils (28). This was also supported by Yao et al. (49), who found that nitrification was driven mainly by AOA in acidic soils and that specific AOA and AOB populations dominated in these low pH soils. These studies provided substantial insight into the correlation between soil pH and AOP community structure in different soils; however, they did not evaluate nitrification and AOP community dynamics under continued acidification.
The objective of this study was to assess the impact of short-term acidification due to nitrification on nitrification rates and nitrifying bacterial community dynamics in soilless growth media. Microcosm experiments were conducted with perlite (an amorphous volcanic glass substrate frequently used as an artificial soil medium in greenhouse agriculture) from an established commercial greenhouse horticulture system incubated in ammonia-amended nonbuffered medium. Time-dependent pH-associated fluctuations in nitrogen species were measured concomitant to microbial community dynamics using molecular fingerprinting (denaturing gradient gel electrophoresis [DGGE]), real-time PCR, and pyrosequencing of bacterial and archaeal amoA and 16S rRNA genes. AOA abundance in the microcosm experiments was significantly lower than AOB; therefore, assessment of AOP diversity and community dynamics relative to pH fluctuations focused on betaproteobacterial ammonia oxidizers.
MATERIALS AND METHODS
Experimental setup.
Two-year-old perlite that served as a soilless growth medium for sweet peppers in an experimental greenhouse (Bsor experimental station, Israel) was used to assess nitrification and nitrifying microbial community dynamics in the bench-scale microcosm experiments described below.
Prior to the experiments, the perlite was irrigated (with water) daily to saturation in a greenhouse at 25°C for a week and then fertigated (irrigated with fertilizer) for an additional week in order to enhance the relative abundance of nitrifying bacteria. The pH and electrical conductivity (EC) of the fertigation solution were 6.5 ± 0.4 and 1.3 ± 0.1 dS m−1, respectively. The N, P, K, Ca, and Mg concentrations in the irrigation solution were 100, 25, 150, 80, and 25 mg liter−1, respectively. The nutrient solutions were prepared from commercial fertilizers: KNO3, KCl, Ca(NO3)2, Mg(NO3)2, H3PO4, and tap water containing Ca, Mg, Na, and Cl at 40, 5, 30, and 30 mg liter−1, respectively. Micronutrients Zn, Fe, Cu, B, and Mo, all EDTA based, were applied at concentrations of 0.25, 1.0, 0.02, 0.2, and 0.02 mg liter−1, respectively. During this period, ammonium was measured daily to verify AOP activity in the mature perlite.
Following the 2 weeks, 10-g perlite aliquots were transferred to 2-liter Erlenmeyer flasks containing 500 ml of tap water supplemented with 1 mm of KH2PO4 and 1 mm of KCl. The bench-scale experiments had four initial solution pH values (4.5 ± 0.1, 5.5 ± 0.1, 6.2 ± 0.2, and 7.4 ± 0.3) and were conducted in four replicates. The Erlenmeyer flasks were incubated at 30°C for 1 week (preequilibration period), during which the pH values were adjusted by adding appropriate amounts of 0.1 m HCI or NaOH as needed. At the end of this period, 50 mg N liter−1 in the form of (NH4)2SO4 was added, and the Erlenmeyer flasks were incubated at 30°C without further intervention. Aqueous and perlite samples were taken for chemical and microbial analyses as described below.
Physiochemical analyses.
Physical and chemical parameters were measured at selected time points in 15-ml subsamples taken from the Erlenmeyer flasks. The pH and EC were measured, and the samples were filtered through a 0.45-μm membrane (Millipore Corporation, Bedford, MA). To stop the nitrification activity, 100 μl 1% methiolate (ethylmercurithio-salicylic acid, sodium salt) was added to the subsamples, and the solutions were frozen and kept for chemical analyses. NH4+, NO3−, and NO2− concentrations were later measured with a Lachat Autoanalyzer (Lachat Instruments, Milwaukee, WI).
DNA extraction, PCR, and DGGE.
Perlite subsamples were periodically removed from the Erlenmeyer flasks using a long, sterile spatula. DNA was extracted from 0.3 g of perlite using the PowerSoil DNA isolation kit (MoBio, Carlsbad, CA). Extracted DNA was visualized by electrophoresis in 1% agarose gels and quantified spectrophotometrically by NanoDrop (NanoDrop Technologies, Wilmington, DE).
PCR amplification of partial bacterial amoA gene sequences for DGGE analysis was performed on ∼10 ng of extracted DNA template using the amoA1F/amoA2R primer set with a 40-bp GC clamp attached to its 5′ end (32, 37). PCR mixtures included, in a final volume of 50 μl, 1.5 U Taq DNA polymerase (DreamTaq; Fermentas Life Science, Vilnius, Lithuania), Taq buffer containing a final magnesium concentration of 2.5 mM, deoxynucleoside triphosphates (dNTPs) (20 nmol each), 12.5 μg bovine serum albumin, and 25 pmol of each primer, and the reactions were carried out as previously described (32). PCR amplification was verified by agarose gel electrophoresis (1%) and staining with ethidium bromide.
DGGE was performed in 6% (wt/vol) acrylamide gels containing a linear urea-formamide gradient ranging from 20 to 60% denaturant (with 100% defined as 7 M urea and 40% [vol/vol] formamide). Gels were run for 17 h at 100 V in the Dcode Universal Mutation system (Bio-Rad Laboratories, Hercules, CA). DNA was visualized after staining with Gelstar (Invitrogen Corporation, Carlsbad, CA) by UV transillumination (302 nm) and photographed using a Kodak KDS digital camera (Kodak Co., New Haven, CT).
qPCR.
Quantitative PCR (qPCR) targeting bacterial and archaeal amoA genes at selected time points of the incubation experiment was accomplished using an MX 3000 Real Time PCR system (Stratagene, La Jolla, CA). Amplification was carried out using the bacterial amoA primer set amoA1F/amoA2R and the archaeal amoA primer set Arch-amoAF/Arch-amoAR (15). Standard curves for the archaeal and bacterial amoA genes were prepared by decimally diluting plasmids containing partial amoA amplicons to range from 109 to 100 copy numbers. Each real-time (RT)-PCR mixture contained 12.5 μl Absolute Blue SYBR green ROX Mix (Thermo Fisher Scientific, Surrey, United Kingdom), 1 μl of each 0.2 mM primer, and 1 μl of DNA template and was brought to a final volume of 25 μl by addition of 9.5 μl ultrapure PCR-grade water. The PCR program began with a hot-start step of 15 min at 95°C, required for the activation of the DNA polymerase, followed by 40 cycles each consisting of 95°C for 40 s, 53°C/57°C for 40 s, and 72°C for 40 s, followed by a final melting curve stage with temperature ramping from 53 to 95°C. Endpoint measurements were made at 72°C for 10 s. Melting curve analysis of the PCR products was conducted following each assay to confirm that the fluorescence signal originated from specific PCR products. All the real-time PCRs were done in triplicate for both the standards and the perlite-extracted DNA samples. Baseline and threshold calculations were performed with the MxPro QPCR software analysis tools (Stratagene, La Jolla, CA).
High-throughput sequencing.
Sequencing was performed by the 454 sequencing method using tag-encoded FLX gene amplicon pyrosequencing (bTEFAP) at the Research and Testing Laboratories (Lubbock, TX). PCR for pyrosequencing targeting general bacterial and archaeal communities was conducted using the 16S rRNA eubacterial primer set Gray 28F and Gray 519R and the archaeal primer set A340F90 and a806R96 as previously described (41), while PCR targeting AOBs was carried out using the amoA primer pair amoA-1F and amoA-2R (37) using the previously defined bTEFAP protocol (10). High-throughput sequencing of perlite-extracted DNA from selected time points (0, 3, 6, 9, 27, 34, and 48 days and 0, 2, 3, 6, 9, 13, 27, 34, and 48 days following ammonia amendment for the amoA and 16S rRNA gene amplicons, respectively) yielded 35,949 raw amoA and 41,319 raw 16S rRNA sequence reads.
Bioinformatic and statistical analyses.
Betaproteobacterial amoA and 16S rRNA amplicons from the different time points were trimmed, aligned, clustered, checked for chimeras (16S rRNA amplicons only), and classified using the MOTHUR software package (39). The amoA amplicons were aligned with 9,188 bacterial amoA genes from the FunGene functional gene pipeline and repository database (http://fungene.cme.msu.edu/index.spr); and 16S rRNA amplicons were aligned using the Silva consensus sequence database on the MOTHUR website (http://www.mothur.org/wiki/Alignment_database). The amoA downloaded reference sequences were aligned using MAFFT Web-based multiple alignment program (MAFFT version 6, http://mafft.cbrc.jp/alignment/server/, FFT-NS-1-Progresive method). The sequences generated by the 454 method were aligned to the reference sequences with the MOTHUR alignment tool using default settings. Following alignment, the sequences were cut to defined lengths of 455 and 967 aligned characters (including gaps), resulting in a total of 15,642 and 5,962 sequences for the amoA and 16S rRNA gene amplicons, respectively. A distance matrix was calculated for all sequences, and operational taxonomic units (OTUs) were defined for 80% and 97% similarity for amoA and 16S rRNA genes, respectively, as suggested by Purkhold et al. (35). Using a group annotation file, a matrix of samples versus OTU was created and further analyzed using multivariate statistics.
Phylogenetic analysis.
The phylogenetic affiliation of the bTEFAP-generated perlite-associated amoA sequences from the selected time points was determined using the Mega5 (Molecular Evolutionary Genetics Analysis) phylogenetic package (44). The evolutionary history was inferred via the neighbor-joining method (38) using amino acid sequences generated from DNA sequences. The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the taxa analyzed (12). Branches corresponding to partitions reproduced in <50% bootstrap replicates are collapsed. The evolutionary distances were computed using the Poisson correction method (50) and are given in units representing the number of amino acid substitutions per site. The analysis involved 47 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 76 positions in the final data set.
Statistical analysis.
Clustering and ordination of the bacterial ammonia-oxidizing community, based on amoA OTUs from the seven analyzed time points defined above, were accomplished via nonmetric multidimensional scaling (NMDS) followed by verification with the multiple-response permutation procedure (MRPP) with a significance set at P values of <0.01 using the PC-ord Multivariate Analysis of Ecological Data software package (MjM Software, Gleneden Beach, OR).
RESULTS
Chemical parameters.
Fluctuations in pH (Fig. 1), ammonium (Fig. 2), and nitrate (see Fig. S1 in the supplemental material) were measured in all four initial pH profiles (4.5, 5.5, 6.2, and 7.4) of the soilless culture microcosm experiment over a period of 90 days subsequent to the initial ammonium amendment. Nitrification (as visualized by ammonium oxidation and the corresponding nitrate formation) in the 5.5, 6.2, and 7.4 initial pH profiles was highest between days 0 and 9, when pH dropped from 7.4 to 4.9 (nitrification was not initially observed in the initial pH 4.5 profile). During this period, nitrite did not accumulate in the vessels (except for day 3 of the initial pH 7.4 profile, when a concentration of 0.04 mM was measured), indicating substantial activity of nitrite-oxidizing bacteria. Ammonium and corresponding nitrate concentrations (Fig. 1; see Fig. S1 in the supplemental material) indicated that nitrification continued at much lower rates at pH values of 3 to 4.8.
Fig 1.
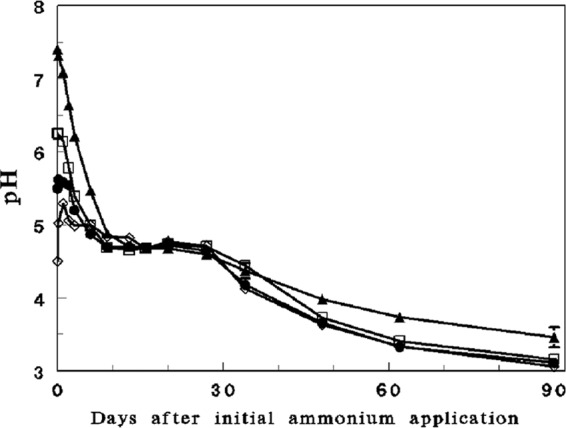
Time-dependent fluctuations in pH in the different initial pH profiles of the microcosm experiment over a period of 90 days of incubation. Initial pHs: 7.4 (filled triangles); pH 6.2 (open squares); 5.5 (filled circles); and 4.5 (open diamonds).
Fig 2.
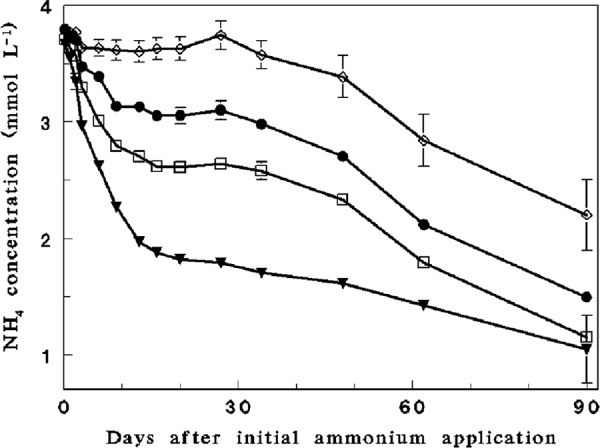
Time-dependent fluctuations in ammonium concentration in the different initial pH profiles of the microcosm experiment over a period of 90 days of incubation. Initial pHs: 7.4 (filled triangles); 6.2 (open squares); 5.5 (filled circles); and 4.5 (open diamonds).
qRT-PCR.
Preliminary qRT-PCRs targeting betaproteobacterial and archaeal amoA genes at five time points of the initial pH 7.4 profile indicated that bacterial ammonia oxidizers were significantly more abundant than their archaeal counterparts (see Fig. S2A in the supplemental material). In addition, pyrosequencing of archaeal 16S rRNA gene amplicons (see Fig. S2B in the supplemental material) resulted in extremely low specificity (70 to 90% of the 3,910 generated sequences were actually bacteria). Previous experiments that used identical pyrosequencing conditions to detect archaea in environmental samples (41; Scot Dowd, personal communication) determined that this phenomenon occurs when ratios of archaea to bacteria in environmental samples are low. Collectively, these results led us to conclude that betaproteobacterial ammonia oxidizers were the dominant AOPs in the soilless culture microcosms, and we therefore focused on this group for the remainder of the analyses described below and did not further investigate AOA community dynamics.
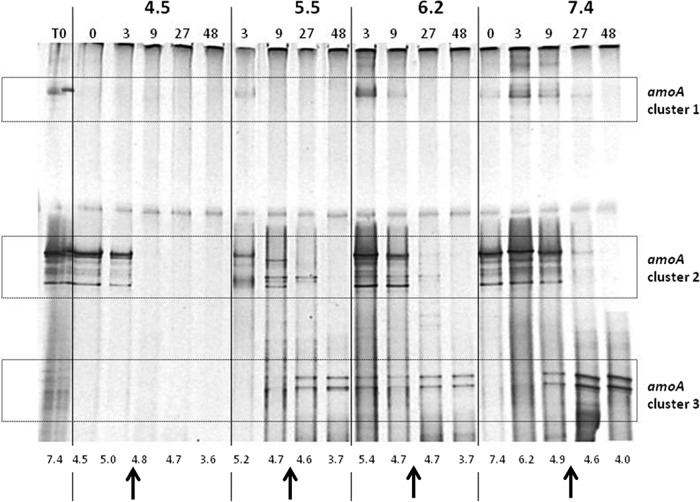
DGGE.
The community composition of betaproteobacterial ammonia-oxidizing organisms in the perlite of the 4 initial pH profiles at selected time points was visualized using DGGE targeting betaproteobacterial amoA amplicons (Fig. 3). Initial analyses of duplicate biological samples (in separate Erlenmeyer flasks) showed almost identical band patterns, and therefore, only one replicate for each time point is shown. Three band clusters characterized by their migration distance in the acrylamide gel were detected. Previous analyses using identical primer sets and PCR and DGGE conditions (6, 32; Dror Minz, personal communication) have constitutively shown that all amoA bands migrating to the upper third of the gel (Fig. 3, clusters 1 and 2) are phylogenetically associated with Nitrosomonas strains, whereas amoA bands migrating to the lower third of the DGGE gel (Fig. 3, cluster 3) were determined to be phylogenetically related to Nitrosospira strains. Using this stipulation, we ascertained that Nitrosomonas strains were more abundant at pH levels above 4.8 and Nitrosospira strains were dominant at pH levels under 4.6 (Fig. 2).
Fig 3.
AOB community dynamics depicted by DGGE of amplified betaproteobacterial amoA gene amplicons in the four initial pH profiles of the microcosm experiment: 4.5, 5.5, 6.2, and 7.4. Bold numbers above the gel indicate the initial pH values. Labels above the gel show the time (in days) following initial ammonia amendment (T0 is the initial preincubation perlite inoculant). Numbers below the gel indicate the actual pH values during the time of sampling. Arrows indicate the critical pH zone (4.6 to 4.8), in which ammonium oxidation rates dropped significantly and the AOB community shifted from Nitrosomonas-like phylotypes (clusters 1 and 2) to Nitrosospira-like phylotypes (cluster 3).
High-throughput DNA pyrosequencing of bacterial amoA genes.
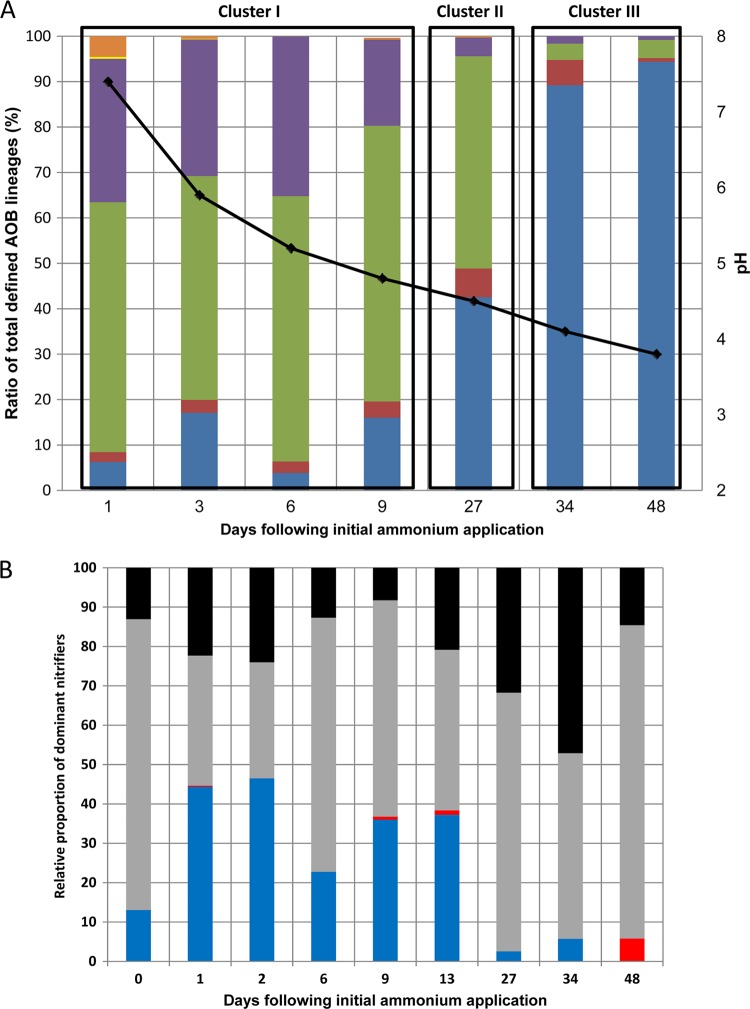
Pyrosequencing of amplified betaproteobacterial partial amoA gene fragments was conducted at selected time points of the initial pH 7.4 profile in order to assess the diversity and relative abundance of ammonia-oxidizing bacteria in the perlite as they correlated to pH (Fig. 1), ammonia (Fig. 2), and nitrate (see Fig. S1 in the supplemental material) values. AmoA amplicons were grouped into 15 taxonomical groups using an 80% nucleotide sequence similarity cutoff criterion and phylogenetically characterized relative to known betaproteobacterial amoA sequences (see Fig. S3 in the supplemental material). The ammonia-oxidizing community composition significantly shifted in response to ammonia oxidation-associated reductions in pH over the course of the incubation experiment. Cluster analysis of the amoA amplicons revealed three time/pH-associated clusters (see Fig. S4 in the supplemental material) that were characterized by significant differences in AOB relative abundance (Fig. 4A). Days 0 to 9 (cluster I) were dominated by Nitrosomonas communis-associated lineages (subgroups A and B), and day 27 (cluster 2) was characterized by relatively equal abundances of Nitrosomonas and Nitrosospira strains, whereas days 34 to 48 were characterized by a significant reduction in the relative abundance of nitrosomonads and a significant increase in the relative abundance of Nitrosospira cluster III-affiliated OTUs (Fig. 4A). These results strongly correlated with the DGGE analyses, wherein pH levels above 4.8 were characterized by higher levels of Nitrosomonas-associated phylotypes and pH values below 4.6 were dominated by Nitrosospira-associated phylotypes. Reduction in pH also resulted in a significant decrease in both ammonia-oxidizing and general bacterial species' richness and diversity (especially after day 27), as visualized by Chao and Shannon indices, respectively (see Fig. S5 in the supplemental material).
Fig 4.
(A) Relative abundance of amoA-defined AOB lineages in relation to time-dependent pH values at selected time points of the initial pH 7.4 profile of the microcosm experiment. Nitrosospira cluster I (blue); Nitrosospira cluster III (red); Nitrosomonas communis lineage subgroup A (green); Nitrosomonas communis lineage subgroup B (purple); Nitrosomonas communis lineage subgroup C (yellow); Nitrosomonas oligotropha lineage (orange). The black line shows time-dependent pH fluctuations (right x axis). (B) Relative proportion of dominant nitrification-associated bacterial genera relative to the total nitrifier abundance at selected time points of the initial pH 7.4 profile of the microcosm experiment based on pyrosequencing of 16S rRNA gene amplicons. Nitrosomonas (blue bars); Nitrosospira (red bars); Nitrospira (gray bars); Nitrobacter (black bars).
High-throughput DNA pyrosequencing of 16S rRNA gene fragments.
Pyrosequencing of general bacterial 16S rRNA gene amplicons was applied to the initial pH 7.4 profile of the microcosm experiment to obtain a broader overview of nitrifying bacterial dynamics in the soilless culture microcosm experiments. Phylogenetic characterization found that nitrifying bacteria made up between 2.5 and 21% of the genus level-defined organisms at the various time points of the incubation experiment. The betaproteobacterial ammonia oxidizers Nitrosomonas and Nitrosospira and the nitrite-oxidizing genera Nitrobacter (alphaproteobacterial class) and Nitrospira (nitrospira class) were the primary nitrifying bacteria detected (Fig. 4B). Nitrosomonas represented between 10 and 45% of the total fraction of bacterial nitrifiers up to day 13, after which its relative abundance significantly dropped, similar to the results of the pyrosequencing analysis targeting amoA amplicons (Fig. 3A). However, although like in the amoA analysis, the Nitrosospira/Nitrosomonas ratio significantly increased at the later time points, the overall AOB abundance from day 27 onward represented less than 6% of the total nitrifying bacterial community (Fig. 4B). It appeared that the nitrite oxidizers were much more resistant to acidification, with Nitrospira and Nitrobacter representing between 30 and 70% and between 8 and 45%, respectively, of the nitrifying bacterial community during the entire duration of the experiment (Fig. 4B).
DISCUSSION
Preliminary qPCR and pyrosequencing of archaeal 16S rRNA gene amplification analyses (see Fig. S2A and B in the supplemental material) indicated that bacteria and not archaea were the prominent AOPs in the soilless medium microcosm experiments. The proportion of AOA to AOB in agricultural soils based on current literature is extremely ambiguous. While several recent studies have determined that AOA dominate the active ammonia-oxidizing microbial community in some agricultural soils (17, 25, 28), other studies have shown that ammonia oxidation activity is primarily associated with AOB (19). It has been suggested that the AOB/AOA ratio is directly correlated to inorganic nitrogen loads. For example, Fan et al. showed that increased inorganic fertilization led to a 6- to 60-fold increase in the AOB/AOA ratio (11). Integration of our results with these previous studies suggests that ammonia oxidation in ammonium-fertigated soilless culture systems is primarily associated with AOB and not AOA.
DGGE and pyrosequencing of amoA and 16S rRNA gene amplicons showed that the soilless medium microcosm was initially dominated by members of the Nitrosomonas communis lineage (Fig. 4A), contrary to most molecular-method-based studies, which have found Nitrosospira to be the major ammonium-oxidizing genus in agricultural soils (3, 4, 21, 22, 26). Fierer et al. hypothesized that N additions may result in a shift from a more oligotrophic bacterial community to one that is more copiotrophic, analogous to a shift from K to r selection, terms often used to describe plant species ecology (13). This is supported by a recent study targeting amoA gene fragments in rice paddy soils, which found that the AOB community was predominantly composed of members of the Nitrosomonas communis cluster and that the relative abundance of Nitrosomonas increased relative to Nitrosospira with the increase of N fertilization, particularly in soils exposed to high oxygen concentrations (47). Ecological analyses of both soil (18, 29, 47) and wastewater (40) treatment environments have indeed supported the notion of Nitrosomonas strains as r strategists, with low substrate affinities and high maximum activity compared with the K strategist Nitrosospira.
The significant decline in Nitrosomonas abundance relative to that of Nitrosospira following medium acidification to pH values below 4.4 to 4.8 (see Fig. S1B in the supplemental material; Fig. 3A and 3B) supports the notion that copiotrophic strains may be less resilient to environmental stress than oligotrophic ones. Although results indicate that Nitrosospira strains are much more resilient to acidic pH than Nitrosomonas strains, the low abundance of AOB in the 16S rRNA gene-targeted pyrosequencing analyses of the perlite fragments (Fig. 3A) indicates that AOB in soilless culture media are significantly more sensitive to acidification than nitrite-oxidizing bacteria. Nonetheless, evidence of continued ammonia oxidation (at much lower rates), which characterized the microcosm profiles from day 27 onward (Fig. 1 and 2; see Fig. S1 in the supplemental material), may imply that the dominant Nitrosospira cluster 10-associated strains were still active under extremely acidic pH values. Several cultivation-based and molecular-method-based studies have determined that Nitrosospira is indeed the dominant AOB genus in acidic soils, where clusters 2, 3, and 4 are generally most abundant (7, 20, 21, 30). Indeed, the dominant Nitrosospira-associated OTU (NSP1) shared high sequence similarity with an uncultured ammonia-oxidizing bacterium (GU048997) from Chinese polytunnel greenhouse vegetable soil that is generally characterized by pH values below 5.5 (47).
This study demonstrated that intensive ammonia-based fertilization in soilless culture media may result in significant differences in both nitrification dynamics and AOP community composition, relative to traditional soil-based agricultural systems. We show that under neutral and slightly acidic pH, r strategists like Nitrosomonas strains are the dominant AOB due to their low substrate affinity and high maximum activity, but that with increasing pH stress K strategists like Nitrosospira strains, characteristically dominant in soils, become more abundant. Intensive horticulture based on growth in soilless media is becoming increasingly popular; therefore, additional studies are required to enhance understanding of the biogeochemical processes within them.
Supplementary Material
ACKNOWLEDGMENTS
We thank Maya Ofek and Max Kolton for technical assistance.
Footnotes
Published ahead of print 6 July 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Arp DJ, Chain PSG, Klotz MG. 2007. The impact of genome analyses on our understanding of ammonia-oxidizing bacteria. Annu. Rev. Microbiol. 61:503–528 [DOI] [PubMed] [Google Scholar]
- 2. Avrahami S, Conrad R. 2003. Patterns of community change among ammonia oxidizers in meadow soils upon long-term incubation at different temperatures. Appl. Environ. Microbiol. 69:6152–6164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Avrahami S, Conrad R, Braker G. 2002. Effect of soil ammonium concentration on N2O release and on the community structure of ammonia oxidizers and denitrifiers. Appl. Environ. Microbiol. 68:5685–5692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Avrahami S, et al. 2011. Active autotrophic ammonia-oxidizing bacteria in biofilm enrichments from simulated creek ecosystems at two ammonium concentrations respond to temperature manipulation. Appl. Environ. Microbiol. 77:7329–7338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boyle Yarwood SA, Bottomley PJ, Myrold DD. 2008. Community composition of ammonia oxidizing bacteria and archaea in soils under stands of red alder and Douglas fir in Oregon. Environ. Microbiol. 10:2956–2965 [DOI] [PubMed] [Google Scholar]
- 6. Briones AM, et al. 2002. Influence of different cultivars on populations of ammonia-oxidizing bacteria in the root environment of rice. Appl. Environ. Microbiol. 68:3067–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burton SAQ, Prosser JI. 2001. Autotrophic ammonia oxidation at low pH through urea hydrolysis. Appl. Environ. Microbiol. 67:2952–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Calvo-Bado LA, et al. 2006. Microbial community responses associated with the development of oomycete plant pathogens on tomato roots in soilless growing systems. J. Appl. Microbiol. 100:1194–1207 [DOI] [PubMed] [Google Scholar]
- 9. Carlile WR, Wilson DP. 1991. Microbial activity in growing media—a brief review. Acta Hort. 294:197–206 [Google Scholar]
- 10. Dowd SE, et al. 2008. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 8:125 doi:10.1186/1471-2180-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fan FL, et al. 2011. Impacts of organic and inorganic fertilizers on nitrification in a cold climate soil are linked to the bacterial ammonia oxidizer community. Microb. Ecol. 62:982–990 [DOI] [PubMed] [Google Scholar]
- 12. Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed] [Google Scholar]
- 13. Fierer N, et al. 2011. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 6:1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 102:14683–14688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Glaser K, et al. 2010. Dynamics of ammonia-oxidizing communities in barley-planted bulk soil and rhizosphere following nitrate and ammonium fertilizer amendment. FEMS Microbiol. Ecol. 74:575–591 [DOI] [PubMed] [Google Scholar]
- 16. Gubry-Rangin C, et al. 2011. Niche specialization of terrestrial archaeal ammonia oxidizers. Proc. Natl. Acad. Sci. U. S. A. 108:21206–21211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gubry Rangin C, Nicol GW, Prosser JI. 2010. Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microb. Ecol. 74:566–574 [DOI] [PubMed] [Google Scholar]
- 18. Hastings RC, et al. 1997. Direct molecular biological analysis of ammonia oxidising bacteria populations in cultivated soil plots treated with swine manure. FEMS Microbiol. Ecol. 23:45–54 [Google Scholar]
- 19. Jia ZJ, Conrad R. 2009. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 11:1658–1671 [DOI] [PubMed] [Google Scholar]
- 20. Jordan FL, Cantera JJL, Fenn ME, Stein LY. 2005. Autotrophic ammonia-oxidizing bacteria contribute minimally to nitrification in a nitrogen-impacted forested ecosystem. Appl. Environ. Microbiol. 71:197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kowalchuk GA, Stienstra AW, Heilig GHJ, Stephen JR, Woldendorp JW. 2000. Changes in the community structure of ammonia-oxidizing bacteria during secondary succession of calcareous grasslands. Environ. Microbiol. 2:99–110 [DOI] [PubMed] [Google Scholar]
- 22. Kowalchuk GA, Stienstra AW, Heilig GHJ, Stephen JR, Woldendorp JW. 2000. Molecular analysis of ammonia-oxidising bacteria in soil of successional grasslands of the Drentsche A (The Netherlands). FEMS Microbiol. Ecol. 31:207–215 [DOI] [PubMed] [Google Scholar]
- 23. Lamont WJ., Jr 2005. Plastics: modifying the microclimate for the production of vegetable crops. Hort. Technol. 15:477–481 [Google Scholar]
- 24. Lang HJ, Elliott GC. 1997. Enumeration and inoculation of nitrifying bacteria in soilless potting media. J. Am. Soc. Hort. Sci. 122:709–714 [Google Scholar]
- 25. Leininger S, et al. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809 [DOI] [PubMed] [Google Scholar]
- 26. Mintie AT, Heichen RS, Cromack K, Myrold DD, Bottomley PJ. 2003. Ammonia-oxidizing bacteria along meadow-to-forest transects in the Oregon Cascade mountains. Appl. Environ. Microbiol. 69:3129–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morimoto S, et al. 2011. Quantitative analyses of ammonia-oxidizing Archaea (AOA) and ammonia-oxidizing Bacteria (AOB) in fields with different soil types. Microbes Environ. 26:248–253 [DOI] [PubMed] [Google Scholar]
- 28. Nicol GW, Leininger S, Schleper C, Prosser JI. 2008. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 10:2966–2978 [DOI] [PubMed] [Google Scholar]
- 29. Nicolaisen MH, Risgaard-Petersen N, Revsbech NP, Reichardt W, Ramsing NB. 2004. Nitrification-denitrification dynamics and community structure of ammonia oxidizing bacteria in a high yield irrigated Philippine rice field. FEMS Microbiol. Ecol. 49:359–369 [DOI] [PubMed] [Google Scholar]
- 30. Nugroho RA, Roling WFM, Laverman AM, Zoomer HR, Verhoef HA. 2005. Presence of Nitrosospira cluster 2 bacteria corresponds to N transformation rates in nine acid Scots pine forest soils. FEMS Microbiol. Ecol. 53:473–481 [DOI] [PubMed] [Google Scholar]
- 31. Ofek M, Hadar Y, Minz D. 2009. Comparison of effects of compost amendment and of single-strain inoculation on root bacterial communities of young cucumber seedlings. Appl. Environ. Microbiol. 75:6441–6450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oved T, Shaviv A, Goldrath T, Mandelbaum RT, Minz D. 2001. Influence of effluent irrigation on community composition and function of ammonia-oxidizing bacteria in soil. Appl. Environ. Microbiol. 67:3426–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pardossi A, Tognoni F, Incrocci L. 2004. Mediterranean greenhouse technology. Chronica Hort. 44:28–34 [Google Scholar]
- 34. Postma J, Geraats BP, Pastoor R, van Elsas JD. 2005. Characterization of the microbial community involved in the suppression of Pythium aphanidermatum in cucumber grown on rockwool. Phytopathology 95:808–818 [DOI] [PubMed] [Google Scholar]
- 35. Purkhold U, et al. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rasmussen PER. 1989. Soil acidification from ammonium-nitrogen fertilization in moldboard plow and stubble-mulch wheat-fallow tillage. Soil Sci. Soc. Am. 53:119 [Google Scholar]
- 37. Rotthauwe JH, Witzel KP, Liesack W. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 39. Schloss PD, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schramm A, De Beer D, Gieseke A, Amann R. 2000. Microenvironments and distribution of nitrifying bacteria in a membrane-bound biofilm. Environ. Microbiol. 2:680–686 [DOI] [PubMed] [Google Scholar]
- 41. Shah V, et al. 2011. Bacterial and archaea community present in the Pine Barrens Forest of Long Island, NY: unusually high percentage of ammonia oxidizing bacteria. PLoS One 6:e26263 doi:10.1371/journal.pone.0026263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Silber A, Bar-Tal A. 2008. Nutrition of substrate-grown plants, p 291–339. In Raviv M, Lieth H. (ed), Soilless culture: theory and practice. Elsevier Science, London, United Kingdom [Google Scholar]
- 43. Sonneveld C, Voogt W. 2009. Plant nutrition of greenhouse crops. Springer Verlag, Dordrecht, Netherlands [Google Scholar]
- 44. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van Lenteren JC. 2000. A greenhouse without pesticides: fact or fantasy? Crop Prot. 19:375–384 [Google Scholar]
- 46. Von Elsner B, et al. 2000. Review of structural and functional characteristics of greenhouses in European Union countries. Part I. Design requirements. J. Agric. Eng. Res. 75:1–16 [Google Scholar]
- 47. Wang Y, Ke X, Wu L, Lu Y. 2009. Community composition of ammonia-oxidizing bacteria and archaea in rice field soil as affected by nitrogen fertilization. Syst. Appl. Microbiol. 32:27–36 [DOI] [PubMed] [Google Scholar]
- 48. Xia WW, et al. 2011. Autotrophic growth of nitrifying community in an agricultural soil. ISME J. 5:1226–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yao H, et al. 2011. Links between ammonia oxidizer community structure, abundance, and nitrification potential in acidic soils. Appl. Environ. Microbiol. 77:4618–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zuckerkandl E, Pauling L. 1965. Evolutionary divergence and convergence in proteins. Evolv. Gen. Prot. 97:166 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.