Abstract
Previous studies reported “mode 1” Bacillus thuringiensis resistance in a colony of diamondback moths (NO-QA), and recently, this resistance has been mapped to an ABC transporter (ABCC2) locus. We report the lack of binding of Cry1Fa to insects derived from this colony and compare our data with those from other insects with ABCC2-associated resistance.
TEXT
Although Bacillus thuringiensis has been used in foliar sprays for more than 30 years, the commercialization in 1996 of the first genetically modified crops protected against insects (Bt crops) increased the importance of B. thuringiensis as a source of insecticidal proteins for the control of insect pests. The extensive use of B. thuringiensis proteins in sprays or in Bt crops has led to the evolution of insect resistance in the field (6, 18). Field resistance to B. thuringiensis sprays was first described in populations of the diamondback moth, Plutella xylostella (5, 6, 12, 19). One of the first resistant colonies (NO-QA) was derived from a Hawaiian population that had evolved resistance to Dipel 2X (Abbott Laboratories, North Chicago, IL) in the field and that was further selected in the laboratory for additional resistance to the same formulated product (20). NO-QA insects showed “mode 1” resistance, which entails strong resistance to at least one Cry1A protein (in this case, Cry1Aa, Cry1Ab and Cry1Ac, all three being found in Dipel), little or nil cross-resistance to Cry1C, recessive inheritance, and reduced binding of at least one Cry1A protein (16). Further characterization showed that resistant insects were cross-resistant to Cry1F and Cry1J (13, 17). Genetic studies indicated that resistance to Cry1A and Cry1F proteins was determined by a single autosomal locus (14) and that in a diet-adapted descendant of NO-QA (NO-QAGE), the Cry1Ac resistance locus was genetically linked to a membrane transporter gene (ABCC2) (3).
For susceptible diamondback moths, an integrated model for the binding sites of Cry1A and Cry1Fa proteins was proposed (2). In this model, at least two binding sites are involved: one which is shared by Cry1Aa, Cry1Ab, Cry1Ac, and Cry1Fa and a second which is specific for Cry1Aa and apparently does not contribute to its toxicity. Binding studies with NO-QA resistant larvae showed extremely reduced binding of Cry1Ab and Cry1Ac but not of Cry1Aa (15). Therefore, it was proposed that resistance in NO-QA is due to a mutation that alters the common binding site for Cry1Aa, Cry1Ab, Cry1Ac, and Cry1F.
The aim of the present study was to check whether binding of Cry1Fa was indeed reduced in NO-QAGE insects and in this way to confirm (or reject) the hypothesis that the multiple resistance in this colony is due to an alteration of the shared binding site affecting binding not only of the Cry1A proteins but of Cry1F as well. To achieve this objective, Cry1Fa and Cry1Aa (as a control) were labeled with 125I and biotin, respectively, and binding to brush border membrane vesicles (BBMV) from susceptible (Lab-V) and resistant (NO-QAGE) larvae was tested.
BBMV were prepared from whole last-instar larvae by the differential magnesium precipitation method (23). For binding studies, Cry1Fa and Cry1Aa proteins were obtained as protein inclusions from recombinant B. thuringiensis and Escherichia coli strains, respectively. Solubilized toxins were prepared as trypsin-activated and chromatography-purified proteins (10). Bioassays were performed using Cry1Fa protein inclusions. While all susceptible insects died at a dose of 10 μg/ml, NO-QAGE insects all survived a dose of 200 μg/ml. Cry1Fa was labeled with 125I by the chloramine-T method according to the conditions described by Hernández-Rodríguez et al. (9), and the specific radioactivity obtained was 0.5 μCi/μg. To test total binding, increasing amounts of BBMV from both colonies were incubated with 27 nM labeled Cry1Fa, in a final volume of 0.1 ml of binding buffer (10 mM Na2HPO4, 1 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4, 0.1% bovine serum albumin [BSA]) for 1 h at 25°C. An excess of unlabeled toxin (15-fold) was used to determine the nonspecific binding. After incubation, samples were centrifuged at 16,000 × g for 10 min and the pellet was washed with cold binding buffer. The results with 125I-Cry1Fa showed an increase in the specific binding (obtained after subtracting nonspecific binding from the total binding) when BBMV from susceptible insects were used (Fig. 1A), whereas no specific binding was observed with BBMV from NO-QAGE insects (Fig. 1B).
Fig 1.
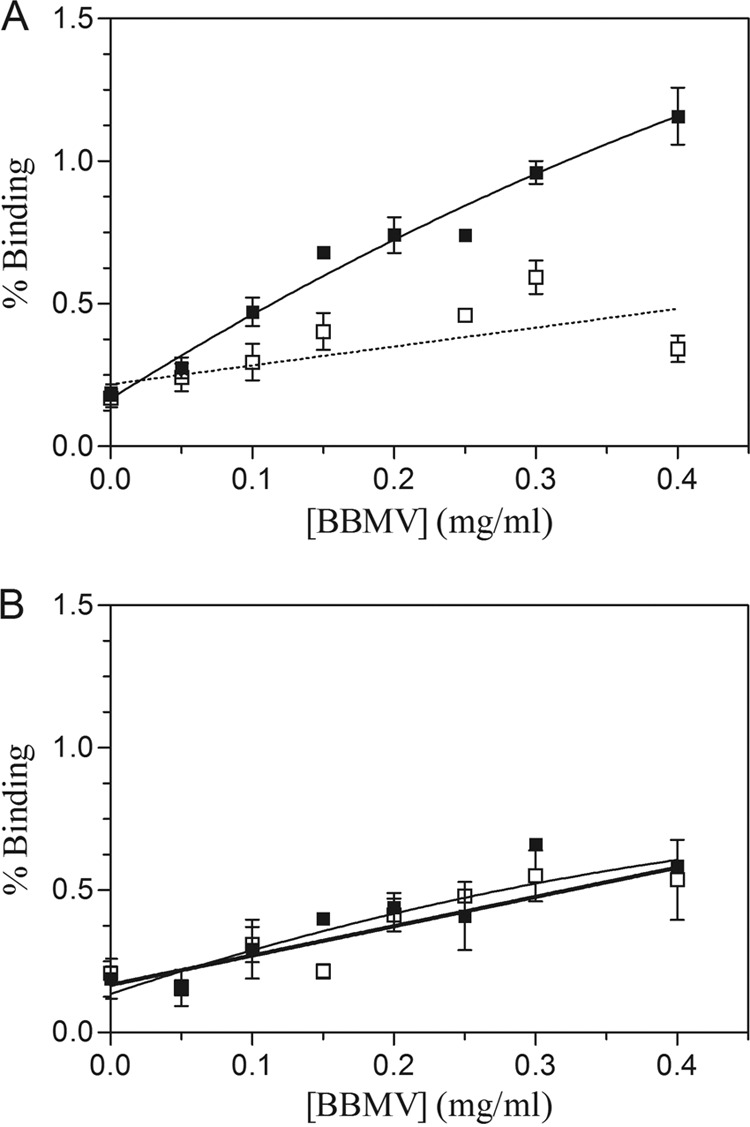
Binding of 125I-labeled Cry1Fa at increasing concentrations of BBMV from susceptible (A) and resistant (B) diamondback moths. ■, total binding; □, nonspecific binding.
As a control, binding of biotin-labeled Cry1Aa was performed with 0.2 mg/ml of BBMV proteins. As expected, biotinylated Cry1Aa bound to BBMV from both susceptible and resistant insects (Fig. 2). Addition of an excess of unlabeled Cry1Aa notably reduced binding of biotinylated Cry1Aa in both cases, indicating that most of the binding observed was specific.
Fig 2.
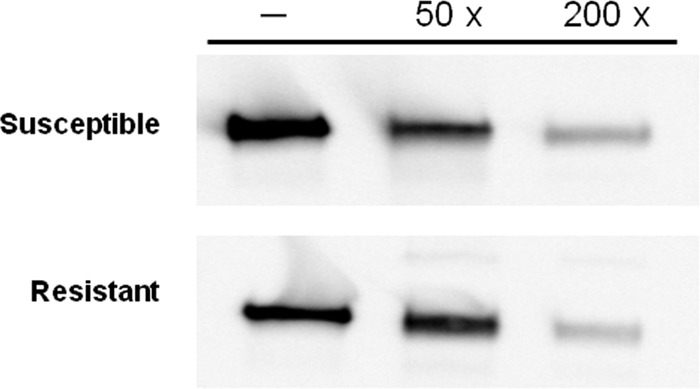
Binding of biotinylated Cry1Aa to BBMV from susceptible and resistant diamondback moths, in the absence of competitor (lanes labeled −) or in the presence of a 50- or 200-fold excess of unlabeled Cry1Aa.
Reduced binding has been described as a primary mechanism of insect resistance to Cry1A family proteins in several insect species (6), to Cry1Fa in Heliothis virescens (11), and, more recently, to Cry2A proteins in two Helicoverpa species (4). However, the alteration of the target site for Cry1Fa was only indirectly proposed for resistant colonies of diamondback moths based on the heterologous competition of this protein with labeled Cry1A proteins (2, 8). Mutations in the gene encoding ABCC2 have been implicated in the Cry1Ac resistance of H. virescens, P. xylostella (NO-QAGE), Trichoplusia ni, and Bombyx mori (1, 3, 7). A resistant colony of H. virescens (YHD2), believed to carry mutations in the genes for both cadherin and ABCC2, was cross-resistant to Cry1Fa and lacked binding of Cry1Fa (11). However, neither toxicity nor binding of this protein has been reported for subsequent colonies in which the two mutations were separated (7). The Cry1Ac-resistant T. ni colony showed only weak cross-resistance to Cry1Fa (22). In this colony, binding of Cry1Ab and Cry1Ac was reduced (22), but no binding data for Cry1Fa were reported. Interestingly, in the B. mori Cry1Ab-resistant colony, no loss of binding of Cry1Ab was observed; the effect of Cry1Fa was not investigated (1). In the current study, we have looked at P. xylostella, the other species in which resistance has been linked to ABCC2. We have found a strong correlation between cross-resistance and loss of binding for Cry1Fa, complementing previous findings for Cry1Aa, Cry1Ab, and Cry1Ac. The lack of Cry1Fa binding in this resistant strain supports the common binding site for Cry1Ab, Cry1Ac, and Cry1Fa in the model proposed by Ballester et al. (2). The T. ni and P. xylostella colonies show a different cross-resistance pattern, despite their resistance phenotypes both being linked to the same ABCC2 locus. This may represent different mutations in ABCC2 affecting susceptibility to the toxins in a different manner (for example, in different toxin binding sites), although the lack of current evidence indicating the functionality of ABCC2 as a binding site for B. thuringiensis toxins should be noted. Alternatively, it is possible that mutations in ABCC2 affect susceptibility to the toxins indirectly. It is noteworthy that in the resistant T. ni colony, a reduction in the expression of aminopeptidase N (APN1), which is a known receptor for B. thuringiensis toxins, was observed via a transregulatory mechanism (21). An unknown, indirect effect may also be able to provide an explanation for the differences in Cry1Ab binding observed with the T. ni, P. xylostella, and B. mori resistant colonies. Thus, while mutations in ABCC2 can confer resistance in a variety of insects, the effect may be indirect and consequently the resistance phenotype may reflect differences in physiology between the species.
ACKNOWLEDGMENTS
We thank Ruud de Maagd (Plant Research International B.V.) and Jeroen Van Rie (Bayer BioScience N.V.) for providing the Cry1 recombinant clones.
This research was supported by the Generalitat Valenciana under the program PROMETEO for research groups of excellence (reference no. 2011/044).
Footnotes
Published ahead of print 6 July 2012
REFERENCES
- 1. Atsumi S, et al. 2012. Single amino acid mutation in an ATP-binding cassette transporter gene causes resistance to Bt toxin Cry1Ab in the silkworm, Bombyx mori. Proc. Natl. Acad. Sci. U. S. A. 109:E1591–E1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ballester V, Granero F, Tabashnik BE, Malvar T, Ferré J. 1999. Integrative model for binding of Bacillus thuringiensis toxins in susceptible and resistant larvae of the diamondback moth (Plutella xylostella). Appl. Environ. Microbiol. 65:1413–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baxter SW, et al. 2011. Parallel evolution of Bacillus thuringiensis toxin resistance in lepidoptera. Genetics 189:675–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caccia S, et al. 2010. Binding site alteration is responsible for field-isolated resistance to Bacillus thuringiensis Cry2A insecticidal proteins in two Helicoverpa species. PLoS One 5:e9975 doi:10.1371/journal.pone.0009975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ferré J, Real MD, Van Rie J, Jansens S, Peferoen M. 1991. Resistance to the Bacillus thuringiensis bioinsecticide in a field population of Plutella xylostella is due to a change in a midgut membrane receptor. Proc. Natl. Acad. Sci. U. S. A. 88:5119–5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ferré J, Van Rie J. 2002. Biochemistry and genetics of insect resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 47:501–533 [DOI] [PubMed] [Google Scholar]
- 7. Gahan LJ, Pauchet Y, Vogel H, Heckel DG. 2010. An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 6:e1001248 doi:10.1371/journal.pgen.1001248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Granero F, Ballester V, Ferré J. 1996. Bacillus thuringiensis crystal proteins Cry1Ab and Cry1Fa share a high affinity binding site in Plutella xylostella (L.). Biochem. Biophys. Res. Commun. 224:779–783 [DOI] [PubMed] [Google Scholar]
- 9. Hernández-Rodríguez CS, Hernández-Martínez P, Van Rie J, Escriche B, Ferré J. 2012. Specific binding of radiolabeled Cry1Fa insecticidal protein from Bacillus thuringiensis to midgut sites in lepidopteran species. Appl. Environ. Microbiol. 78:4048–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Herrero S, González-Cabrera J, Ferré J, Bakker PL, de Maagd RA. 2004. Mutations in the Bacillus thuringiensis Cry1Ca toxin demonstrate the role of domains II and III in specificity towards Spodoptera exigua larvae. Biochem. J. 384:507–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jurat-Fuentes JL, Gould FL, Adang MJ. 2002. Altered glycosylation of 63- and 68-kilodalton microvillar proteins in Heliothis virescens correlates with reduced Cry1 toxin binding, decreased pore formation, and increased resistance to Bacillus thuringiensis Cry1 toxins. Appl. Environ. Microbiol. 68:5711–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kirsch K, Schmutterer H. 1988. Low efficacy of a Bacillus thuringiensis (Berl.) formulation in controlling the diamondback moth, Plutella xylostella (L.), in the Philippines. J. Appl. Entomol. 105:249–255 [Google Scholar]
- 13. Tabashnik BE, Finson N, Johnson MW, Heckel DG. 1994. Cross-resistance to Bacillus thuringiensis toxin CryIF in the diamondback moth (Plutella xylostella). Appl. Environ. Microbiol. 12:4627–4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tabashnik BE, Liu YB, Finson N, Masson L, Heckel DG. 1997. One gene in diamondback moth confers resistance to four Bacillus thuringiensis toxins. Proc. Natl. Acad. Sci. U. S. A. 94:1640–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tabashnik BE, et al. 1997. Global variation in the genetic and biochemical basis of diamondback moth resistance to Bacillus thuringiensis. Proc. Natl. Acad. Sci. U. S. A. 94:12780–12785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tabashnik BE, et al. 1998. Insect resistance to Bacillus thuringiensis: uniform or diverse? Philos. Trans. R. Soc. Lond. B Biol. Sci. 353:1751–1756 [Google Scholar]
- 17. Tabashnik BE, et al. 1996. Cross-resistance of the diamondback moth indicates altered interactions with domain II of Bacillus thuringiensis toxins. Appl. Environ. Microbiol. 62:2839–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tabashnik BE, Van Rensburg JB, Carrière Y. 2009. Field-evolved insect resistance to Bt crops: definition, theory, and data. J. Econ. Entomol. 102:2011–2025 [DOI] [PubMed] [Google Scholar]
- 19. Tabashnik BE, Cushing NL, Finson N, Johnson MW. 1990. Field development of resistance to Bacillus thuringiensis in diamondback moth (Lepidoptera: Plutellidae). J. Econ. Entomol. 83:1671–1676 [Google Scholar]
- 20. Tabashnik BE, Finson N, Johnson MW. 1991. Managing resistance to Bacillus thuringiensis: lessons from the diamondback moth (Lepidoptera: Plutellidae). J. Econ. Entomol. 84:49–55 [Google Scholar]
- 21. Tiewsiri K, Wang P. 2011. Differential alteration of two aminopeptidases N associated with resistance to Bacillus thuringiensis toxin Cry1Ac in cabbage looper. Proc. Natl. Acad. Sci. U. S. A. 108:14037–14042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang P, et al. 2007. Mechanism of resistance to Bacillus thuringiensis toxin Cry1Ac in a greenhouse population of the cabbage looper, Trichoplusia ni. Appl. Environ. Microbiol. 73:1199–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wolfersberger MG, et al. 1987. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp. Biochem. Physiol. 86A:301–308 [DOI] [PubMed] [Google Scholar]


