Abstract
Rhodococcus jostii RHA1, a catabolically diverse soil actinomycete, is highly resistant to long-term nutrient starvation. After 2 years of carbon starvation, 10% of the bacterial culture remained viable. To study the molecular basis of such resistance, we monitored the abundance of about 1,600 cytosolic proteins during a 2-week period of carbon source (benzoate) starvation. Hierarchical cluster analysis elucidated 17 major protein clusters and showed that most changes occurred during transition to stationary phase. We identified 196 proteins. A decrease in benzoate catabolic enzymes correlated with benzoate depletion, as did induction of catabolism of alternative substrates, both endogenous (lipids, carbohydrates, and proteins) and exogenous. Thus, we detected a transient 5-fold abundance increase for phthalate, phthalate ester, biphenyl, and ethyl benzene catabolic enzymes, which coincided with at least 4-fold increases in phthalate and biphenyl catabolic activities. Stationary-phase cells demonstrated an ∼250-fold increase in carbon monoxide dehydrogenase (CODH) concurrent with a 130-fold increase in CODH activity, suggesting a switch to CO or CO2 utilization. We observed two phases of stress response: an initial response occurred during the transition to stationary phase, and a second response occurred after the cells had attained stationary phase. Although SigG synthesis was induced during starvation, a ΔsigG deletion mutant showed only minor changes in cell survival. Stationary-phase cells underwent reductive cell division. The extreme capacity of RHA1 to survive starvation does not appear to involve novel mechanisms; rather, it seems to be due to the coordinated combination of earlier-described mechanisms.
INTRODUCTION
In most natural environments, and particularly in soil, microbes spend most of their existence starving (46). Organic compounds are the primary nutrients for heterotrophic organisms, providing both the reductant and carbon source for catabolism. Therefore, adaptation to deprivation of organic substrates must have been one of the key selective pressures during evolution of soil bacteria. Limitation of organic substrates is frequently referred to as C-limitation, but reductant for respiration, not C, is probably the limiting factor in these situations. To date, diverse strategies to exploit scarce nutrients and survive starvation have been discovered in different organisms. They include reductive cell division, scavenging endogenous substrates, dormancy, and spore formation, as well as programmed cell death and increased mutation rate (52). Some of these major modifications to cell physiology are mediated by different regulatory responses, including (i) a nonspecific, general stress response (regulated by SigB in many bacteria) providing cells with multiple stress resistance mechanisms; (ii) the ppGpp-dependent stringent response including decrease of growth-related functions (transcription, translation, nucleotide biosynthesis, cell envelope metabolism, and cell division); (iii) specific regulation of catabolic processes (central metabolism, alternative carbon source utilization, and use of endogenous storage compounds) and modified expression of transporters (5, 63).
Physiological responses to nutrient starvation have been studied in a number of organisms; however, most studies have focused on Escherichia coli (52). Far less is known of the molecular basis of starvation survival in organisms that better tolerate starvation, such as actinomycetes, although this group has attracted some recent attention. For example, models using oxygen, organic substrate, or nitrogen deprivation and stationary-phase survival demonstrated that mycobacteria are able to survive the extended periods in a nongrowing state by activating a SigB- and SigH-regulated general stress response as well as a ppGpp-regulated stringent response (7, 22, 28, 39, 56, 61, 67). One central hypothesis about starvation resistance is focused on the role of lipid metabolism, which generally maintains mycobacterial viability in the absence of growth and particularly allows persistence in lung lesions (10).
Rhodococci are mycolate-containing soil actinomycetes, related to mycobacteria, that are abundant in soil environments. Rhodococci have great catabolic diversity, which permits them to play important roles in nutrient cycling and to have numerous important commercial applications (3, 69). Other rhodococci have evolved for pathogenicity in humans, animals, and plants (18, 45). Despite the critical role of starvation survival in these important processes mediated by rhodococci, very few studies are reported (17, 64), and the molecular mechanisms of nutrient starvation survival in rhodococci are not defined. To better understand the physiology and molecular basis of starvation survival in rhodococci, we have selected Rhodococcus jostii RHA1. The diverse catabolic capabilities of RHA1 were studied extensively (20, 24, 30, 49, 55, 59, 60, 66), and its genome was sequenced and annotated (44). Previous studies of RHA1 have investigated global responses to heat shock, high salinity (15), and desiccation (35).
The physiological responses to a stress condition primarily rely on changes in protein synthesis. Therefore, to characterize the molecular basis of starvation tolerance in RHA1, we employed a global quantitative proteomic approach, utilizing high-resolution, large-format two-dimensional gel electrophoresis (2DGE) in combination with mass spectrometry (MS)-based protein identification. This approach allowed monitoring of temporal abundance profiles of a large number of cytosolic proteins and thus identification of the RHA1 carbon starvation proteome and its temporal behavior. These studies were followed by targeted gene disruption and resting-cell biotransformation and enzyme activity assays. The results provide new insights into the strategies rhodococci employ to survive carbon starvation.
MATERIALS AND METHODS
Chemicals.
Pharmalyte 3-10 and Immobiline Dry-Strips were purchased from GE Healthcare (Baie d'Urfé, Canada). Iodoacetamide and 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) were from Acros Organics (Fair Lawn, NJ) and MP Biomedicals (Aurora, OH), respectively. Mini Complete protease inhibitor cocktail was purchased from Roche (Laval, Canada). Sypro Ruby was from Bio-Rad Laboratories Ltd. (Mississauga, Canada). 3-[N,N-Dimethyl(3-myristoylaminopropyl)-ammonio]propanesulfonate (ABS-14) and dithiothreitol (DTT) were from Fluka (Switzerland) and Sigma, respectively. Nucleotides were purchased from Qiagen (Mississauga, Canada). All chemicals were of analytical grade and used without further purification.
Strains and media.
Rhodococcus jostii strain RHA1 was isolated prior to 1995 from soil contaminated with lindane in Japan (41) and kindly provided by M. Fukuda. RHA1 and the mutant RHA1_013 (Table 1) were grown at 30°C on W minimal salt medium containing KH2PO4 (1.7 g/liter), Na2HPO4 (9.8 g/liter), (NH4)2SO4 (1.0 g/liter), MgSO4 · 7H2O (0.1 g/liter), FeSO4 · 7H2O (0.95 mg/liter), MgO (10.75 mg/liter), CaCO3 (2.0 mg/liter), ZnSO · 7H2O (1.44 mg/liter), CuSO4 · 5H2O (0.25 mg/liter), CoSO4 · 7H2O (0.28 mg/liter), H3BO3 (0.06 mg/liter), and concentrated HCl (51.3 μl/liter) (32) supplemented with 10 mM benzoate and 20 mM ammonium sulfate. These amounts of exogenous carbon and nitrogen were adjusted to ensure that it is carbon limitation that causes RHA1 to enter stationary phase. For proteomic studies, RHA1 cells were first precultured in 100-ml Erlenmeyer flasks to mid-log phase (optical density at 600 nm [OD600], 1.0), then used to inoculate (1%) fresh medium in 500-ml flasks, and further cultured to a growth phase of interest and harvested. We selected seven time points representing different growth phases (Fig. 1). Growth data were collected using cultures inoculated with normalized quantities of cells based on OD and were based on at least 3 independent experiments, each consisting of 3 biological replicates. For gene replacement, Escherichia coli and Rhodococcus strains were grown using media and culture conditions described previously (23). E. coli BW25113 was used to propagate pKD46 and fosmid RF00131J24. E. coli DH10B/pUZ8002 was the fosmid donor strain for intergeneric conjugation. Strains were grown on LB broth supplemented with ampicillin (100 μg/ml), apramycin (Apra, 50 g/ml), chloramphenicol (12.5 μg/ml), kanamycin (50 μg/ml), and hygromycin (50 and 150 μg/ml for RHA1 and E. coli, respectively) as required.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype/comments | Source |
|---|---|---|
| Plasmids | ||
| pKD46 | λ-RED (gam bet exo) araC rep101(Ts) Ampr | 10 |
| pIJ773 | λ-RED (gam bet exo) araC rep101(Ts) Ampr | 23 |
| pUZ8002 | tra neo RP4, Kmr | 57 |
| pUC-Hy | Source of hyg gene | 38 |
| RF00131J24 | Fosmid clone carrying sigG, Cmr | 71 |
| RFMD5 | RF00131J24 with cat replaced with hyg, Hmr | This study |
| RFMD6 | RFMD5 with sigG replaced with Apra cassette, Hmr Aprar | This study |
| Strains | ||
| E. coli BW25113 | K-12 derivative: ΔaraBAD, ΔrhaBAD Ampr | 23 |
| E. coli DH10B | Host for pUZ8002 and RFMD4, RFMD6, RFMD8, RFMD10, Kmr | 23 |
| RHA1 | Rhodococcus jostii RHA1, wild type | 65 |
| RHA1_013 | Derivative of RHA1; ΔsigG; Aprr | This study |
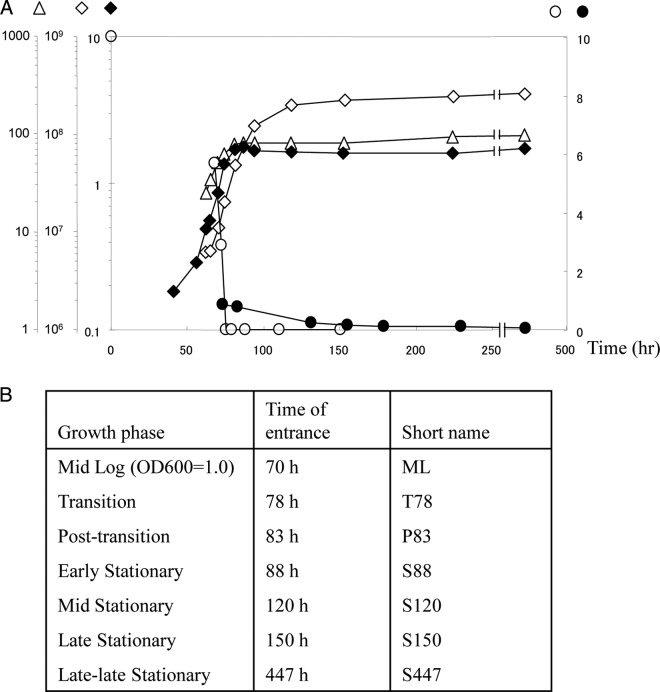
Fig 1.
(A) Growth of R. jostii RHA1 and entry into benzoate-limited stationary phase: changes in OD600 (⧫); in benzoate (mM) (○) and muconate (mM) (●) concentration; in cell viability (CFU/ml) (◇); and in total cell protein concentration (μg/ml) (△). (B) Description of the time points representing different growth phases when the samples were taken for proteomic analyses.
Preparation of cell extracts.
Cell extracts were prepared essentially as described previously (59) with some modifications. Briefly, cell pellets were washed once with 0.14 M NaCl and once with Tris-EDTA (TE) buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and then stored as aliquots at −80°C. The pelleted cells were suspended in lysis buffer (2% ABS-14, 4% CHAPS, 30 mM DTT, 30 mM Tris [pH 7.5], Mini Complete protease inhibitor cocktail [1:100, vol/vol]) and disrupted using a Fast Prep Bio 101 Thermo Savant bead beater (5 cycles of 20 s). Protein concentration was determined using the 2D Quant kit (GE Healthcare). For enzyme assays, cell-free protein extracts were prepared in the same manner, except that the cell pellets were washed using 20 ml of 50 mM Tris-HCl, pH 7.5, and suspended in 1.0 ml of the same buffer, and the cell extracts were used immediately. The protein concentrations of the cell extracts were quantified using the bicinchoninic acid (BCA) method.
Proteomic analyses.
The cytosolic proteomes were resolved using 2DGE essentially as described previously (59) with minor modifications. Briefly, aliquots of cell extracts containing 90 μg protein were separated in the first dimension by isoelectric focusing (IEF) for a total of 88 kVh at 20°C using 24-cm, nonlinear immobilized pH gradient (IPG) strips and a pH gradient of 3 to 7. The rehydration solution contained 9 M urea, 3 M thiourea, 35 mM DTT, 4% CHAPS, and Pharmalyte pH 3-10. Proteins were separated in the second dimension using 12% SDS-polyacrylamide gels and the ETTAN DALTtwelve System (GE Healthcare). Gels were stained using Sypro Ruby and digitally imaged using a Typhoon 9400 (GE Healthcare). Spot detection, matching, abundance quantification, and normalization were performed using Progenesis Workstation software (Nonlinear Dynamics, Durham, NC). The relative abundance of each protein was determined from the signal intensity and normalized against total signal intensity detected on a gel. Only spots with a minimum normalized volume of 0.002 or greater were analyzed. Protein patterns of each growth condition were based on gels from three independent cultures. Protein spots of interest were identified using a Voyager DESTR matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometer (Applied Biosystems, Foster City, CA) based on peptide mass fingerprint (PMF) analysis combined with the MASCOT search engine (Matrix Science) and the RHA1 protein database at the Proteomics Centre, University of Victoria. Identified proteins fulfilled four criteria: the MASCOT search score was above 55, a minimum of 4 peptides were matched, the protein sequence coverage was at least 20%, and the predicted molecular weight (MW) and pI values were consistent with the experimentally observed ones. We clustered the detected proteins based on their temporal abundance profiles. For this, the normalized signal intensities of the proteins were subjected to hierarchical cluster analysis by Gene Spring using Pearson's correlation coefficient.
CODH assay.
The carbon monoxide dehydrogenase (CODH) enzyme activity assay was performed using a Varian Cary IE UV-visible spectrophotometer equipped with a cuvette holder with a thermostat. The CODH activity of RHA1 cells grown to M70 or S120 was measured by monitoring the CO-dependent reduction of 2-(4-indophenyl)-3-(4-nitrophenyl)-2H-tetrazolium chloride (INT) (ε = 17.981 mM−1 min−1) as described in reference 34 with some modifications. Briefly, samples of 20 ml were harvested from three 100-ml cultures by centrifugations at 4°C at 10,000 × g. The cell extracts were prepared as described above. We added 20 μl of each cell extract to 100 μl of CO-saturated phosphate buffer per assay, and A562 was followed over time for 30 min. One unit of enzyme activity was defined as the amount of enzyme required to reduce 1 nmol of INT per minute at 30°C.
Resting-cell assay.
RHA1 cells were collected by centrifugation at different growth phases and washed with, and resuspended in, W medium to an OD600 of 1.0. The resultant cell suspensions (1 ml) were supplied with 0.2 mM biphenyl or 0.2 mM phthalate and incubated for 120 min at 30°C with shaking in sealed 4.5-ml glass vials. To stop the reaction, 0.1 ml of 6 N HCl was added to the mix. As an internal standard, 30 ppm of 2,6-dichlorobiphenyl or 40 ppm terephthalic acid dimethyl ester was added. To extract biphenyl or phthalate, we added hexane or ethyl acetate, respectively, and the mixtures were mixed vigorously using a vortex mixer for 1 min. The supernatant was recovered and dehydrated with sodium sulfate. One microliter of extract was analyzed by gas chromatography-MS. The analysis was done in triplicate. Controls were performed using heat-killed cells.
HPLC.
The high-pressure liquid chromatography (HPLC) system consisted of a Waters 2695 separations module and 2996 photodiode array detector (Waters, MA). Chromatographic data were collected and compiled using Empower software (Waters). Cell culture supernatants were filtered through 0.2-μm syringe filters. Chromatographic separation of benzoate was performed using an Aqua C18 125-Å (250 by 4.6 mm, 5-μm particle size) column (Phenomenex, CA) at a flow rate of 1.0 ml/min at room temperature. The mobile phase consisted of two solutions: A, 0.5% phosphoric acid; and B, 100% methanol. The gradient profile was 0 to 5 min 25% (vol/vol) B; 5 to 6 min linear gradient to 60% (vol/vol) B; 6 to 11 min 60% (vol/vol) B; 11 to 12 min linear gradient to 25% (vol/vol) B; and 12 to 18 min 25% (vol/vol) B. The wavelengths of UV detection were set at 230 nm, 273 nm, and 260 nm to detect benzoic acid, catechol, and cis,cis-muconic acid, respectively.
Gene replacement.
The sigG gene was deleted and replaced by an apramycin cassette using the λ-RED-based methodology as described previously (59). Fosmid RF00131J24 from the RHA1 fosmid library, which contained 36.5 kb of RHA1 genomic DNA, including sigG, was used. The cassette used to replace the sigG gene was constructed by PCR amplification and contained the Aprar gene flanked by flippase recognition target (FRT) sequences. The fosmid with the replaced gene was transferred to RHA1 by intergeneric conjugation as described in reference 31. Aprar Hms exconjugants were selected as double-crossover mutants. The mutant genotypes were verified using PCR with three sets of primers: SigGtestF and SigGtestR, SigGtestF and SigGapraR, and SigGintF and SigGintR (Table 2).
Table 2.
PCR primers used in this studya
| Primer | Sequence (5′–3′) | Comments |
|---|---|---|
| SigGapraF | CCG ACA TCT GTC GGA CGG CCG CAG TAA GCT CCA ACC ATG att ccg ggg atc cgt cga cc | Aprar cassette amplification for sigG replacement, forward |
| SigGapraR | CAT GTA CGG ACG CGT TTT CCG CGC CAT CGG TAG GAG TCA tgt agg ctg gag ctg ctt c | Aprar cassette amplification for sigG replacement, reverse |
| SigGtestF | CGGAATGAAGACTCTGCTC | External sigG primer, forward |
| SigGtestR | GTGTGGGACACGGTAGAAC | External sigG primer, reverse |
| SigGintF | GTGTGGGACACGGTAGAAC | Internal sigG primer, forward |
| SigGintR | ATCAGGGTGCCGATCGAAG | Internal sigG primer, reverse |
The 5′ end of SigGapraF contained 39 nucleotides (uppercase letters) corresponding to the sense strand upstream of sigG ending in the start codon. The 5′ end of SigGapraR contained 39 nucleotides (uppercase letters) corresponding to the antisense strand ending in the stop codon. The 3′ ends of the PCR primers (lowercase letters) matched the 20-nucleotide and 19-nucleotide extensions, respectively, of the aprA-containing disruption cassette sequence.
Phylogenetic analysis.
Sequences of various CODHs were aligned by using ClustalX (version 2.0.9) with all parameters set at their default values. Phylogenetic analyses were performed using the neighbor-joining algorithm of PHYLIP (version 3.66). Neighbor joining was performed by using a random order of sequences on input. For bootstrap analyses, the Seqboot program of the PHYLIP package was used to generate 100 data sets, which were jumbled three times and analyzed as described above. Trees generated were visualized and printed by using TreeView (version 1.6.6).
RESULTS
Benzoate-limited growth of RHA1 and reductive division.
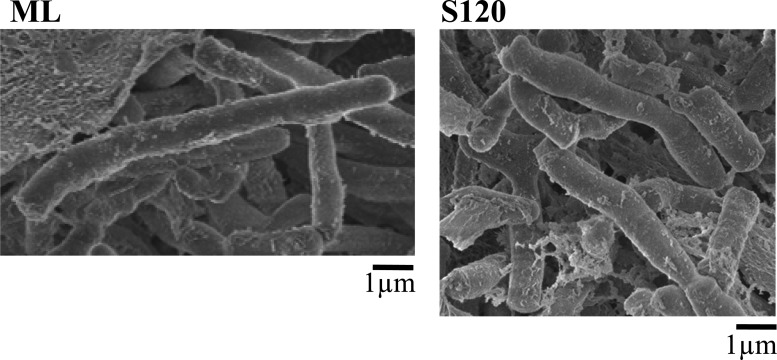
As benzoate was depleted from the medium, RHA1 ended logarithmic growth, passed through a short transition phase (TP), and entered stationary phase (ST) (Fig. 1). The point of entry into ST was defined as the point when the biomass stopped increasing, which corresponded to the beginning of a plateau in both total protein and optical density. Although both protein and OD measurements stopped increasing in ST, CFU continued to increase until late stationary phase (S150). During TP, a metabolite of benzoate, muconate, was detected, and its concentration decreased gradually throughout stationary phase. The low level of muconate in the medium (less than 1 mM) was unlikely to support the increase in CFU during stationary phase, and this increase was probably primarily due to reductive division. Accordingly, scanning electron microscopy showed a decrease in cell length from 7.5 μm (±2 μm) for exponentially growing cells to 3.5 μm (±1.5 μm) during ST (Fig. 2). Further, the total protein calculated per CFU increased during mid-log phase (ML) and then sharply decreased in TP and stationary phases (not shown).
Fig 2.
Scanning electron micrographs of R. jostii RHA1 cells collected during mid-log (ML) and stationary (S120) phases of growth.
To estimate a survival capacity of RHA1, cells continued to be incubated for a total of about 2 years. After plateauing, protein, OD, and CFU remained essentially constant for 83 days. After 125 days, both OD and CFU decreased 2-fold, and after 500 days we detected about 30% of the initial maximum number of cells remaining viable. Finally, after 700 days of incubation 10% of the cells were still viable (data not shown).
Changes in the RHA1 proteome during starvation.
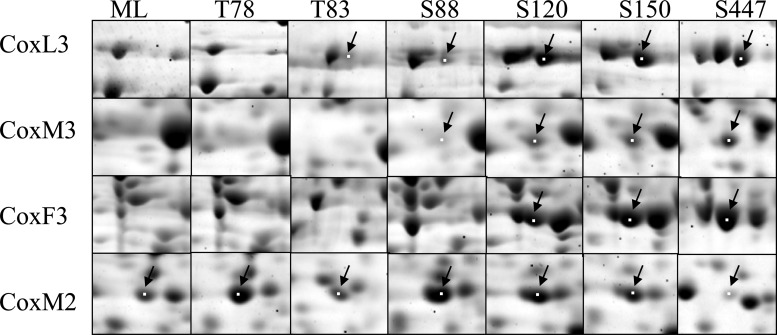
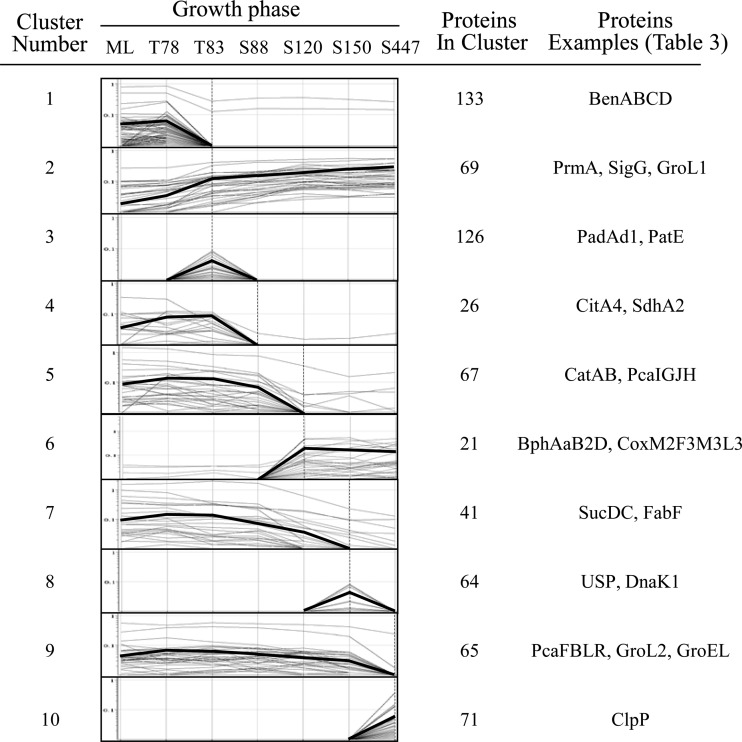
To understand the RHA1 starvation response at the molecular level, we classified more than 1,600 proteins based on their abundance profile. This constitutes 18% of the total number of predicted proteins encoded by the RHA1 genome. Considering that the starvation response develops over time and depends on the severity of starvation, we monitored protein abundance profiles over seven time points representing different growth phases: ML, transition after 78 and 83 h of incubation (T78 and T83, respectively), and stationary phase after 88, 120, 150, and 447 h of incubation (S88, S120, S150, and S447, respectively) (Fig. 1). The total number of detected proteins remained approximately constant throughout all the time points, while the relative abundance of a large proportion of proteins changed (Fig. 3). Compared to the ML proteome, the total proportion of proteins with increased abundance (including the unique spots) was 21% in the T78 proteome and increased to 41% in the S120, S150, and S447 samples. The total proportion of proteins with decreased abundance (including the absent spots) was 16% in P78 cells and increased to 40% in the T83 proteome and further to 47 and 53% in the S150 and S447 proteomes, respectively. Assuming that proteins displaying similar patterns of abundance often participate in related processes, we clustered the detected proteins based on their temporal expression patterns. This analysis clustered 1,090 proteins, which constitute about 66% of the total detected proteins, and elucidated 17 clusters, of which the most distinct and represented 10 are shown in Fig. 4. The highest number (30%) of the abundance changes (including increase and decrease) among the clustered proteins occurred during the relatively brief posttransition phase (T78 to T83) of growth, indicating that the switch between exponentially growing and nongrowing states requires more significant adjustments in cell physiology.
Fig 3.
Sections of 2D gels showing parts of the cytosolic proteomes of R. jostii RHA1 grown on benzoate and collected at different time points: ML, T78, T83, S88, S120, S150, and S447. CO dehydrogenase subunits CoxM2, CoxF3, CoxM3, and CoxL3 are shown with arrows.
Fig 4.
Hierarchical cluster analysis of protein spot signal intensities for the RHA1 proteomes. Protein signal intensities from averaged triplicate samples were obtained by using Progenesis and exported into GeneSpring software for cluster analysis. Proteins with similar signal intensity patterns were grouped together into 17 clusters, of which 10 are shown. The thick lines represent the average profiles.
Based on MALDI-TOF, we identified 196 proteins differentially expressed in at least one of the seven proteomes, each representing one time point of growth. These constitute 16% of the proteins detected in an average proteome and represent each of the elucidated clusters. The averaged normalized signal intensity of each identified protein at each time point, as well as protein MS identification parameters, is presented in Table S1 in the supplemental material. Of these proteins, 55 are shown in Table 3.
Table 3.
Identification of the proteins differentially expressed during carbon starvationa
| Protein name | Gene name | Ro no. | Genetic context | Cluster | Normalized signal intensityb |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ML | T78 | T83 | S88 | S120 | S150 | S447 | |||||
| Benzoate catabolism | |||||||||||
| Benzoate 1,2-dioxygenase alpha subunit | benA | 02384 | ben | 1 | 1.70 | 1.30 | 0.61 | 0.43 | 0.15 | 0.11 | 0.07 |
| Benzoate 1,2-dioxygenase beta subunit | benB | 02385 | ben | 1 | 1.09 | 0.97 | 0.59 | 0.46 | 0.18 | 0.17 | 0.18 |
| Benzoate 1,2-dioxygenase reductase subunit | benC | 02386 | ben | 1 | 0.98 | 0.81 | 0.35 | 0.21 | 0.02 | N | N |
| cis-1,6-Dihydroxycyclohexa-3,5-diene-1-carboxylate dehydrogenase | benD | 02387 | ben | 1 | 0.56 | 0.50 | 0.33 | 0.20 | 0.02 | 0.04 | 0.01 |
| Catechol 1,2-dioxygenase | catA | 02373 | cat | 5 | 3.50 | 3.28 | 2.40 | 1.82 | 0.67 | 0.36 | 0.19 |
| Muconate cycloisomerase | catB | 02372 | cat | 5 | 0.42 | 0.35 | 0.23 | 0.11 | 0.02 | 0.01 | 0.01 |
| 3-Oxoacid CoA-transferase, subunit A | pcaI | 01334 | pca | 5 | 0.35 | 0.32 | 0.30 | 0.27 | 0.10 | 0.04 | 0.02 |
| Protocatechuate dioxygenase alpha subunit | pcaG | 01336 | pca | 5 | 0.26 | 0.28 | 0.25 | 0.18 | 0.04 | 0.05 | 0.05 |
| 3-Oxoacid CoA-transferase | pcaJ | 01333 | pca | 5 | 0.10 | 0.12 | 0.10 | 0.02 | 0.02 | 0.02 | 0.02 |
| Protocatechuate dioxygenase beta subunit | pcaH | 01335 | pca | 5 | 0.14 | 0.20 | 0.08 | 0.08 | 0.02 | 0.01 | 0.01 |
| Acetyl-CoA C-acyltransferase | pcaF | 01340 | pca | 9 | 0.28 | 0.39 | 0.42 | 0.39 | 0.30 | 0.20 | 0.02 |
| 3-Carboxy-cis,cis-muconate cycloisomerase | pcaB | 01337 | pca | 9 | 0.10 | 0.11 | 0.07 | 0.08 | 0.08 | 0.07 | N |
| 3-Oxoadipate enol-lactone hydrolase/4-carboxymuconolactone decarboxylase | pcaL | 01338 | pca | 9 | 0.21 | 0.20 | 0.10 | 0.06 | 0.08 | 0.06 | 0.03 |
| Transcriptional regulator, IclR family | pcaR | 01339 | pca | 9 | 0.03 | 0.04 | 0.05 | 0.06 | 0.05 | 0.03 | 0.02 |
| Other catabolic enzymes | |||||||||||
| Biphenyl 2,3-dioxygenase alpha subunit | bphAa | 08060 | bph | 6 | N | N | N | N | 0.48 | 0.54 | 0.15 |
| cis-3-Phenylcyclohexa-3,5-diene-1,2-diol dehydrogenase | bphB2 | 10126 | bph/etb | 6 | N | N | N | N | 0.03 | 0.03 | 0.13 |
| 2,6-Dioxo-6-phenylhexa-3-enoate hydrolase | bphD1 | 10136 | bph/etb | 6 | N | N | N | N | 0.10 | 0.08 | 0.03 |
| Ethylbenzene dioxygenase alpha subunit | etbAa1 | 10133 | bph/etb | 6 | N | N | N | N | 0.47 | 0.45 | 0.26 |
| 2-Hydroxy-6-oxohepta-2,4-dienoate hydrolase | etbD1 | 08044 | bph/etb | 6 | 0.01 | 0.01 | 0.02 | 0.01 | 0.24 | 0.29 | 0.18 |
| 2,6-Dioxo-6-phenylhexa-3-enoate hydrolase | etbD2 | 10146 | bph/etb | 6 | 0.01 | 0.01 | 0.02 | 0.02 | 0.27 | 0.26 | 0.10 |
| Phthalate 3,4-dioxygenase, ferredoxin reductase subunit | padAd1 | 08162/12013 | pad | 3 | 0.02 | 0.03 | 0.07 | 0.09 | 0.03 | 0.02 | N |
| Phthalate ester hydrolase | patE | 08169 | pat | 3 | 0.04 | 0.04 | 0.09 | 0.12 | 0.07 | 0.06 | 0.03 |
| Propane monooxygenase hydroxylase large subunit | prmA | 00441 | Cluster with phenol hydroxylase and GroEL | 2 | 0.02 | 0.03 | 0.03 | 0.04 | 0.13 | 0.68 | 0.38 |
| Probable 1,3-propanediol dehydrogenase | 06057 | ATP-dependent protease | 2 | 0.05 | 0.03 | 0.15 | 0.24 | 0.24 | 0.56 | 0.29 | |
| Other catabolic enzymes | |||||||||||
| CODH, medium subunit | coxM2 | 04975 | CODH | 6 | 0.09 | 0.11 | 0.08 | 0.12 | 0.22 | 0.20 | 0.19 |
| CODH, maturation factorc | coxF3 | 05229 | CODH | 6 | N | N | N | N | 0.17 | 0.38 | 0.49 |
| CODH, medium subunitc | coxM3 | 05230 | CODH | 6 | 0.04 | 0.07 | 0.03 | 0.04 | 0.18 | 0.20 | 0.43 |
| CODH, large subunitc | coxL3 | 05232 | CODH | 6 | N | N | 0.05 | 0.04 | 0.17 | 0.20 | 0.46 |
| Central metabolism | |||||||||||
| 6-Phosphofructokinase | pfkA | 06479 | Amidotransferase gat | 5 | 0.02 | 0.04 | 0.03 | 0.03 | 0.01 | N | N |
| Pyruvate kinase | pyk1 | 01007 | Glutamate synthase | 5 | 0.29 | 0.29 | 0.29 | 0.27 | 0.30 | 0.18 | 0.19 |
| Dihydrolipoyl dehydrogenasee | dldH1 | 02140 | Hypothetical | 1 | 0.09 | 0.04 | N | N | N | N | N |
| Citrate synthase | 04998 | Peptidylprolyl isomerase | 7 | 0.03 | 0.05 | 0.05 | 0.05 | 0.03 | 0.02 | N | |
| Citrate synthase | citA4 | 04993, 08812 | Phosphoserine transaminase | 4 | 0.09 | 0.09 | 0.03 | 0.01 | 0.02 | 0.01 | 0.004 |
| Isocitrate dehydrogenasee | 06238 | Arabinose efflux | 4 | 0.16 | 0.10 | 0.02 | N | N | N | N | |
| Succinate dehydrogenase | sdhA2 | 06246 | sdh | 4 | 0.60 | 0.45 | 0.33 | 0.31 | 0.32 | 0.19 | 0.27 |
| Succinate-CoA ligase, alpha subunit | sucD | 05573 | suc | 7 | 1.00 | 0.85 | 0.85 | 0.80 | 0.67 | 0.58 | 0.42 |
| Succinate-CoA ligase, beta subunit | sucC | 05574 | suc | 7 | 1.07 | 0.82 | 0.79 | 1.03 | 0.92 | 0.58 | 0.50 |
| Malate dehydrogenase | fabF | 06000 | Single gene | 7 | 0.20 | 0.24 | 0.25 | 0.16 | 0.06 | 0.02 | 0.06 |
| Isocitrate lyase | 02122 | Lipid metabolism | 2 | 0.20 | 0.22 | 0.74 | 0.75 | 0.59 | 0.38 | 0.46 | |
| Fumarate hydratasee | 08824 | Succinate dehydrogenase | 2 | N | N | 0.01 | 0.01 | 0.01 | 0.02 | 0.02 | |
| Glucose-6-phosphate 1-dehydrogenase | zwf4 | 07184 | Pentose phosphate pathway | 2 | 0.03 | 0.02 | 0.03 | 0.04 | 0.05 | 0.05 | 0.11 |
| Phosphogluconate dehydrogenase | gnd3 | 07246 | RNA helicase | 2 | 0.08 | 0.06 | 0.15 | 0.23 | 0.39 | 0.43 | 0.50 |
| Stress response | |||||||||||
| Chaperonin GroL2c,e | groL2 | 02146 | Universal stress protein | 9 | 0.83 | 0.69 | 0.60 | 0.60 | 0.65 | 0.63 | N |
| Chaperone protein GroELc,e | groEL | 06190 | 10-kDa chaperonin | 9 | 0.81 | 0.67 | 1.20 | 0.93 | 0.71 | 0.63 | 0.34 |
| Chaperonin GroL1c,e | groL1 | 00448 | Propane monooxygenase | 2 | N | N | 0.61 | 0.37 | 0.11 | 1.45 | N |
| Heat shock protein Hsp70 DnaK1c,e | dnaK1 | 05497 | Chaperones: grpE1, dnaJ2 | 8 | 1.01 | 1.10 | 0.71 | 0.72 | 1.07 | 1.99 | 1.36 |
| Universal stress proteinc,e | 02263 | Universal stress protein, oxidoreductase | 3 | 0.03 | 0.03 | 0.06 | 0.06 | 0.05 | 0.03 | 0.03 | |
| Universal stress proteinc,e | 04893 | Pseudouridylate synthase | 8 | 0.08 | 0.06 | 0.06 | 0.12 | 0.12 | 0.19 | 0.18 | |
| Catalased,e | katE | 04309 | Fe(3+) uptake regulator | 7 | 0.02 | 0.05 | 0.05 | 0.07 | 0.04 | 0.01 | 0.02 |
| Superoxide dismutasee | sodA | 04009 | Amidotransferase | 2 | 1.08 | 0.90 | 1.10 | 1.19 | 1.40 | 1.38 | 1.62 |
| Heme superoxide dismutasec | 06860 | Heme biosynthesis, oxidoreductase | 10 | 0.19 | 0.19 | 0.18 | 0.26 | 0.25 | 0.31 | 0.39 | |
| ATP-dependent Clp proteased,e | clpP | 01371 | clpX, FKBP-type bacterial trigger factor | 10 | 0.25 | 0.25 | 0.40 | 0.41 | 0.30 | 0.43 | 0.50 |
| Prolyl aminopeptidasec,e | 01171 | panB, coenzyme metabolism | 8 | 0.01 | 0.03 | 0.03 | 0.02 | 0.01 | 0.09 | N | |
| Cell division initiation protein | 01080 | Cell division | 6 | 0.002 | 0.005 | 0.01 | 0.07 | 0.11 | 0.08 | 0.03 | |
| Transcription | |||||||||||
| RNA polymerase sigma factor G | sigG | 05361 | ArsR | 2 | 0.07 | 0.07 | 0.11 | 0.13 | 0.17 | 0.14 | 0.18 |
Protein identification was accepted if the MASCOT-generated probability-based Mowse score in the RHA1 protein database was greater than 55 (P < 0.05) and at least 3 peptides were identified. When the MASCOT score was below the threshold, the protein ID was accepted only if the theoretical and experimental pI and MW agreed. N, not detected.
Spot signal intensities were normalized and averaged over three biological replicates.
Proteins reported to be regulated by MglA (21).
Proteins reported to be regulated by SigB (4).
Catabolism of aromatics.
Fourteen enzymes responsible for the catabolism of benzoate (Ben), catechol (Cat), and protocatechuate (Pca) were identified (Table 3). The abundance of the upper benzoate pathway enzymes (BenABCD) started declining in T83 (cluster 1), whereas the abundance of the lower benzoate pathway enzymes (CatAB and PcaIGJH) decreased in S120 (cluster 5). This decline correlated with the depletion of benzoate and muconate (Fig. 1). The abundance of PcaBLF and the regulatory protein PcaR remained essentially constant until a reduction of at least 2-fold in the S447 cells (cluster 9). This supports differences observed earlier in PcaBLFR and PcaIGJH abundance profiles (59) and further suggests their independent regulation.
Once benzoate and its intermediates were depleted, starving RHA1 cells increased capacity to use alternative exogenous substrates. We observed the activation of several aromatic catabolic pathways. The abundance of enzymes responsible for the catabolism of phthalate (PadAd1) and phthalate esters (PatE) increased up to 5-fold in T83 (cluster 3). These abundances correspond to 5 to 10% of those previously measured in phthalate-grown ML phase cells (59). Later, during S120 and S150, six Bph and Etb enzymes responsible for utilization of biphenyl, ethylbenzene, benzene, and styrene appeared (cluster 6). The abundances of the large catalytic subunits BphAa and EtbAa1 reached 28 to 35% of those in biphenyl-grown ML phase cells (60). Further, we identified 6 other catabolic enzymes whose abundance increased in starving RHA1 cells (ro08833, ro02718, ro08019, ro02204, ro00441, and ro06057) (see Table S1 in the supplemental material). Two of them (propane monooxygenase and propanediol dehydrogenase) were up to 40-fold more abundant during S150 than ML (cluster 2) (Table 3).
To test whether the abundance increase correlates with the catalytic activities of the Bph and Pad enzymes, we measured phthalate and biphenyl degradation in resting cells of benzoate-limited RHA1. As summarized in Table 4, phthalate degradation activity was greatest during T83, when PadAd1 expression reached its maximum. Biphenyl degradation activity was greatest in S150, in agreement with BphAa and EtbAa1 abundance profiles. Although increased, the maximum biphenyl degradation rate of benzoate-limited cells was 150-fold lower than that of ML cells grown on biphenyl (data not shown).
Table 4.
Degradation activities in RHA1 cells during different growth phasesa
| Substrate | Degradation activity, 10−3 μmol/min/mg (SD) |
|||
|---|---|---|---|---|
| ML | T83 | S88 | S150 | |
| Phthalate | 0.23 (0.10) | 0.58 (0.05) | 0.26 (0.004) | 0.36 (0.007) |
| Biphenyl | 0.33 (0.12) | ND | 0.15 (0.10) | 1.37 (0.07) |
The assays were performed in duplicate. The degradation activities were measured by monitoring the concentration of the substrates. ND, not detected.
CO oxidation.
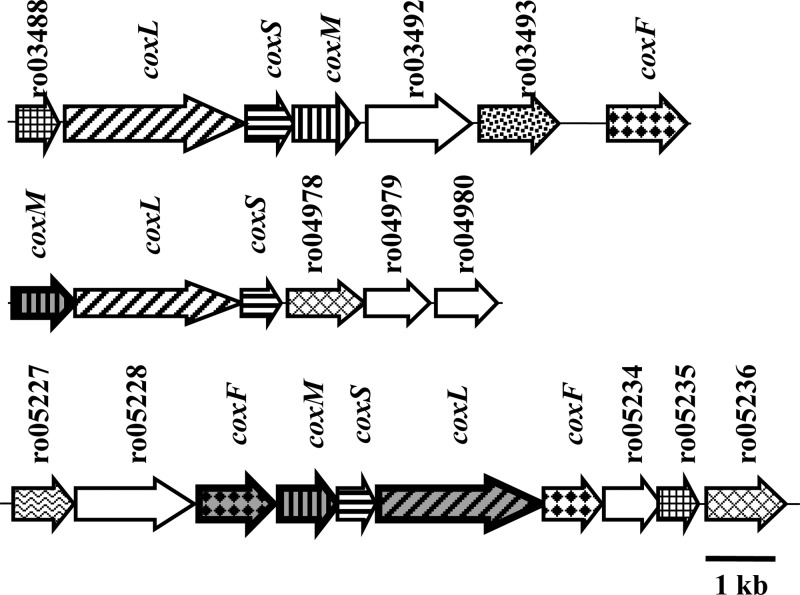
In addition to aromatic catabolism, we observed the activation of CO oxidation systems during S120 and S447 phases. The RHA1 genome encodes three CODHs, whose structural and accessory genes are organized in 3 separate chromosomal clusters (Fig. 5). Each CODH enzyme is comprised of three subunits: small (CoxS), medium (CoxM), and large (CoxL). We observed the expression of four of these proteins (Fig. 5). Expression of these proteins increased up to 245-fold in S150 compared to the expression of those in ML cells (Fig. 3, cluster 6). We also measured the CODH activity in ML and S120 cells, which increased 130 times during starvation (not shown).
Fig 5.
Genetic organization of CO oxidation genes in R. jostii RHA1. The numbers above the arrows indicate CDS (Ro) numbers from the RHA1 genome assembly (http://www.rhodococcus.ca). These gene clusters are located on the chromosome. Patterns represent different types of genes. Structural CODH genes: diagonally striped, CODH large subunit; vertically striped, CODH medium subunit; horizontally striped, CODH small subunit (ferredoxin). Accessory CODH genes: diamonds, CODH maturation factor; hatched diamonds, CoxE homologs; squares, CoxG homologs. Other genes: stippled, transcriptional regulator; white, miscellaneous; and waved, hypothetical. Gray background and bold border indicate genes whose products were identified in this study, as listed in Table 3.
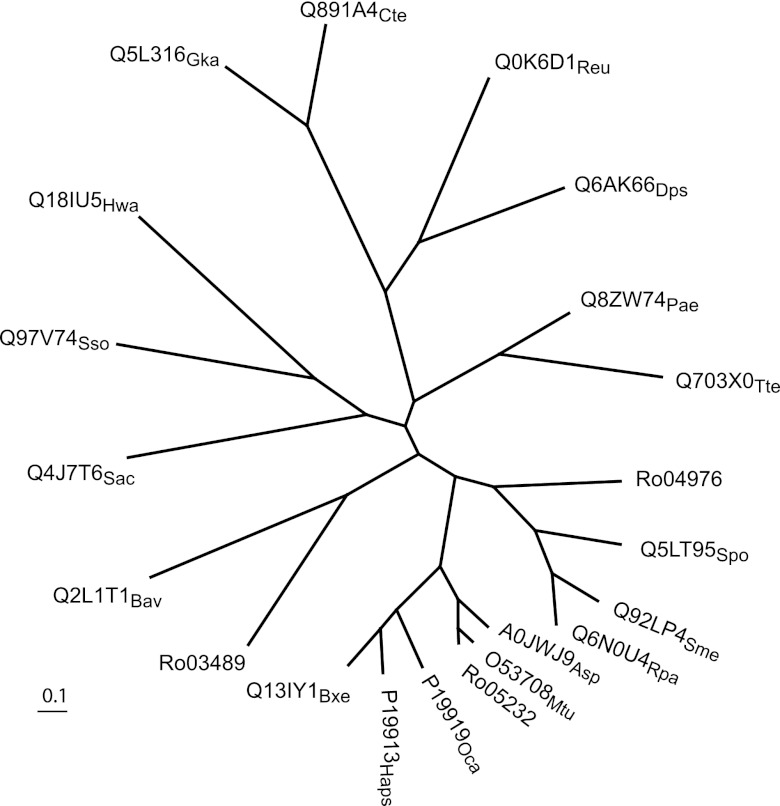
The three CODH-encoding gene clusters in the RHA1 genome differ in the organization of structural as well as accessory genes (Fig. 5). Furthermore, the comparative sequence analysis revealed that the large subunits, CoxL, of the three CODHs share only 33 to 43% sequence identity and cluster with distinct sets of homologs. Phylogenetic analysis using the best-studied CODHs confirmed that the RHA1 CODHs belong to three distinct subclades (Fig. 6). CoxL3 (Rjo_ro05232) is most similar to CODHs from actinomycetes Mycobacterium tuberculosis and Arthrobacter sp. FB24, whereas CoxL1 (Rjo_ro03489) and CoxL2 (Rjo_ro04976) cluster with CODHs from alphaproteobacteria and betaproteobacteria, respectively. The fact that CoxL3 closely clusters with CODHs from Mycobacterium species was also demonstrated in a recent study (68).
Fig 6.
Phylogenetic analysis of CODH. The unrooted neighbor-joining tree was calculated from an alignment of the amino acid sequences of the large subunits of three predicted CODHs from R. jostii RHA1 (Ro03489, Ro04976, and Ro05232) and the following proteins from the PIRSF (strain followed by accession number with a subscript indicating the strain): Oligotropha carboxidovorans P19919Oca; Hydrogenophaga pseudoflava P19913Hps; Sulfolobus solfataricus Q97V74Sso; Pyrobaculum aerophilum Q8ZW74Pae; Sulfolobus acidocaldarius Q4J7T6Sac; Thermoproteus tenax Q703X0Tte; Haloquadratum walsbyi Q18IU5Hwa; Arthrobacter sp. FB24 A0JWJ9Asp; Burkholderia xenovorans LB400 Q13IY1Bxe; Ralstonia eutropha H16 Q0K6D1Reu; Geobacillus kaustophilus Q5L316Gka; Bordetella avium 197N Q2L1T1Bav; Mycobacterium tuberculosis O53708Mtu; Clostridium tetani Q891A4Cte; Rhodopseudomonas palustris Q6N0U4Rpa; Desulfotalea psychrophila Q6AK66Dps; Sinorhizobium meliloti Q92LP4Sme; Silicibacter pomeroyi Q5LT95Spo.
Central metabolism.
Four central metabolic pathways, i.e., glycolysis, tricarboxylic acid (TCA), glyoxylate shunt, and pentose phosphate pathways, responded to starvation (Table 3). Two glycolysis enzymes (6-phosphofructokinase and pyruvate kinase) decreased abundance in S120 (cluster 5). Dihydrolipoyl dehydrogenase (pyruvate dehydrogenase complex) was reduced in T78 and not detected during starvation (cluster 1). The abundance of seven TCA cycle enzymes (two homologs of citrate synthase, isocitrate dehydrogenase, α and β subunits of succinate-CoA ligase, succinate dehydrogenase, and malate dehydrogenase) decreased in S88 and S150 (clusters 4 and 7). In contrast, the abundance of the glyoxylate shunt enzyme (isocitrate lyase) and fumarate hydratase as well as two enzymes of the pentose phosphate pathway (glucose-6-phosphate 1-dehydrogenase and phosphogluconate dehydrogenase) increased in T83 (cluster 2).
Protein, lipid, and carbohydrate metabolism.
Consistent with reductive cell division, the abundance of the cell division initiation protein (Ro01080) responsible for cell division and chromosome partitioning was increased up to 56-fold during starvation (cluster 6) (Table 3); however, ribosomal proteins (RpsAIB, AlaS, and Tsf) were less abundant upon entering T83 phase (see Table S1 in the supplemental material). Further, ClpP protease (Ro01371, cluster 10) and peptidase (Ro01171, cluster 8) (Table 3) were more abundant. Biosynthesis of individual amino acids also showed significant changes (see Table S1). This includes IlvC, involved in biosynthesis of branched amino acids, which increased in abundance 500-fold during stationary phase.
Further, several enzymes required for incorporation of inorganic nitrogen into cell material changed abundance. Glutamate dehydrogenase and glutamine synthetase GlnA1 decreased in abundance (see Table S1 in the supplemental material); however, the alternative pathway of l-glutamate production that includes prolyl aminopeptidase, 1-pyrroline-5-carboxylate dehydrogenase, and 5-oxoprolinase showed up to 300-fold-higher abundance.
A number of proteins responsible for metabolism of carbohydrates increased in abundance in starving cells (see Table S1 in the supplemental material). These included glycosyltransferases and sugar epimerases, as well as glucan and glycoside hydrolases, which increased in abundance up to 13-fold in stationary-phase cells.
We detected 21 proteins involved in lipid metabolism, whose abundance was changed during starvation (see Table S1 in the supplemental material). More abundant proteins included acyl carriers and carboxylases, likely involved in lipid degradation. Among proteins with decreased abundance, we identified acyl and mycolyl carriers, fatty acid synthases, and desaturases, as well as enzymes involved in de novo synthesis and modification of fatty and mycolic acids.
Sigma factor G.
We identified 9 transcriptional regulators whose abundance changed during starvation (see Table S1 in the supplemental material). Sigma factor SigG (cluster 2) doubled in abundance during stationary phase (Table 3). To elucidate a possible physiological role of SigG in RHA1 starvation response, we investigated the growth phenotype of a gene disruption mutant lacking sigG (Table 1). No drastic changes in the phenotype (OD, CFU, total cell protein) were observed in the ΔsigG strain RHA1_013 during growth on benzoate. The strain showed 40% loss in viability after sudden starvation and an ∼5-h delay in, and 20% lower, recovery after benzoate was provided back to the medium (data not shown).
Stress response during starvation.
We identified 23 proteins whose functions may be associated with stress response and whose abundance changed under starvation (Table 3; see Table S1 in the supplemental material). All three GroEL chaperones encoded by the RHA1 genome and chaperone DnaK were identified and belong to the 5 most abundant proteins in the detected proteomes. Interestingly, their abundance profiles differed but covered all the tested points of growth. GroL2 (ro02146) was highly abundant at all the time points except S447, where it was not detected (cluster 9). Chaperonin GroL1 (ro00448) was first detected in T83 cells, which coincided with the abundance increase of GroEL (ro06190), and then increased in abundance 723-fold in S150 cells, which coincided with the abundance increase of DnaK1 (ro05497).
Two universal stress proteins (USPs) were detected. Similarly to the GroEL proteins, the USPs had different abundance profiles, falling into 2 phases: ro02263 was 2-fold more abundant in T83 to S120 cells (cluster 3), and ro04893 was 2-fold more abundant in S120 to S447 cells (cluster 8). Three oxidative stress response proteins, superoxide dismutase (ro04009), heme superoxide dismutase (ro06860), and catalase KatE (ro04309), were more abundant during starvation. Notably, we again see 2 phases in the abundance profiles: KatE was more abundant during T78 to S120 phases of growth; in contrast, the dismutases were more abundant in S150 to S447.
DISCUSSION
This study demonstrated the remarkable ability of a soil actinomycete, Rhodococcus jostii RHA1, to survive carbon source starvation for at least 2 years, retaining 10% of cells viable. The detailed proteomics study identified two groups of processes responding to starvation. The first group included a transition from growing to nongrowing cells, a reductive division, a shutdown of benzoate catabolism, and a decrease in transcription, translation, de novo biosynthesis, glycolysis, and TCA cycle. The second group represented induction of the two-phase stress response and a switch to utilization of alternative endogenous and exogenous carbon sources including the activation of multiple aromatic catabolic pathways and CO utilization. These mechanisms, in combination with other detected rearrangements in cell physiology, likely contribute to the extreme capacity of RHA1 to survive long-term starvation.
The ability to survive long-term starvation groups RHA1 with mycobacterial strains known to survive for extended periods of time (51, 67, 72). We observed that RHA1 stationary-phase cells underwent reductive cell division, which generally results from completion of DNA replication and cell division in the absence of growth. It is also known as dwarfing. This strategy has been proposed to be beneficial during early starvation (33, 52). To understand the RHA1 starvation response at the molecular level, we characterized the temporal abundance profiles of seven cytosolic proteomes collected at different points of growth and starvation and showed that the highest number of protein changes occurred during the switch between exponentially growing and nongrowing states. We identified alternative catabolic pathways that transiently increased abundance levels at different time points during starvation, which may indicate that RHA1 cells attempt to exploit different alternative substrates at different times. Such a strategy may allow starving RHA1 a quicker recovery from a dormant state once substrates become available. Examples of this survival strategy have been seen in other bacteria. For example, adaptation of Lactobacillus lactis to carbon starvation involves induction of pathways for metabolism of alternative exogenous carbon sources, such as sugars, citrate, and glycerol (63). The choice of alternative carbon sources may be indicative of catabolic specialization of a starving organism.
A particularly intriguing result is the increased abundance of CODHs, which reversibly oxidize CO to CO2 and permit use of CO as a reductant for energy metabolism (62). Organisms oxidizing CO usually can also use CO as the sole carbon source (62). The induction of CODHs during starvation suggests that RHA1 is switching from heterotrophic to autotrophic metabolism. Since CODH interconverts CO and CO2, it enables assimilation of CO2 or CO into cellular building blocks (62). Interestingly, it has recently been shown that glycerol-limited M. tuberculosis is able to fix CO2 into biomass through carboxylation reactions (6). The phylogenetic differences between the three CODHs encoded by the RHA1 genome coincide with the detected differences in their abundance profiles. For example, the CODH related to CODH in actinomycetes (CoxF3M3L3) was not detected in ML to T83 but showed a significant abundance spike in S447, whereas CoxM2 was detected during all phases and doubled in abundance in stationary cells. Thus, there may be a meaningful link between the phylogeny and physiology of the individual RHA1 CODH isozymes.
Among central metabolic pathways, the enzymes of glycolysis, pyruvate oxidation, and the TCA cycle reduced in abundance during starvation; however, the glyoxylate shunt and pentose phosphate pathway enzymes became more abundant during transition phase. The glyoxylate shunt is required to assimilate fatty acids via acetyl-coenzyme A (acetyl-CoA), whereas the pentose phosphate pathway provides precursors for nucleic acids. Interestingly, in M. tuberculosis, glyoxylate shunt enzymes were shown to be essential for fatty acid catabolism, virulence, and survival in the host (47). Most recently, isocitrate lyase has been shown to be essential for M. tuberculosis growth under glycerol-limited conditions and proposed to be a key enzyme of a novel metabolic route for carbohydrate metabolism (6).
The decreased production of protein synthesis machinery and the increased abundance of ClpP protease and peptidase suggest an arrest of de novo protein synthesis and a switch to protein turnover. This is consistent with the stringent response, which occurs in response to starvation in a wide range of bacteria, including E. coli (42, 73). In addition to protein scavenging, ClpP proteases degrade short-lived regulatory proteins as well as misfolded and damaged proteins, which occur more often under stress (19), thus maintaining cellular homeostasis (75). Biosynthesis of branched amino acids was significantly induced during stationary phase as exemplified by IlvC. Intriguingly, its 25% identical homolog in yeast was suggested to be a bifunctional protein, whose second role is to be involved in DNA packaging required for maintenance of stable mitochondrial DNA (76), which may be of higher demand under starvation stress.
Synthesis of glutamate and glutamine is required for incorporating inorganic nitrogen into cell material. Although carbon-starved RHA1 cells were provided with nitrogen, the enzymes responsible for nitrogen uptake decreased in abundance. This suggests that carbon starvation may trigger the nitrogen starvation response. Such coregulation of the responses to carbon and nitrogen starvation has been reported in E. coli (43, 48). Simultaneously, RHA1 increased the level of the alternative pathway of l-glutamate production using proline cleaved from peptides. This indicates that starving RHA1 reduces inorganic nitrogen uptake, but in order to provide amino groups to amino acid synthesis, it continues to maintain a cellular glutamate pool using cellular peptides.
Starvation increased the production of a series of enzymes necessary for metabolizing glycogen, a storage compound in RHA1 (26). In addition to glycogen, lipids (triacylglycerols, wax esters, and polyhydroxyalkanoates) are important storage compounds in actinomycetes (1). They can be deposited as insoluble inclusions in the cytoplasm, accounting for up to 70% of the cellular dry matter (1). Therefore, in the absence of an available carbon source, the products of lipid degradation, fatty acids, become an important source of carbon and energy. We detected a massive rearrangement in the biosynthesis of enzymes involved in lipid metabolism as well as metabolism of fatty and mycolic acids, major components of the cell envelope of the mycolic acid bacteria. It is noteworthy that lipid metabolism has been shown to play a central role in maintaining long-term mycobacterial viability in the absence of growth (10). Thus, starving RHA1 scavenges cellular glycogen and lipids as endogenous sources of carbon.
Overall, the detected reduction in biosynthesis and the switch to utilizing the endogenous sources of carbon and energy have been reported in other organisms as the first response to carbon and energy deprivation (52, 63). This allows cells to save and redirect the intracellular carbon and energy resources to essential and strategically important processes. Reductive cell division accompanied by dwarfing involves self-digestion, resulting in degradation of endogenous material including storage and membrane lipids, carbohydrates, and proteins.
We aimed to elucidate a possible physiological role for SigG in RHA1 starvation response. The protein belongs to the most phylogenetically divergent group 4 of σ70 family of sigma factors with extracytoplasmic function (ECF), which regulates genes in response to environmental signals (58). SigG shares 64% amino acid sequence identity with SigG from Mycobacterium tuberculosis strain H37Rv, whose expression was also highly increased in stationary phase (40) and during macrophage infection (9). We think that the subtle changes in the phenotype of the sigG deletion mutant do not necessarily negate a role for SigG in the starvation response, and the role of SigG in RHA1 starvation remains unresolved. One possibility is that, similarly to the M. tuberculosis SigG, it may play a role in the SOS response (36).
We detected a significant stress response in starving RHA1. Three GroEL and DnaK chaperones were more abundant at different points during starvation, indicating their differential regulation. A high abundance of GroL2 in all except one proteome is consistent with the housekeeping role suggested for GroL2 homologs (Cpn60.2) in actinobacteria (37). GroEL and DnaK protein chaperones are commonly induced under stress and assist in proper assembly of misfolded or unfolded polypeptides generated under stress conditions (8, 13). The highest level of induction of the homologous chaperones in M. tuberculosis was detected in cells exposed to heat shock, but induction was also seen under hyperosmolarity, oxidative stress, and starvation conditions (29). Similarly, two USPs had different abundance profiles, falling into two phases. Intriguingly, the RHA1 genome encodes 21 predicted USPs (reference 44 and this study). This number stands out drastically from one USP predicted in the Xanthomonas campestris genome, 8 in the M. tuberculosis genome, and 12 in the Streptomyces coelicolor genome (56). The detected USPs, ro02263 and ro04893, share only 36% sequence identity. Although induction of USPs was repeatedly associated with stress response, their biochemical function remains largely unknown. In E. coli, UspA was shown to be required for survival of prolonged growth arrest, and its mutation caused global changes in protein synthesis (53, 54).
Starvation also induced a two-phase oxidative stress response. Oxidative damage was suggested to be the Achilles' heel of stationary-phase bacterial cells (52) and therefore requires a protective response. As a part of this response, respiratory activity is reduced during early starvation. This prevents uncontrolled drainage of endogenous carbon reserves and protects cells against the damaging effects of reactive oxygen species (ROS) produced by the electron transport chain (52). Higher resistance to oxidative stress was also demonstrated for Mycobacterium smegmatis (67). We also detected the increased abundance of several other stress-related proteins. They include methyltransferase of the type I restriction-modification system (ro11151), known to protect bacterial cells against invasion of foreign DNA (74), and two subunits of succinate-semialdehyde dehydrogenase from the glutamate catabolic pathway, a homolog of which has been proposed to buffer redox changes in the cytoplasm of Saccharomyces cerevisiae (11). They further include two predicted DPS (DNA protection during starvation) family miniferritins (ro04028 and ro04043) known to protect bacterial DNA from oxidants during starvation (27) and diaminobutyrate-pyruvate aminotransferase, responsible for biosynthesis of ectoine, a primary compatible solute in RHA1, whose production had been shown to be induced during heat shock, osmotic, and desiccation stresses (15, 35). NADPH:quinone reductase was also more abundant and, similarly to the NADPH quinone reductase in Helicobacter pylori, may play an important role in coping with oxidative stress by managing the cell quinone pool (70).
Most of the detected stress-related proteins followed 2 phases of expression: the first phase was transiently induced during transition phase and included catalase KatE, GroEL chaperones, and USP ro02263. The second phase was induced later during stationary phase and included superoxide dismutases, DnaK chaperone, thiocyanate hydrolase, USP ro04893, ClpP, and a type I restriction-modification system. This two-phase stress response is consistent with the early stationary expression of general stress proteins (2) and catalase KatE (16) and the late-starvation-induced ROS-specific proteins MrgA (Dps homolog), catalase KatA, and alkyl hydroxyperoxide hydrolase AhpCF (12) observed in Bacillus subtilis.
Overall, this study reports the complex rearrangements in cytosolic protein biosynthesis in response to starvation in Rhodococcus jostii strain RHA1, a catabolically diverse soil actinomycete. We conclude that the exceptional ability of RHA1 to survive starvation does not involve novel mechanisms but rather is due to the coordinated combination of many previously described mechanisms.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from Genome Canada and Genome BC.
Footnotes
Published ahead of print 13 July 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Alvarez HM, Steinbuchel A. 2002. Triacylglycerols in prokaryotic microorganisms. Appl. Microbiol. Biotechnol. 60:367–376 [DOI] [PubMed] [Google Scholar]
- 2. Antelmann H, et al. 1997. Expression of a stress- and starvation-induced dps/pexB-homologous gene is controlled by the alternative sigma factor sigmaB in Bacillus subtilis. J. Bacteriol. 179:7251–7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bell KS, Philp JC, Aw DW, Christofi N. 1998. The genus Rhodococcus. J. Appl. Microbiol. 85:195–210 [DOI] [PubMed] [Google Scholar]
- 4. Bernhardt J, et al. 1997. Specific and general stress proteins in Bacillus subtilis—a two-dimensional protein electrophoresis study. Microbiology 143:999–1017 [DOI] [PubMed] [Google Scholar]
- 5. Bernhardt J, Weibezahn J, Scharf C, Hecker M. 2003. Bacillus subtilis during feast and famine: visualization of the overall regulation of protein synthesis during glucose starvation by proteome analysis. Genome Res. 13:224–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beste DJ, et al. 2011. C Metabolic flux analysis identifies an unusual route for pyruvate dissimilation in Mycobacteria which requires isocitrate lyase and carbon dioxide fixation. PLoS Pathog. 7:e1002091 doi:10.1371/journal.ppat.1002091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717–731 [DOI] [PubMed] [Google Scholar]
- 8. Bukau B, Horwich AL. 1998. The Hsp70 and Hsp60 chaperone machines. Cell 92:351–366 [DOI] [PubMed] [Google Scholar]
- 9. Cappelli G, et al. 2006. Profiling of Mycobacterium tuberculosis gene expression during human macrophage infection: upregulation of the alternative sigma factor G, a group of transcriptional regulators, and proteins with unknown function. Res. Microbiol. 157:445–455 [DOI] [PubMed] [Google Scholar]
- 10. Cole ST, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544 [DOI] [PubMed] [Google Scholar]
- 11. Coleman ST, Fang TK, Rovinsky SA, Turano FJ, Moye-Rowley WS. 2001. Expression of a glutamate decarboxylase homologue is required for normal oxidative stress tolerance in Saccharomyces cerevisiae. J. Biol. Chem. 276:244–250 [DOI] [PubMed] [Google Scholar]
- 12. Dowds BC. 1994. The oxidative stress response in Bacillus subtilis. FEMS Microbiol. Lett. 124:255–263 [DOI] [PubMed] [Google Scholar]
- 13. Duchene AM, Thompson CJ, Mazodier P. 1994. Transcriptional analysis of groEL genes in Streptomyces coelicolor A3(2). Mol. Gen. Genet. 245:61–68 [DOI] [PubMed] [Google Scholar]
- 14. Dukan S, Nystrom T. 1998. Bacterial senescence: stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes Dev. 12:3431–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ekpanyaskun P. 2006. Transcriptomic analysis of Rhodococcus sp. RHA1 responses to heat shock and osmotic stress. Master thesis. University of British Columbia, Vancouver, BC, Canada [Google Scholar]
- 16. Engelmann S, Lindner C, Hecker M. 1995. Cloning, nucleotide sequence, and regulation of katE encoding a sigma B-dependent catalase in Bacillus subtilis. J. Bacteriol. 177:5598–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fanget NV, Foley S. 2011. Starvation/stationary-phase survival of Rhodococcus erythropolis SQ1: a physiological and genetic analysis. Arch. Microbiol. 193:1–13 [DOI] [PubMed] [Google Scholar]
- 18. Finnerty WR. 1992. The biology and genetics of the genus Rhodococcus. Annu. Rev. Microbiol. 46:193–218 [DOI] [PubMed] [Google Scholar]
- 19. Frees D, Ingmer H. 1999. ClpP participates in the degradation of misfolded protein in Lactococcus lactis. Mol. Microbiol. 31:79–87 [DOI] [PubMed] [Google Scholar]
- 20. Goncalves ER, et al. 2006. Transcriptomic assessment of isozymes in the biphenyl pathway of Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 72:6183–6193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guina T, et al. 2007. MglA regulates Francisella tularensis subsp. novicida (Francisella novicida) response to starvation and oxidative stress. J. Bacteriol. 189:6580–6586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gupta S, Pandit SB, Srinivasan N, Chatterji D. 2002. Proteomics analysis of carbon-starved Mycobacterium smegmatis: induction of Dps-like protein. Protein Eng. 15:503–512 [DOI] [PubMed] [Google Scholar]
- 23. Gust B, Challis GL, Fowler K, Kieser T, Chater KF. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hara H, Eltis LD, Davies JE, Mohn WW. 2007. Transcriptomic analysis reveals a bifurcated terephthalate degradation pathway in Rhodococcus sp. strain RHA1. J. Bacteriol. 189:1641–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Helloin E, Jansch L, Phan-Thanh L. 2003. Carbon starvation survival of Listeria monocytogenes in planktonic state and in biofilm: a proteomic study. Proteomics 3:2052–2064 [DOI] [PubMed] [Google Scholar]
- 26. Hernandez MA, et al. 2008. Biosynthesis of storage compounds by Rhodococcus jostii RHA1 and global identification of genes involved in their metabolism. BMC Genomics 9:600 doi:10.1186/1471-2164-9-600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hintze KJ, Theil EC. 2006. Cellular regulation and molecular interactions of the ferritins. Cell. Mol. Life Sci. 63:591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu Y, Coates AR. 1999. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J. Bacteriol. 181:469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu Y, et al. 2008. A Mycobacterium tuberculosis mutant lacking the groEL homologue cpn60.1 is viable but fails to induce an inflammatory response in animal models of infection. Infect. Immun. 76:1535–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iwasaki T, et al. 2007. Characterization of two biphenyl dioxygenases for biphenyl/PCB degradation in a PCB degrader, Rhodococcus sp. strain RHA1. Biosci. Biotechnol. Biochem. 71:993–1002 [DOI] [PubMed] [Google Scholar]
- 31. Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 32. Kimbara K, et al. 1989. Cloning and sequencing of two tandem genes involved in degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. J. Bacteriol. 171:2740–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kjelleberg S, Humphrey BA, Marshall KC. 1983. Initial phases of starvation and activity of bacteria at surfaces. Appl. Environ. Microbiol. 46:978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kraut M, Hugendieck I, Herwig S, Meyer O. 1989. Homology and distribution of CO dehydrogenase structural genes in carboxydotrophic bacteria. Arch. Microbiol. 152:335–341 [DOI] [PubMed] [Google Scholar]
- 35. Leblanc JC, Goncalves ER, Mohn WW. 2008. Global response to desiccation stress in the soil actinomycete Rhodococcus jostii RHA1. Appl. Environ. Microbiol. 74:2626–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee JH, Geiman DE, Bishai WR. 2008. Role of stress response sigma factor SigG in Mycobacterium tuberculosis. J. Bacteriol. 190:1128–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lund PA. 2009. Multiple chaperonins in bacteria—why so many? FEMS Microbiol. Rev. 33:785–800 [DOI] [PubMed] [Google Scholar]
- 38. Mahenthiralingam E, et al. 1998. Site-directed mutagenesis of the 19-kilodalton lipoprotein antigen reveals no essential role for the protein in the growth and virulence of Mycobacterium intracellulare. Infect. Immun. 66:3626–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Manabe YC, Chen JM, Ko CG, Chen P, Bishai WR. 1999. Conditional sigma factor expression, using the inducible acetamidase promoter, reveals that the Mycobacterium tuberculosis sigF gene modulates expression of the 16-kilodalton alpha-crystallin homologue. J. Bacteriol. 181:7629–7633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Manganelli R, Dubnau E, Tyagi S, Kramer FR, Smith I. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715–724 [DOI] [PubMed] [Google Scholar]
- 41. Masai E, et al. 1995. Characterization of biphenyl catabolic genes of gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:2079–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matin A. 1991. The molecular basis of carbon-starvation-induced general resistance in Escherichia coli. Mol. Microbiol. 5:3–10 [DOI] [PubMed] [Google Scholar]
- 43. Maurizi MR, Rasulova F. 2002. Degradation of L-glutamate dehydrogenase from Escherichia coli: allosteric regulation of enzyme stability. Arch. Biochem. Biophys. 397:206–216 [DOI] [PubMed] [Google Scholar]
- 44. McLeod MP, et al. 2006. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. U. S. A. 103:15582–15587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Meijer WG, Prescott JF. 2004. Rhodococcus equi. Vet. Res. 35:383–396 [DOI] [PubMed] [Google Scholar]
- 46. Morita RY. 1993. Bioavailability of energy and the starvation state, p 1–23 In Kjelleberg S. (ed), Starvation in bacteria. Plenum Press, New York, NY [Google Scholar]
- 47. Munoz-Elias EJ, McKinney JD. 2005. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 11:638–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mutalik VK, Venkatesh KV. 2007. A theoretical steady state analysis indicates that induction of Escherichia coli glnALG operon can display all-or-none behavior. Biosystems 90:1–19 [DOI] [PubMed] [Google Scholar]
- 49. Navarro-Llorens JM, et al. 2005. Phenylacetate catabolism in Rhodococcus sp. strain RHA1: a central pathway for degradation of aromatic compounds. J. Bacteriol. 187:4497–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Novotna J, et al. 2003. Proteomic studies of diauxic lag in the differentiating prokaryote Streptomyces coelicolor reveal a regulatory network of stress-induced proteins and central metabolic enzymes. Mol. Microbiol. 48:1289–1303 [DOI] [PubMed] [Google Scholar]
- 51. Nyka W. 1974. Studies on the effect of starvation on mycobacteria. Infect. Immun. 9:843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nystrom T. 2004. Stationary-phase physiology. Annu. Rev. Microbiol. 58:161–181 [DOI] [PubMed] [Google Scholar]
- 53. Nystrom T, Neidhardt FC. 1994. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol. Microbiol. 11:537–544 [DOI] [PubMed] [Google Scholar]
- 54. Nystrom T, Neidhardt FC. 1993. Isolation and properties of a mutant of Escherichia coli with an insertional inactivation of the uspA gene, which encodes a universal stress protein. J. Bacteriol. 175:3949–3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Okamoto S, Eltis LD. 2007. Purification and characterization of a novel nitrile hydratase from Rhodococcus sp. RHA1. Mol. Microbiol. 65:828–838 [DOI] [PubMed] [Google Scholar]
- 56. O'Toole R, Williams HD. 2003. Universal stress proteins and Mycobacterium tuberculosis. Res. Microbiol. 154:387–392 [DOI] [PubMed] [Google Scholar]
- 57. Paget MS, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. 1999. Evidence that the extracytoplasmic function sigma factor sigmaE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Paget MS, Helmann JD. 2003. The sigma70 family of sigma factors. Genome Biol. 4:203 doi:10.1186/gb-2003-4-1-203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Patrauchan MA, et al. 2005. Catabolism of benzoate and phthalate in Rhodococcus sp. strain RHA1: redundancies and convergence. J. Bacteriol. 187:4050–4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Patrauchan MA, et al. 2008. Roles of ring-hydroxylating dioxygenases in styrene and benzene catabolism in Rhodococcus jostii RHA1. J. Bacteriol. 190:37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Primm TP, et al. 2000. The stringent response of Mycobacterium tuberculosis is required for long-term survival. J. Bacteriol. 182:4889–4898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ragsdale SW. 2004. Life with carbon monoxide. Crit. Rev. Biochem. Mol. Biol. 39:165–195 [DOI] [PubMed] [Google Scholar]
- 63. Redon E, Loubiere P, Cocaign-Bousquet M. 2005. Transcriptome analysis of the progressive adaptation of Lactococcus lactis to carbon starvation. J. Bacteriol. 187:3589–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sanin SL. 2003. Effect of starvation on resuscitation and the surface characteristics of bacteria. J. Environ. Sci. Health A Tox. Hazard. Subst. Environ. Eng. 38:1517–1528 [DOI] [PubMed] [Google Scholar]
- 65. Seto M, et al. 1995. A novel transformation of polychlorinated biphenyls by Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:3353–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sharp JO, et al. 2007. An inducible propane monooxygenase is responsible for N-nitrosodimethylamine degradation by Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 73:6930–6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Smeulders MJ, Keer J, Speight RA, Williams HD. 1999. Adaptation of Mycobacterium smegmatis to stationary phase. J. Bacteriol. 181:270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Song T, et al. 2010. Cloning and expression analysis of the duplicated genes for carbon monoxide dehydrogenase of Mycobacterium sp. strain JC1 DSM 3803. Microbiology 156(Pt 4): 999–1008 [DOI] [PubMed] [Google Scholar]
- 69. van der Geize R, Dijkhuizen L. 2004. Harnessing the catabolic diversity of rhodococci for environmental and biotechnological applications. Curr. Opin. Microbiol. 7:255–261 [DOI] [PubMed] [Google Scholar]
- 70. Wang G, Maier RJ. 2004. An NADPH quinone reductase of Helicobacter pylori plays an important role in oxidative stress resistance and host colonization. Infect. Immun. 72:1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Warren R, et al. 2004. Functional characterization of a catabolic plasmid from polychlorinated-biphenyl-degrading Rhodococcus sp. strain RHA1. J. Bacteriol. 186:7783–7795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wayne LG, Hayes LG. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Weichart D, Querfurth N, Dreger M, Hengge-Aronis R. 2003. Global role for ClpP-containing proteases in stationary-phase adaptation of Escherichia coli. J. Bacteriol. 185:115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wilson GG, Murray NE. 1991. Restriction and modification systems. Annu. Rev. Genet. 25:585–627 [DOI] [PubMed] [Google Scholar]
- 75. Yu AY, Houry WA. 2007. ClpP: a distinctive family of cylindrical energy-dependent serine proteases. FEBS Lett. 581:3749–3757 [DOI] [PubMed] [Google Scholar]
- 76. Zelenaya-Troitskaya O, Perlman PS, Butow RA. 1995. An enzyme in yeast mitochondria that catalyzes a step in branched-chain amino acid biosynthesis also functions in mitochondrial DNA stability. EMBO J. 14:3268–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.