Abstract
Mercury (Hg) resistance (mer) by the reduction of mercuric to elemental Hg is broadly distributed among the Bacteria and Archaea and plays an important role in Hg detoxification and biogeochemical cycling. MerA is the protein subunit of the homodimeric mercuric reductase (MR) enzyme, the central function of the mer system. MerA sequences in the phylum Aquificae form the deepest-branching lineage in Bayesian phylogenetic reconstructions of all known MerA homologs. We therefore hypothesized that the merA homologs in two thermophilic Aquificae, Hydrogenobaculum sp. strain Y04AAS1 (AAS1) and Hydrogenivirga sp. strain 128-5-R1-1 (R1-1), specified Hg resistance. Results supported this hypothesis, because strains AAS1 and R1-1 (i) were resistant to >10 μM Hg(II), (ii) transformed Hg(II) to Hg(0) during cellular growth, and (iii) possessed Hg-dependent NAD(P)H oxidation activities in crude cell extracts that were optimal at temperatures corresponding with the strains' optimal growth temperatures, 55°C for AAS1 and 70°C for R1-1. While these characteristics all conformed with the mer system paradigm, expression of the Aquificae mer operons was not induced by exposure to Hg(II) as indicated by unity ratios of merA transcripts, normalized to gyrA transcripts for hydrogen-grown AAS1 cultures, and by similar MR specific activities in thiosulfate-grown cultures with and without Hg(II). The Hg(II)-independent expression of mer in the deepest-branching lineage of MerA from bacteria whose natural habitats are Hg-rich geothermal environments suggests that regulated expression of mer was a later innovation likely in environments where microorganisms were intermittently exposed to toxic concentrations of Hg.
INTRODUCTION
Microbes must have been exposed to toxic heavy metals since the beginning of life on Earth and have evolved diverse mechanisms to live in the presence of high concentrations of toxic metal ions (42). These mechanisms, such as efflux, intra- or extracellular precipitation, and enzyme-mediated transformations, control intracellular concentrations of heavy metal ions that may be inhibitory to physiological functions and form nonspecific complex compounds in the cell (28). While much of our existing knowledge of these resistance mechanisms has arisen from research motivated by metal contamination from the perspective of human and environmental health (5, 9, 11, 26, 29), a cosmopolitan distribution of metal-resistant microorganisms inhabiting environments that are enriched with metals of geological origin suggests evolution of metal ion resistance prior to industrial release of metal contaminants (2, 13, 50).
Mercury (Hg) is a potent neurotoxic substance and the heavy metal most toxic to microorganisms due to its high affinity to sulfur (27). Globally distributed Hg (3) is toxic to humans and wildlife, mostly due to the accumulation of methylmercury (MeHg) in aquatic and terrestrial food webs (7). Microbial activities are central in modulating environmental Hg toxicity and mobility. Resistance to inorganic Hg [Hg(II)] is controlled by the activities of the enzyme mercuric reductase (MR), an NAD(P)H-dependent flavin oxidoreductase which catalyzes the reduction of Hg(II) to the elemental form, Hg(0). The gene encoding MR, merA, is part of the Hg resistance (mer) operon, which is widespread among both Bacteria and Archaea (2, 3, 43), allowing these organisms to survive in the presence of elevated Hg concentrations (3, 4). At a minimum, Hg resistance systems are comprised of transport, enzymatic, and regulatory functions. MerT and a number of alternative transporters are involved in the transport of thiolated Hg(II) into the cytoplasm for reduction by MR (3). MerR regulates expression of the mer operon, binding to the operator/promoter (O/P) region to repress transcription in the absence of Hg(II). When present, Hg(II) binds to the MerR-mer O/P and RNA polymerase complex, prompting the DNA to unwind, inducing transcription of the operon's functional genes (3, 17).
A recent body of literature supports the hypothesis that microbial resistance to Hg evolved in geothermal environments where microbial life has perhaps been exposed to Hg since the beginning of life on Earth (2, 35, 48). Mercury-resistant microbes were readily isolated from deep-sea hydrothermal vents (48) and terrestrial hot springs (10, 43), and their distribution suggested a role in adaptation to Hg toxicity. Culture-independent techniques detected mer genes in Yellowstone National Park (YNP) (50) and Coso Hot Springs, CA (43). The large-scale sequencing of microbial genomes has resulted in an increased availability of merA sequences and allowed for a robust analysis of gene evolution, further supporting an origin and early evolution of Hg resistance among thermophilic microbes from geothermal environments (2).
To date, functional mer operons have been characterized in mesophilic Actinobacteria, in Firmicutes, among the Beta- and Gammaproteobacteria (2), and in one thermophilic bacterium representing an early bacterial lineage, Thermus thermophilus HB27 (51). The phylum Aquificae contains primary producers which are dominant in many geothermal environments (46) and represents the deepest-branching bacterial lineage (23). Furthermore, merA homologs in the genomes of Hydrogenobaculum sp. strain Y04AAS1 (AAS1) and Hydrogenivirga sp. strain 128-5-R1-1 (R1-1) (34) form the deepest-branching lineage in a MerA phylogeny (2). Strain AAS1 was isolated from a small channel proximal to Obsidian Pool Prime, YNP, and R1-1 was isolated from the Eastern Lau Spreading Center, South Pacific (34), both of which are geothermal environments similar to those where elevated Hg concentrations were reported (12, 21, 31). The basal position of the Aquificae loci in the MerA phylogenetic reconstructions suggests that merA originated in an ancestor common to deep-branching thermophilic bacteria. Here, we report on the activity and characteristics of the Hg resistance systems of two Aquificae strains representing chemotrophic primary producers in many geothermal environments.
MATERIALS AND METHODS
MerA phylogeny and bioinformatic analyses.
MerA sequences were compiled in April 2011 by tblastx searches of the Entrez Protein database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=protein) using the MerA amino acid sequences of Tn501 (accession number CAA77323.1) and Sulfolobus solfataricus P2 (AAK42805.1) as queries. These searches identified two open reading frames (ORFs), HY04AAS1_1213 and HG1285_05690, as merA homologs in Hydrogenobaculum sp. Y04AAS1 and Hydrogenivirga sp. 128-R1-1, respectively. The alignment block of bacterial and archaeal MerA sequences, corresponding to positions 8 to 472 of Streptomyces lividans (P30341), were used in a Bayesian inferred phylogenetic reconstruction, performed as described by Wang et al. (50). Of the 284 gene homologs available, 99 were selected for reconstruction; the selected homologs represented all major clusters in the MerA phylogeny. Putative promoter regions were determined using the BPROM tool (Softberry Inc., Mt. Kisco, NY).
Bacterial strains and growth conditions.
Strains Hydrogenobaculum sp. Y04AAS1 (AAS1), Hydrogenivirga sp. 128-5-R1-1 (R1-1), and Persephonella marina EX-H1 were generously provided by Anna-Louise Reysenbach (Portland State University); major characteristics of these strains are summarized in Table S1 in the supplemental material. All growth media were prepared under a CO2 headspace, microaerophilic conditions were then created by the postautoclaving addition of O2 (to 4%, vol/vol), and the tubes were pressurized with either H2 or CO2 after inoculation. Strain AAS1 was grown at 55°C in modified DSMZ 743 medium (Boone's medium no. 5) as described by Shima and Suzuki (41), supplemented with 0.5 ml liter−1 of a trace element stock solution (adapted from Ferguson and Mah [15]). Elemental sulfur was replaced in this medium with S2O32− or H2 as the energy source, and the pH was adjusted to 4.5 with 6 N HCl prior to autoclaving. Strains R1-1 and EX-H1 were grown at 70°C in modified MSH medium (Boone's medium no. 2) (6) supplemented with 10 ml liter−1 of trace element stock solution, with pH adjusted to 6.0 with 6 M H2SO4. The headspace consisted of 96:4 CO2/O2 and 80:16:4 CO2/H2/O2 when grown on S2O32− and H2 as electron donors, respectively. In both media, S2O32− was omitted when H2 was provided as the sole electron donor, whereas no H2 was added to the headspace when S2O32− was used. Strain R1-1, which grew poorly with H2, was tested only in S2O32−-amended medium. Unless specifically described, all culture maintenance and growth experiments were performed in a final volume of 5 ml in 26-ml Balch tubes (Bellco, Vineland, NJ) fitted with crimp-sealed Teflon stoppers.
Growth measurements.
Growth was determined by measuring the optical density at a wavelength of 660 nm (A660) (Genesys 20; Thermo Spectronic Instruments, Waltham, MA) or by acridine orange direct counts (49). For direct count preparations of strain Hydrogenobaculum sp. Y04AAS1, acridine orange staining was carried out following cell filtration onto polycarbonate filters (General Electric, Feasterville-Trevose, PA) to minimize staining of medium precipitates. Cell numbers were obtained using an Olympus BX60 microscope with an oil immersion objective lens (Uplan F1 100×/1.3) and determined using Olympus Microsuite Basic (version 3.2) (Olympus Corp, Center Valley, PA).
Modeling of Hg speciation.
The chemical equilibrium speciation model MINEQL+ (version 4.5) (38) was used to determine the speciation of Hg in growth media. Input parameters were obtained from the MINEQL+ and the National Institute of Standards and Technology databases (24). Dissociation constants used as input parameters are shown in Table S2 in the supplemental material. Hg speciation in each growth medium was modeled at a HgCl2 concentration range of 5 to 60 μM.
Resistance to Hg(II).
Mid-log-phase cultures were diluted 100-fold into fresh growth medium to a final volume of 5 ml at an A660 of 0.010, HgCl2 was added to a final concentration of 0, 5, 10, 20 or 40 μM, and tubes were incubated at each organism's optimal growth temperature in the dark without shaking. Growth was monitored every 4 to 8 h until commencement of stationary phase. The effect of Hg on growth was expressed as cell density at a given HgCl2 concentration as a percentage of cell density of the zero-HgCl2 control when the control culture approached late exponential growth, with 5.3 × 107 and 2.8 × 107 cells ml−1 for strain AAS1 growing on S2O32− and H2, respectively, and 2.6 × 107 cells ml−1 for S2O32−-grown cultures of strain R1-1.
Loss of Hg(II) during culture growth.
Fresh growth media were inoculated with mid-log-phase cultures and were spiked with 5 or 10 μM 203HgCl2 (specific activity, 0.5 to 0.12 nCi [μmol Hg]−1; kindly provided by Christie Bridges [Mercer University, GA]) for cultures grown on H2 or S2O32−, respectively. 203Hg remaining in growth media was monitored by removing 250-μl aliquots from growing cultures every 4 to 8 h to 3 ml of Scinti-Verse scintillation fluid (Thermo Fisher Scientific, Waltham, MA), and samples were counted in a Beckman LS 6500 liquid scintillation counter (Beckman-Coulter, Brea, CA). Growth was measured in parallel cultures containing unlabeled HgCl2 at similar concentrations.
Maximum apparent specific Hg loss rates [fmol Hg(II) lost h−1 cell−1] were calculated as ([Hgt1−Hgt2]/[t2−t1])N−1, where N is the average number of cells at t1 and t2, Hg is fmol of Hg(II) lost between t1 and t2 as calculated from the concentration of Hg that remained in the growth medium at t1 and t2, and t1 and t2 are the times (h) bracketing the interval at which the loss of 203Hg from the medium was the greatest (37). Controls included autoclaved cells (105°C, 30 min) with identical inoculum sizes to test cultures as well as uninoculated media. Significance (P < 0.05) of differences in Hg(II) loss rates for each treatment was calculated using Student's t test.
Production of Hg(0) by growing cultures.
Cultures were grown with 203HgCl2 as described above to stationary phase when the headspace of the incubation vessels was flushed with sterile air for 40 min to drive 203Hg(0) that accumulated during growth into a Hg-trapping solution consisting of 0.75% KMnO4, 0.40% K2S2O3, 3.5% HNO3, and 5% H2SO4. At the conclusion of the 40 min, the remaining growth medium was acidified to 0.5 N HCl and mixed by vortexing. Aliquots of 250 μl were removed from the acidified stationary-phase culture and the trapping solution for scintillation counting. Control treatments included autoclaved cells and uninoculated growth medium.
Mercuric reductase (MR) assays.
Mid-log-phase cultures of Hydrogenobaculum sp. Y04AAS1 and Hydrogenivirga sp. 128-R1-1 were diluted 100-fold into 250 ml of medium in rubber-capped 2-liter Pyrex medium bottles (Corning, Lowell, MA) with a microaerophilic headspace as described under “Bacterial strains and growth conditions” above. Cultures were grown to mid-log phase, and cells were harvested by centrifugation for 10 min at 5,750 × g at 4°C in a prerefrigerated Sorvall RC-5B centrifuge (Thermo Scientific, Waltham, MA). Pelleted cells were washed in phosphate-buffered saline and stored at −20°C until used for further analysis.
Cell-free lysates were prepared by following protocols described by Vetriani et al. (48). Cells were resuspended to a concentration of 200 mg ml−1 (wet weight) in a buffer consisting of 20 mM sodium phosphate (pH 7.5), 0.5 mM EDTA, and 1 mM β-mercaptoethanol and were lysed by intermittent sonication using a Misonex S-4000 sonicator (Misonex, Newtown, CT) for a total of 3 min on ice. MR assays were performed in 80 mM sodium phosphate buffer (pH 7.4) with 1 mM β-mercaptoethanol, 200 μM NAD(P)H, and 50 μM HgCl2 in a final volume of 800 μl (16). Mercury-dependent NAD(P)H oxidation was monitored as the decrease in A340 using a UV-visible (UV-Vis) spectrophotometer (Cary 300; Agilent Technologies, Budd Lake, NJ). Assay temperatures were controlled using a water-jacketed cuvette holder connected to a water bath. Cell extracts and test buffer were preincubated at the assay temperature for 10 min prior to the addition of 1 to 4 μl of extract to the assay buffer. Initial rates of NAD(P)H oxidation were determined in the first 10 s, when the A340 decreased linearly with time. At least 3 different extract concentrations were tested, once with and once without the addition of HgCl2. Specific Hg-dependent NAD(P)H oxidation rates, expressed as units mg protein−1 [1 U = 1 μmol of NAD(P)H oxidized min−1], were calculated by subtracting the slope of the curve obtained in the absence of HgCl2 from that observed following HgCl2 addition. Protein concentrations in crude cell extracts were determined using the Bradford assay (Bio-Rad Laboratories Inc., Hercules, CA).
The effect of growth with and without Hg on MR levels of AAS1 and R1-1 was determined as described above except that treated cultures were grown with 10 μM HgCl2 on S2O32−. An additional 10 μM HgCl2 was added at mid-log phase, followed by continued incubation for 2 doubling times prior to cell harvesting. Significance (P < 0.05) of differences in MR activities among treatments was calculated by Student's t test.
Induction of merA transcription.
Cultures of R1-1 and AAS1 were grown in 25 ml of medium in 125-ml serum bottles (Wheaton Scientific, Millville, NJ). When the culture approached mid-log phase, 1 μM HgCl2 was added (t = 0), and 1.5-ml samples were withdrawn at 0, 10, 30, and 60 min. Cells were harvested by centrifugation (1 min, 13,000 × g) and immediately frozen at −80°C. Control cultures (zero Hg) were included to determine basal levels of merA transcription. Total RNA was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA) and diluted to 20 μg ml−1 prior to DNase treatment with the Turbo DNA-free kit (Applied Biosystems, Carlsbad, CA) as recommended in the manufacturer's instructions. cDNA was synthesized using the High-Capacity cDNA reverse transcription (RT) kit (Applied Biosystems) using a GeneAmp PCR System 9700 (Applied Biosystems) thermocycler. Relative merA fold induction was measured by quantitative real-time PCR (qPCR) using cDNA as a template and was normalized to transcription levels of gyrA, a constitutively expressed gene. No RT controls were performed to rule out the presence of DNA contamination.
For Hydrogenobaculum sp. Y04AAS1, primers RT-HBmerA(f/r) and HBgyrA(f/r) were used to amplify 71- and 66-bp regions of merA- and gyrA-specific products, respectively (see Table S3 in the supplemental material). For Hydrogenivirga sp. 128-R1-1, RT-HVmerA(f/r) and HVgyrA(f/r) were used to produce 70-bp merA-specific and 107-bp gyrA-specific products, respectively. Primers were designed using default parameters within Primer Express (version 3.0) (Applied Biosystems), using merA and gyrA sequences in the genomes of AAS1 (gyrA locus HY04AAS1_0371) and R1-1 (gyrA locus HG1285_00715). The Power SYBR green PCR master mix (Applied Biosystems) was used in all qPCRs. Amplifications were performed in triplicate for both merA and gyrA transcripts using the StepOne Plus PCR machine running StepOne software (version 2.1) (Applied Biosystems). qPCR conditions included an initial denaturation step of 90°C for 10 min and then 45 cycles of 90°C for 15 s followed by 1 min at 55°C for all primer sets used. Upon completion, a melt curve was performed to verify identity of the amplification products. Relative induction levels were calculated by the comparative threshold cycle (CT) method (30), whereby expression of merA was related to that of gyrA in cultures grown with and without Hg. ΔCT values were obtained for each time point following the addition of HgCl2 to exposed cultures.
RESULTS AND DISCUSSION
Identification of putative mer operons in the genomes of Hydrogenobaculum sp. Y04AAS1 and Hydrogenivirga sp. 128-5-R1-1.
An updated phylogeny that included all MerA homologs in databases as of April 2011 confirmed (2) the basal position of the Aquificae sequences in a sister position to all archaeal sequences with posterior probability values of 100. This cluster shared a common, likely bacterial, ancestor with a large cluster consisting of the remaining bacterial MerA sequences (see Fig. S1 in the supplemental material). These results highlight the important position of the Aquificae MerA, potentially representing an ancestral state of the protein among all prokaryotes.
The MerA sequences of both Aquificae loci contained signature motifs known to be required for MerA activity (Fig. 1A) (3). In Hydrogenobaculum sp. Y04AAS1, MerA is 464 amino acids (aa) long, while in Hydrogenivirga sp. 128-R1-1, it is 545 residues long due to the presence of NmerA, the ∼70-aa N-terminal extension heavy metal-associated (HMA) domain that is a part of more than half of all MerA sequences (2). Mercuric reductase activities were documented for both variants of MerA (51), and NmerA is not essential for MerA activity (22).
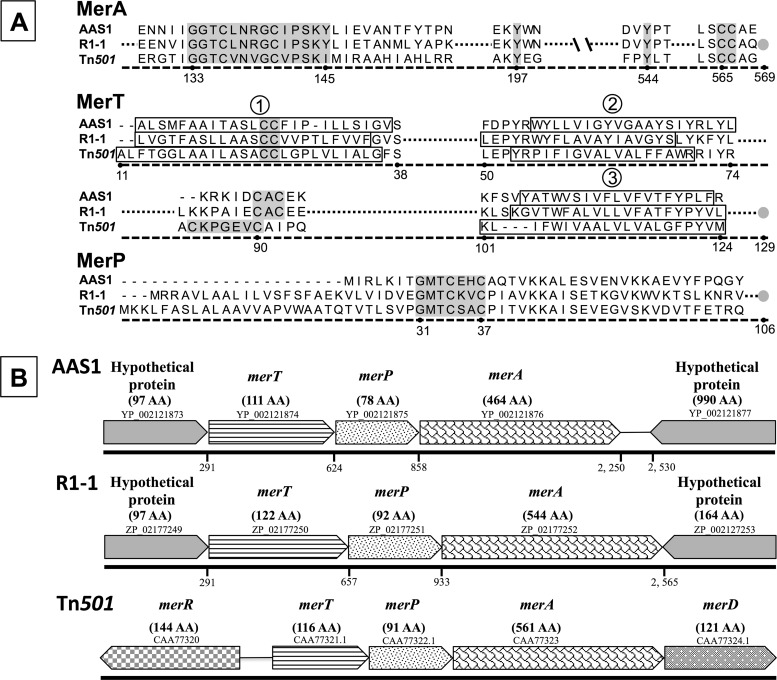
Fig 1.
(A) Alignment of putative Mer proteins, including MerA, MerT, and MerP from Hydrogenobaculum sp. Y04AAS1 (AAS1) and Hydrogenivirga sp. 128-5-R1-1 (R1-1), with mer sequences from Pseudomonas aeruginosa Tn501 as a reference. Highlighted conserved regions of functional importance (3) are as follows: for MerA, the redox active site, two downstream tyrosines, and the carboxy terminus-vicinal CC pair; for MerT, the conserved C residues in the first membrane-spanning domain and in the cytoplasmic loop between the second and third membrane-spanning domains; and for MerP, the metal binding motif GMTCxxC. The membrane-spanning domains (determined using TMpred [http://www.ch.embnet.org/software/TMPRED_form.html]) are boxed (numbered 1 to 3) in MerT. Gray circles indicate the carboxy termini of the proteins. Numbering indicates position in the Tn501 sequence. (B) mer operon gene order. Arrowed boxes indicate each ORF and the direction of transcription; accession numbers are included above each box. Names of putative gene products and corresponding numbers of amino acids (AA) are given above the boxes. The numbered line below each ORF represents the nucleotide position marking the start of the gene counted from the transcription start nucleotide upstream of the hypothetical proteins in each operon.
In both Aquificae strains, two ORFs upstream of the putative merA code for proteins that bear homology to other Mer functions (Fig. 1B). The merA-proximal ORFs, loci YP_002121875 (78 aa) and ZP_02177251 (92 aa) in the genomes of strains AAS1 and R1-1, respectively, may encode the Hg-scavenging protein MerP, as they include the signature metal binding motif sequence GMTCxxC (Fig. 1A). However, the ∼19-residue Sec-type signal known to direct proteobacterial MerP to the periplasmic space (3, 20) is missing in both Aquificae loci, leaving in question the transport of their gene products to the periplasmic space and thus their function as MerP.
ORFs encoding MerT homologs are found upstream of the putative merP genes in both genomes (loci HY04AAS1_1211 and ZP_02177250 [Fig. 1A]). Homology includes three hydrophobic inner membrane-spanning sequences as determined by TMpred (18), the vicinal C pair in the first membrane-spanning sequence, and a C pair located in the cytoplasmic loop between the second and the third membrane-embedded sequences (36).
Homologs of the mer operon regulator, MerR, were not found in the genomes of strains AAS1 and R1-1, though several homologs of ArsR/SmtB, a regulator reported in mer systems of the Archaea and the Actinobacteria (2, 3), were identified using tBLASTn searches. Based on homology searches, putative promoter regions (33) were identified 26 and 15 bp upstream of the ATG start codon of the ORFs preceding merT in strains Hydrogenobaculum sp. Y04AAS1 and Hydrogenivirga sp. 128-R1-1, respectively. These included −35 and −10 regions (separated by 16 and 17 nucleotides, rather than by 19 nucleotides as is common to promoters that are regulated by MerR [8]), Shine-Dalgarno ribosomal binding sites, and translation initiation signals. No other possible promoter sequences were identified upstream of any of the mer homologs of these strains.
Mercury resistance in Hydrogenobaculum sp. AAS1, Hydrogenivirga R1-1, and Persephonella marina EX-H1.
The technology to genetically manipulate bacteria that belong to the Aquificae is not yet available, and thus, obtaining merA mutants of strains AAS1 and R1-1 was not possible. We therefore compared Hg tolerance in strains AAS1 and R1-1 to that of P. marina EX-H1 as a control. Mercury forms complexes with medium components, which control its bioavailability and therefore toxicity (14). Employed growth media included either S2O32− or H2 as a sole energy source, leading to very different Hg(II) speciations. MINEQL+ calculations revealed that in the presence of 10 μM HgCl2 and 8 mM S2O32−, Hg(II) speciated as negatively charged Hg-S2O3 complexes, with 84% as Hg(S2O3)22− and 16% as Hg(S2O3)34− (see Table S4 in the supplemental material). In the H2 growth media, Hg speciated as uncharged HgCl2 and HgCl(OH) complexes. The model suggested similar Hg speciation in the range of 2 to 60 μM added HgCl2 (data not shown).
Using H2 and S2O32− as electron donors, the lowest tested HgCl2 concentrations, 2 μM and 5 μM, respectively, completely inhibited growth of the control P. marina EX-H1, while Hydrogenobaculum sp. Y04AAS1 grew in the presence of 10 and 20 μM HgCl2 in the H2 and S2O32− media, respectively, and Hydrogenivirga sp. 128-R1-1 grew in 20 μM HgCl2 in S2O32− medium (see Fig. S2 in the supplemental material). While it is clear that the two strains with putative mer operons had a higher tolerance to Hg than the control strain, it was impossible to quantitatively compare their responses or to assess the effects of electron donor and Hg speciation on the level of Hg resistance due to variable Hg loss from uninoculated S2O32−-amended media (see Fig. S3 in the supplemental material; see also below).
Loss of Hg(II) during growth of strains AAS1 and R1-1.
To determine if growth of strains AAS1 and R1-1 was related to the removal of Hg(II) from growth media, changes in Hg(II) concentrations were monitored during growth to stationary phase (Fig. 2). Growth experiments were performed using 5 or 10 μM HgCl2 in media containing H2 or S2O32−, respectively—concentrations that inhibited, though did not abolish, growth in either medium (see Fig. S2 in the supplemental material).
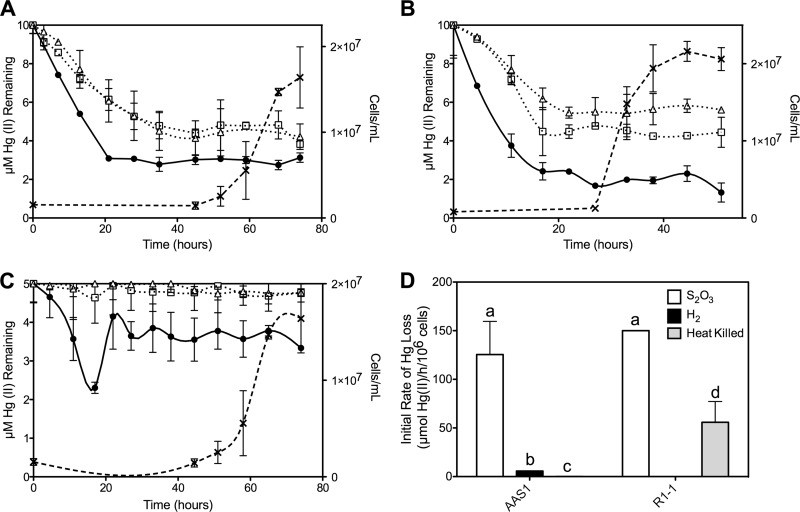
Fig 2.
203Hg(II) remaining in media during growth of strains AAS1 (A and C) and R1-1 (B) using S2O32− (A and B) and H2 (C) as electron donors. Cell density (×) is shown, as is loss of Hg(II) (in μM) from growing cultures (●), heat-killed controls (□), and uninoculated controls (△). (D) Initial rates of Hg(II) loss [μmol Hg(II)/h/106 cells] calculated for strains AAS1 and R1-1 in cultures grown on S2O32− or H2 and for heat-killed controls. Different letters indicate statistical significance (P < 0.05). All data represent the mean Hg(II) loss of triplicate growing cultures ± 1 standard deviation (SD) after subtracting loss rates of uninoculated controls.
Loss of 203Hg(II) from all growth media occurred well before the commencement of growth by strains R1-1 and AAS1. Initial rates of Hg loss from inoculated media were calculated after subtracting loss from uninoculated controls (see below). For strain AAS1, loss was approximately 25-fold faster when strains were grown on S2O32− rather than H2, with initial rates of Hg(II) loss of 125.5 ± 34.1 and 5.7 ± 0.03 μmol Hg(II) lost h−1 106 cells−1, respectively (P < 0.02 [Fig. 2D]). This is not surprising considering the higher energy yield from the oxidation of S2O32− (ΔG° = −818.3 kJ/reaction) than from that of H2 (ΔG° = −237 kJ/reaction) and the dependency of Hg(II) reduction by MR on availability of reducing equivalents (4). Strain R1-1 lost Hg(II) at a rate of 150.0 ± 0.01 μmol Hg(II) h−1 106 cells−1, statistically indistinguishable from the rate of strain AAS1 grown on S2O32−. Interestingly, more Hg was lost from the heat-killed controls of strain R1-1 than from the uninoculated controls (P < 0.02) at a rate that was slightly, though significantly (P = 0.05), higher. Since autoclaving (105°C for 30 min) resulted in cell death, as indicated by failure to grow following such treatment (data not shown), the MR of strain R1-1 must be highly tolerant to heat.
In the S2O32−-amended media, significant amounts of 203Hg(II) were lost (P < 0.05) from the heat-killed and uninoculated controls (see Fig. S3 in the supplemental material) that were incubated at 55°C (strain AAS1 controls [Fig. 2A]) and 70°C (strain R1-1 controls [Fig. 2B]). At the commencement of growth, medium-only controls of strain AAS1 (at >40 h postinoculation) and R1-1 (at >20 h) had only 4.8 ± 1.3 and 5.4 ± 1.4 μM Hg(II) remaining in the media, respectively, approximately half of the starting concentration. No such loss was observed in the H2-supplemented medium. Since loss did not occur when S2O32− medium-only controls were incubated at room temperature (data not shown), this abiotic loss resulted from elevated temperatures. This suggests that uncharged Hg(II) complexes are less volatile at elevated temperatures than the charged Hg-S2O3 complexes. Nevertheless, the live growing cultures of both strains lost a significantly greater proportion of 203Hg(II) than all abiotic controls (P < 0.05).
The Hg concentration in H2-amended AAS1 cultures initially declined, then increased at 20 h following inoculation, and then remained stable at about 3 μM (Fig. 2C). Since incubations were carried out in closed systems where the formed gaseous Hg(0) likely partitioned between the medium and the headspace according to Henry's coefficient (at 35°C, kH′ = 0.4 dimensionless [1]), it is possible that some of the initially formed Hg(0) was oxidized to Hg(II) by the H2-metabolizing culture. Bacterial Hg(0) oxidation has been reported for Escherichia coli, Bacillus subtilis, and Streptomyces venezuelae and attributed to the enzymes catalase and hydroperoxidase (45). The results clearly show that cultures of Hydrogenivirga sp. 128-R1-1 and Hydrogenobaculum sp. Y04AAS1 removed Hg from their growth media and that most of this activity took place during the lag phase of growth.
Production of Hg(0) by growing cultures of AAS1 and R1-1.
Endpoint mass balance experiments determined that the Hg(II) which was lost during growth of strains AAS1 and R1-1 was converted to Hg(0) (Table 1). When grown to early stationary phase on S2O32−, strains AAS1 and R1-1 produced 3.8 ± 2.4 and 3.1 ± 1.3 μmol Hg(0), respectively. When grown on H2, strain AAS1 produced 2.2 ± 0.8 μmol Hg(0), statistically similar to the amount produced by S2O32−-grown cultures. Production of Hg(0) by heat-killed controls was approximately 10-fold lower than that by growing cultures for all treatments. Mass balance calculations showed recoveries of 85 to 122% of the added 203HgCl2.
Table 1.
Reduction of Hg(II) to Hg(0) by Aquificae culturesa
| Strain | Electron donor | Treatment | Medium Hg (μM)b | Headspace Hg (μM) |
|---|---|---|---|---|
| AAS1 | H2 | Growing cultures | 3.91 ± 0.18A | 2.19 ± 0.79D |
| Heat killed | 4.32 ± 0.80A | 0.21 ± 0.06E | ||
| S2O32− | Growing cultures | 5.02 ± 1.47A | 3.80 ± 2.38D | |
| Heat killed | 10.87 ± 2.60B | 0.34 ± 0.03F | ||
| R1-1 | S2O32− | Growing cultures | 6.69 ± 0.48C | 3.09 ± 1.34D |
| Heat killed | 8.13 ± 0.48B | 0.32 ± 0.01F |
Different letters indicate statistical significance (P < 0.05) by Student's t test.
HgCl2 was added to initial concentrations of 5 and 10 μM to H2- and S2O32−-containing growth media, respectively.
MR activities by crude cell extracts of strains AAS1 and R1-1.
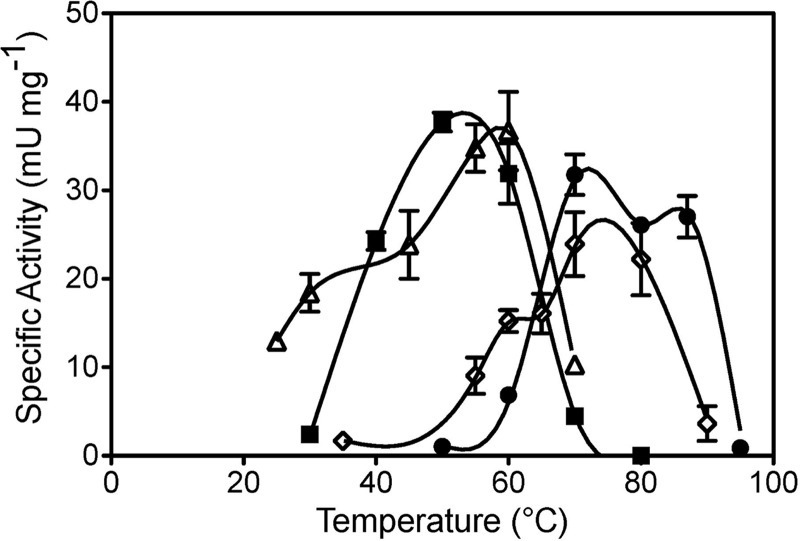
Specific rates of MR activity were measured to determine this enzyme's role in the formation of Hg(0) by Hydrogenobaculum sp. Y04AAS1 and Hydrogenivirga sp. 128-R1-1. Preliminary experiments indicated a preference for NADH by R1-1 and for NADPH by AAS1 crude cell extracts (data not shown), and therefore all further experiments were performed with each strain's preferred reductant. Given the developing understanding of the importance of mer in thermophilic microbes (2, 40, 51), MR activities of strains AAS1 and R1-1 were determined at a range of temperatures. Results showed a correspondence between optimal temperatures for growth and for Hg-dependent NAD(P)H oxidation with maximum apparent specific MR activity for AAS1 (37.7 ± 1.1 mU mg protein−1) at 50°C and for R1-1 (3.2 ± 0.2 mU mg protein−1) at 70°C (Fig. 3). Strain AAS1's MR was active at a temperature range of 30 to 70°C and that of strain R1-1 was active from 60 to 87°C, and both extracts had very low activities below 40°C. MR activity was not detected in crude extracts of the control strain P. marina EX-H1 when tested at its optimal growth temperature, 70°C. These results suggest that strains AAS1 and R1-1 possessed thermophilic MR.
Fig 3.
Effect of temperature on MR activities. Specific activities of crude cell extracts were determined for Hydrogenobaculum sp. Y04AAS1 (■) and Hydrogenivirga sp. 128-5-R1-1 (●). Activities of R1-1 extracts are expressed as those measured, multiplied by 10. Previously reported data for T. thermophilus (♢) and Tn501 (△) (51) are included for comparison. Averages of three to five replicate assays ± 1 SD are presented. One unit of MR activity = 1 μmol NAD(P)H oxidized min−1.
Induction of merA transcription and MR activity.
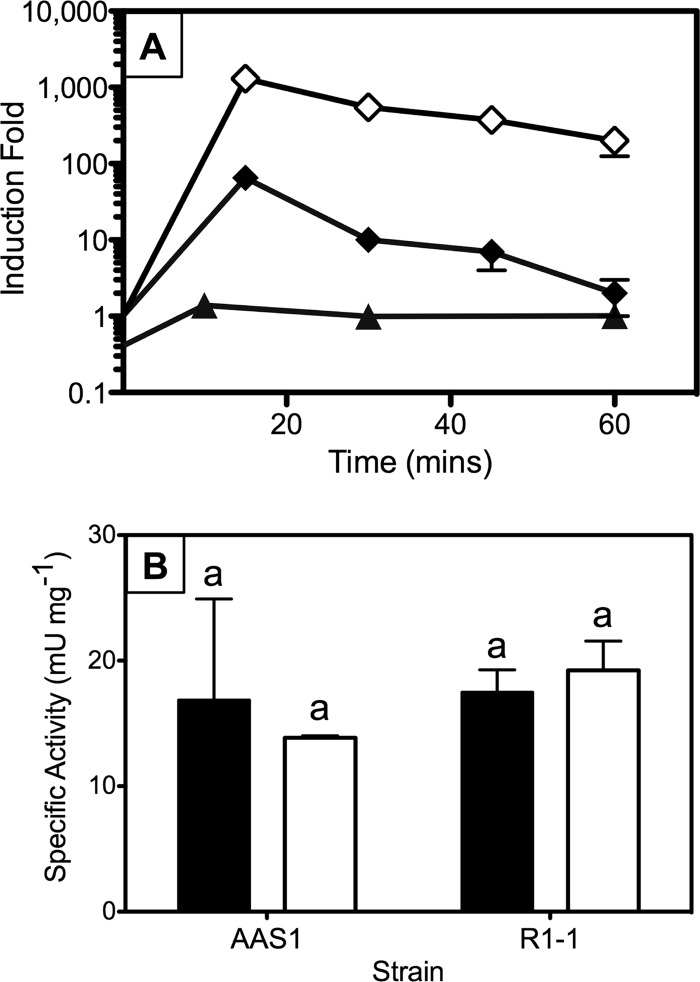
mer operon expression is regulated in both Bacteria (47) and Archaea (39) by the regulatory protein MerR and, less frequently, by ArsR/SmtB-like regulators; most mer operons code for these regulators (3). Because gene homologs of these regulators were not found in proximity to merA of strains AAS1 and R1-1, we determined the effects of growth in the presence of HgCl2 on levels of merA transcription and on MR specific activities. Results for H2-grown Hydrogenobaculum sp. Y04AAS1 clearly showed that exposure to subtoxic concentrations of Hg (1 μM) did not induce merA expression, as indicated by a relative fold induction of 1 for the first 60 min following exposure (Fig. 4A). In contrast, merA expression in T. thermophilus HB27 and in E. coli carrying a chromosomal insertion of Tn501 was induced 65- and 1,300-fold, respectively, relative to expression of the 16S rRNA gene (51). Unfortunately, transcript quantitation was not possible for thiosulfate-grown cultures due to the low quality of cDNA preparations. In such cultures, however, similar levels of MR activity were observed in crude cell extracts of cultures that had been grown with or without HgCl2, with values of 17.5 ± 1.8 and 19.2 ± 1.3 mU mg protein−1 and 16.9 ± 8.0 and 13.6 ± 0.2 mU mg protein−1 for Hg-exposed and unexposed R1-1 and AAS1 cultures, respectively (Fig. 4B).
Fig 4.
merA expression in Hydrogenobaculum sp. Y04AAS1 and Hydrogenivirga sp. 128-5-R1-1. (A) HgCl2-dependent (1 μM) transcription fold induction in strain AAS1 (▲) grown on H2 compared with induction of merA of HB27 (⧫) and Tn501 (◇). Fold induction was calculated as described in Materials and Methods, except that merA expression in Tn501 and HB27 was normalized to expression of 16S rRNA genes rather than to gyrA (data from reference 51). (B) Effect of growth in the presence of Hg on MR specific activities in crude cell extracts of strains AAS1 and R1-1. Cell extract activities were determined for cultures grown with (filled column) or without (clear column) 10 μM HgCl2 in S2O32−-amended media. The means ± 1 SD of triplicate determinations are shown. The same letters above columns indicate no significant difference by Student's t test (P > 0.05).
We conclude that unlike the majority of previously described mer systems (3), expression of mer in strains AAS1 and R1-1 was not induced by exposure to Hg(II). The mer operon of the plasmid pMERPH does not include an adjacent merR, though its expression is inducible, likely associated with a regulatory gene located elsewhere on the plasmid (29a). A constitutively expressed Hg-dependent NADH oxidation was recently described for the marine methylotroph Methylococcus capsulatus (Bath) (5a). Mercury-independent expression of mer among the Aquificae may not be surprising considering that members of this phylum often occupy sulfide-rich geothermal environments (19) where Hg levels were reported at nanomolar to micromolar concentrations (21, 50). Obviously, an elaborate regulatory system would be superfluous in environments with persistent exposure to Hg.
Evolutionary implications.
Phylogenetic reconstructions consistently place Aquificae sequences in a basal position to all bacterial and archaeal MerA proteins (see Fig. S1 in the supplemental material) (2). Thus, our results suggest that the expression of the ancestral Hg detoxification system, which likely originated among thermophilic bacteria in geothermal environments, is not regulated by exposure to Hg. If so, Hg-dependent regulation of this system was likely acquired later, possibly among organisms only intermittently exposed to toxic levels of Hg. We have previously pointed out the increased number of mer functions with MerA evolution (2); the present study further clarifies this system's evolutionary path from simple operons, whose expression does not depend on exposure to Hg, to more complex and finely regulated systems (9a, 17, 45a).
Furthermore, the correspondence of optimal growth and MR activity temperatures of the two thermophilic Aquificae strains (Fig. 3) supports the conclusion that merA originated among thermophiles (2, 51). In contrast, the optimal temperature of Tn501's enzyme was >20°C higher than the optimal growth temperature of Pseudomonas aeruginosa (Fig. 3), the mesophilic Hg-resistant bacterium from which Tn501 was isolated (46a). It has been previously proposed that this discrepancy between the optimal growth and enzyme activity temperatures was a relic of MR evolution in high-temperature environments (51). Our results, therefore, strengthen this conclusion and the identification of high-temperature environments as the place for the origin and early evolution of the microbial Hg detoxification system (2, 48, 51).
Supplementary Material
ACKNOWLEDGMENTS
We thank Anna-Louise Reysenbach, Yitai Liu, and Gilbert Flores (Portland State University) and Eric Boyd and Seth D'Imperio (Montana State University) for help regarding culture maintenance and growth, Costatino Vetriani (Rutgers University) for providing advice, lab space, and materials, Christie Bridges (Mercer University, GA) for providing 203HgCl2, Elisabetta Bini (Rutgers) and Anne Summers (University of Georgia) for help deciphering promoter sequences, and Bill Belden and Allison Isola (Rutgers) for advice and assistance in merA expression experiments. Gratitude is also extended to three anonymous reviewers whose comments helped us improve the manuscript.
The financial support of a graduate student travel award by the Thermal Biology Institute (Montana State University), small grants from the Department of Biochemisty and Microbiology, and the Graduate Program in Ecology and Evolution (Rutgers University) is acknowledged, as is grant NSF-PEET-DEB-0328326 to Anna-Louise Reysenbach.
Footnotes
Published ahead of print 6 July 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Andersson ME, Gardfeldt K, Wangberg I, Stromberg D. 2008. Determination of Henry's law constant for elemental mercury. Chemosphere 73:587–592 [DOI] [PubMed] [Google Scholar]
- 2. Barkay T, Kritee K, Boyd E, Geesey GG. 2010. A thermophilic bacterial origin and subsequent constraints by redox, light and salinity on the evolution of the microbial mercuric reductase. Environ. Microbiol. 12:2904–2917 [DOI] [PubMed] [Google Scholar]
- 3. Barkay T, Miller SM, Summers AO. 2003. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 27:355–384 [DOI] [PubMed] [Google Scholar]
- 4. Barkay T, Wagner-Döbler I. 2005. Microbial transformations of mercury: potentials, challenges, and achievements in controlling mercury toxicity in the environment. Adv. Appl. Microbiol. 57:1–52 [DOI] [PubMed] [Google Scholar]
- 5. Ben-David EA, Holden PJ, Stone DJ, Harch BD, Foster LJ. 2004. The use of phospholipid fatty acid analysis to measure impact of acid rock drainage on microbial communities in sediments. Microb. Ecol. 48:300–315 [DOI] [PubMed] [Google Scholar]
- 5a. Boden R, Murrell JC. 2011. Response to mercury (II) ions in Methylococcus capsulatus (Bath). FEMS Microbiol. Lett. 324:106–110 [DOI] [PubMed] [Google Scholar]
- 6. Boone DR, Johnson RL, Liu Y. 1989. Diffusion of the interspecies electron carriers H2 and formate in methanogenic ecosystems and its implications in the measurement of Km for H2 or formate uptake. Appl. Environ. Microbiol. 55:1735–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boyd ES, et al. 2009. Methylmercury enters an aquatic food web through acidophilic microbial mats in Yellowstone National Park, WY. Environ. Microbiol. 11:950–959 [DOI] [PubMed] [Google Scholar]
- 8. Brown NL, Stoyanov JV, Kidd SP, Hobman JL. 2003. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27:145–163 [DOI] [PubMed] [Google Scholar]
- 9. Bruce KD, Osborn AM, Pearson AJ, Strike P, Ritchie DA. 1995. Genetic diversity within mer genes directly amplified from communities of noncultivated soil and sediment bacteria. Mol. Ecol. 4:605–612 [DOI] [PubMed] [Google Scholar]
- 9a. Champier L, Duarte V, Michaud-Soret I, Coves J. 2004. Characterization of the MerD protein from Ralstonia metallidurans CH34: a possible role in bacterial mercury resistance by switching off the induction of the mer operon. Mol. Microbiol. 52:1475–1485 [DOI] [PubMed] [Google Scholar]
- 10. Chatziefthimiou AD, Crespo-Medina M, Wang Y, Vetriani C, Barkay T. 2007. The isolation and initial characterization of mercury resistant chemolithotrophic thermophilic bacteria from mercury rich geothermal springs. Extremophiles 11:469–479 [DOI] [PubMed] [Google Scholar]
- 11. Craw D. 2005. Potential anthropogenic mobilisation of mercury and arsenic from soils on mineralised rocks, Northland, New Zealand. J. Environ. Manage. 74:283–292 [DOI] [PubMed] [Google Scholar]
- 12. Crespo-Medina M, et al. 2009. Adaptation of chemosynthetic microorganisms to elevated mercury concentrations in deep-sea hydrothermal vents. Limnol. Oceanogr. 54:41–49 [Google Scholar]
- 13. Cuebas M, Sannino D, Bini E. 2011. Isolation and characterization of arsenic resistant Geobacillus kaustophilus strain from geothermal soils. J. Basic Microbiol. 51:364–371 [DOI] [PubMed] [Google Scholar]
- 14. Farrell RE, Germida JJ, Huang PM. 1990. Biotoxicity of mercury as influenced by mercury(II) speciation. Appl. Environ. Microbiol. 56:3006–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ferguson TJ, Mah RA. 1983. Isolation and characterization of an H2 oxidizing thermophilic methanogen. Appl. Environ. Microbiol. 45:265–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fox B, Walsh CT. 1982. Mercuric reductase: purification and characterization of a transposon-encoded flavoprotein containing an oxidation-reduction-active disulfide. J. Biol. Chem. 257:2498–2503 [PubMed] [Google Scholar]
- 17. Guo HB, et al. 2010. Structure and conformational dynamics of the metalloregulator MerR upon binding of Hg(II). J. Mol. Biol. 398:555–568 [DOI] [PubMed] [Google Scholar]
- 18. Hofmann K, Stoffel W. 1993. TMbase—a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374:166 [Google Scholar]
- 19. Huber R, Eder W. 2006. Aquificales, p 925–938 Dworkin M, Falkow S. (ed), The prokaryotes, vol 7 Springer Reference, New York, NY [Google Scholar]
- 20. Jackson WJ, Summers AO. 1982. Biochemical characterization of HgCl2-inducible polypeptides encoded by the mer operon of plasmid R100. J. Bacteriol. 151:962–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. King SA, et al. 2006. Mercury in water and biomass of microbial communities in hot springs of Yellowstone National Park, USA. Appl. Geochem. 21:1868–1879 [Google Scholar]
- 22. Ledwidge R, et al. 2005. NmerA, the metal binding domain of mercuric ion reductase, removes Hg(II) from proteins, delivers it to the catalytic core, and protects cells under glutathione-depleted conditions. Biochemistry 44:11402–11416 [DOI] [PubMed] [Google Scholar]
- 23. Madigan MT, Martinko JM, Stahl DA, Clark DP. 2010. Brock biology of microorganisms, 13th ed. Pearson/Benjamin Cummings, San Francisco, CA [Google Scholar]
- 24. Martell AE, Smith RM. 1974. Critical stability constants. Plenum Press, New York, NY [Google Scholar]
- 25.Reference deleted. [Google Scholar]
- 26. Ní Chadhain SM, Schaefer J, Crane S, Zylstra GJ, Barkay T. 2006. Analysis of mercuric reductase (merA) gene diversity in an anaerobic mercury-contaminated sediment enrichment. Environ. Microbiol. 8:1746–1752 [DOI] [PubMed] [Google Scholar]
- 27. Nies DH. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27:313–339 [DOI] [PubMed] [Google Scholar]
- 28. Nies DH. 1999. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51:730–750 [DOI] [PubMed] [Google Scholar]
- 29. Øregaard G, Sørensen SJ. 2007. High diversity of bacterial mercuric reductase genes from surface and sub-surface floodplain soil (Oak Ridge, USA). ISME J. 1:453–467 [DOI] [PubMed] [Google Scholar]
- 29a. Osborn AM, Bruce KD, Ritchie DA, Strike P. 1996. The mercury resistance operon of the IncJ plasmid pMERPH exhibits structural and regulatory divergence from other Gram-negative mer operons. Microbiology 142:337–345 [DOI] [PubMed] [Google Scholar]
- 30. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45 doi: 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Phelps D, Buseck PR. 1980. Distribution of soil mercury and the development of soil mercury anomalies in the Yellowstone geothermal area, Wyoming. Econ. Geol. 75:730–741 [Google Scholar]
- 32.Reference deleted. [Google Scholar]
- 33. Reese MG. 2001. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 26:51–56 [DOI] [PubMed] [Google Scholar]
- 34. Reysenbach A-L, et al. 2009. Complete and draft genome sequences of six members of the Aquificales. J. Bacteriol. 191:1992–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rosen BP. 2002. Transport and detoxification systems for transition metals, heavy metals and metalloids in eukaryotic and prokaryotic microbes. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 133:689–693 [DOI] [PubMed] [Google Scholar]
- 36. Rossy E, et al. 2004. Is the cytoplasmic loop of MerT, the mercuric ion transport protein, involved in mercury transfer to the mercuric reductase? FEBS Lett. 575:86–90 [DOI] [PubMed] [Google Scholar]
- 37. Schaefer J, Letowski J, Barkay T. 2002. mer-mediated resistance and volatilization of Hg(II) under anaerobic conditions. Geomicrobiol. J. 19:87–102 [Google Scholar]
- 38. Schecher WD, McAvoy D. 1994. MINEQL+: a chemical equilibrium program for personal computers. Environmental Research Software, Howell, ME [Google Scholar]
- 39. Schelert J, et al. 2004. Occurrence and characterization of mercury resistance in the hyperthermophilic archaeon Sulfolobus sulfataricus by use of gene disruption. J. Bacteriol. 186:427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schelert J, Drozda M, Dixit V, Dillman A, Blum P. 2006. Regulation of mercury resistance in the crenarchaeote Sulfolobus solfataricus. J. Bacteriol. 188:7141–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shima S, Suzuki KI. 1993. Hydrogenobacter acidophilus sp. nov., a thermoacidophilic, aerobic, hydrogen-oxidizing bacterium requiring elemental sulfur for growth. Int. J. Syst. Evol. Microbiol. 43:703–708 [Google Scholar]
- 42. Silver S, Phung LT. 2005. A bacterial view of the periodic table: genes and proteins for toxic inorganic ions. J. Ind. Microbiol. Biotechnol. 32:587–605 [DOI] [PubMed] [Google Scholar]
- 43. Simbahan J, et al. 2005. Community analysis of a mercury hot spring supports occurrence of domain-specific forms of mercuric reductase. Appl. Environ. Microbiol. 71:8836–8845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reference deleted. [Google Scholar]
- 45. Smith T, Pitts K, McGarvey JA, Summers AO. 1998. Bacterial oxidation of mercury metal vapor, Hg(0). Appl. Environ. Microbiol. 64:1328–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45a. Song LY, Teng Q, Phillips RS, Brewer JM, Summers AO. 2007. 19F-NMR reveals metal and operator-induced allostery in MerR. J. Mol. Biol. 371:79–92 [DOI] [PubMed] [Google Scholar]
- 46. Spear JR, Walker JJ, McCollom TM, Pace NR. 2005. Hydrogen and bioenergetics in the Yellowstone geothermal system. Proc. Natl. Acad. Sci. U. S. A. 102:2555–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46a. Stanisich VA, Bennett PM, Richmond MH. 1977. Characterization of a translocation unit encoding resistance to mercuric ions that occurs on a nonconjugative plasmid in Pseudomonas aeruginosa. J. Bacteriol. 129:1227–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Summers AO. 1986. Organization, expression, and evolution of genes for mercury resistance. Annu. Rev. Microbiol. 40:604–634 [DOI] [PubMed] [Google Scholar]
- 48. Vetriani C, et al. 2005. Mercury adaptation among bacteria from a deep-sea hydrothermal vent. Appl. Environ. Microbiol. 71:220–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vetriani C, Speck MD, Ellor SV, Lutz RA, Starovoytov V. 2004. Thermovibrio ammonificans sp. nov., a thermophilic, chemolithotrophic, nitrate-ammonifying bacterium from deep-sea hydrothermal vents. Int. J. Syst. Evol. Microbiol. 54:175–181 [DOI] [PubMed] [Google Scholar]
- 50. Wang Y, et al. 2011. Environmental conditions constrain the distribution and diversity of archaeal merA in Yellowstone National Park, Wyoming, USA. Microb. Ecol. 62:739–752 [DOI] [PubMed] [Google Scholar]
- 51. Wang Y, Freedman Z, Lu-Irving P, Kaletsky R, Barkay T. 2009. An initial characterization of the mercury resistance (mer) system of the thermophilic bacterium Thermus thermophilus HB27. FEMS Microbiol. Ecol. 67:118–129 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.