The characterization and role of conjugation-specific small RNAs in the protozoan Tetrahymena are discussed in this Perspective on the study by Mochizuki and colleagues (published in the August 1, 2012, issue). Parallels are also drawn with metazoan small RNAs.
Keywords: DNA elimination, Tetrahymena, epigenetics, small RNA
Abstract
There is growing evidence to support the notion that small RNAs derived from noncoding RNAs (ncRNAs) are mobile carriers of epigenetic information in diverse eukaryotic systems. However, challenges remain in defining what messages are being sent and how. In the August 1, 2012, issue of Genes & Development, Schoeberl and colleagues (pp. 1729–1742) reported a detailed analysis of the turnover of small RNAs during the sexual reproduction of the ciliated protozoan Tetrahymena. The results revealed surprisingly complicated roles played by small RNAs in shaping the communication between the germline and the soma.
The central dogma of molecular biology focuses on the protein-coding potential of nucleic acids, a point of view by which the vast majority of the DNA sequence in the human genome (and most other genomes) is denigrated to “junk DNA” status and RNA is seen as merely the middleman. From this perspective, the advent of noncoding RNAs (ncRNAs) as a major carrier of information as well as an effector playing direct roles in multiple aspects of molecular biology is undoubtedly a paradigm shift. Since its discovery more than a decade ago, RNAi has been implicated in a wide range of epigenetic phenomena (Ghildiyal and Zamore 2009). In the canonical RNAi pathway, small RNAs 20–30 nucleotides (nt) in length are generated by Dicer-dependent processing of double-stranded ncRNAs (Liu and Paroo 2010). Small RNAs are then loaded onto Argonaute proteins (those catalytically active ones also referred to as Slicers), forming the RNA-induced silencing complex (RISC), which targets translation inhibition and post-translational modifications of the associated chromatin (Moazed 2009). Both the Dicer and Argonaute family proteins are highly conserved and ubiquitously distributed in eukaryotic systems. Recent advances in next-generation high-throughput sequencing technologies and the availability of many sequenced genomes have led to the identification and categorization of numerous small RNA species in protozoa, fungi, metazoa, and plants, with siRNA, microRNA (miRNA), and piwi-interacting RNA (piRNA) prominent among them (Ghildiyal and Zamore 2009). Additionally, there are often multiple RNAi-related pathways in one organism, sharing components, exhibiting cross-talk, and playing critical roles in regulating gene expression, cell cycle, and cell differentiation and development (Ketting 2011). All of these demonstrate the versatility and flexibility of the RNAi mechanism to adapt to widely different challenges encountered in eukaryotic systems.
A positive feedback loop that connects ncRNA transcripts, small RNAs, and heterochromatin can be formed, providing an epigenetic switch regulating the access to specific genetic information. Extensive studies, most notably in Schizosaccharomyces pombe, have established RNAi-dependent H3K9 methylation as a central process for the formation of constitutive heterochromatin, by which genes are silenced in pericentromeric regions and other regions containing repetitious DNA sequences (Grewal and Elgin 2007; Moazed 2009). In this pathway, siRNAs are derived from processing of ncRNA transcripts by a Dicer homolog (Dcr1) and associated with an Argonaute homolog (Ago1). Mostly through the actions of two complexes, the RNAi-induced transcriptional silencing complex (RITS) (Noma et al. 2004; Verdel et al. 2004) and the RNA-directed RNA polymerase complex (RDRC) (Motamedi et al. 2004), these siRNAs recruit histone-modifying activities to chromatin regions with homologous sequences (Sugiyama et al. 2007). In particular, Clr4, a histone methyltransferase (HMT), methylates H3K9 (Nakayama et al. 2001). Methylated H3K9 is recognized by Swi6 and Chp1 through direct interaction with the chromodomains, eventually leading to the formation of condensed heterochromatin structures (Cam et al. 2005). Paradoxically, heterochromatin formation promotes ncRNA transcription, particularly during the S phase (Chen et al. 2008; Kloc et al. 2008), thereby propagating the epigenetic state over cell divisions.
Tetrahymena thermophila is one of the model systems in which a molecular connection between the RNAi and heterochromatin formation pathways was first described (Mochizuki et al. 2002; Taverna et al. 2002). Like most ciliated protozoa, Tetrahymena contains in the same cytoplasmic compartment a somatic macronucleus that is transcriptionally active and a germline micronucleus that is transcriptionally inert in asexually dividing cells (Karrer 2012). Dramatic genome rearrangement occurs when the micronucleus differentiates into the macronucleus during conjugation, the sexual phase of the Tetrahymena life cycle. About 15% of the micronuclear genome, mostly in the >6000 DNA elements referred to as internally eliminated sequences (IESs), is heterochromatinized and subsequently removed from the mature macronucleus (Yao and Chao 2005). During conjugation, the meiotic micronucleus is transcriptionally activated, generating bidirectional ncRNA transcripts (Chalker and Yao 2001). A special class of small RNAs are derived from the ncRNA transcripts (Mochizuki et al. 2002) after processing by Dcl1p (a Dicer homolog) (Malone et al. 2005; Mochizuki and Gorovsky 2005). These small RNAs are referred to as scan RNAs (scnRNAs) (Mochizuki et al. 2002) for their proposed epigenetic role in transferring information from the parental macronucleus to the developing macronucleus of the sexual progeny (see below). scnRNAs share many features with piRNAs (which interact specifically with the piwi clade proteins of the Argonaute family) characterized in higher eukaryotes (Malone and Hannon 2009; Thomson and Lin 2009). Importantly, scnRNAs associate with Twi1p (Mochizuki and Gorovsky 2004a), a Tetrahymena piwi homolog required for H3K27 and H3K9 methylation (Liu et al. 2004, 2007). In Tetrahymena, both H3K27 and H3K9 methyl marks are deposited by Ezl1p (Liu et al. 2007), an HMT homologous to the Drosophila Polycomb group (PcG) protein E(z). These heterochromatin marks are subsequently recognized by chromodomain-containing effectors like Pdd1p and Pdd3p (Taverna et al. 2002; Liu et al. 2007), which serve to compact the chromatin regions into cytologically distinct heterochromatic structures containing the germline-restricted DNA sequences destined for elimination (Yao and Chao 2005).
No consensus has been found for all of the IESs, which are often repetitive and derived from transposons (Yao and Chao 2005). There are also strong maternal influences in determining whether a particular DNA sequence is deleted from the developing macronucleus (IES) or retained (MDS [macronuclear-destined sequence]) (Yao and Chao 2005). Simply put, sequences present in the micronucleus but absent from the parental macronucleus have a strong tendency to be lost in the new macronucleus of the conjugation progeny. Indeed, a heterologous DNA fragment introduced into the germline micronucleus but not the somatic macronucleus can be removed from the somatic macronucleus of the sexual progeny (Yao et al. 2003; Liu et al. 2005). This recognition of “self” and “nonself” DNA constitutes an effective trans-generational genome defense system. How the diverse IES elements are specifically targeted with maternal influence was a mystery until the discovery of scnRNAs. scnRNAs are generated from the germline micronucleus and trafficked first to the parental macronucleus and then to the developing macronucleus, in complex with Twi1p (Mochizuki et al. 2002). In the scnRNA model originally proposed by Mochizuki et al. (2002), scnRNAs, as a carrier of sequence-specific information, mediate the comparison of the micronuclear and macronuclear genome by a “scanning” process, resulting in the enrichment of micronuclear-specific species that are subsequently used to target DNA elimination in the developing macronuclei. Even though the teleological causes are obvious, key questions concerning the mechanism for the RNAi-guided genome defense remain to be answered: What information is being passed between the germline micronucleus and the somatic macronucleus, and, more importantly, how?
With the recently available Tetrahymena micronuclear genome sequence as the reference, a detailed deep-sequencing analysis of scnRNAs and their ncRNA precursors was performed by Mochizuki's laboratory (Schoeberl et al. 2012). By comparing scnRNAs from different conjugation time points as well as the wild-type and RNAi-deficient strains, the study provides definitive support for the selective degradation of scnRNAs homologous to MDS, a key prediction by the scnRNA model. Surprisingly, a strong bias toward IES regions is uncovered in scnRNAs in the absence of the scanning process, which is further traced back to biased ncRNA transcription of IES regions in the micronucleus. This result strongly suggests that IES regions in the germline are premarked, most likely by histone modifications, to direct ncRNA transcription and scnRNA biogenesis. Therefore, the scnRNA-guided DNA elimination results from the integration of the germline and somatic signals. More generally, this study and other works in this area have also raised the intriguing possibility for exchange of information between the germline and soma, particularly the soma-to-germline feedback. Testing these tantalizing ideas and unraveling the underlying mechanism will be an exciting and rewarding challenge for the field.
Small RNA turnover: adding or subtracting
The new study by Mochizuki and colleagues (Schoeberl et al. 2012) has systematically addressed the turnover of scnRNAs in early to mid-conjugation, during which scnRNAs are first generated by Dcl1p-dependent processing of ncRNA transcripts from the meiotic micronucleus and then enriched for IES homologous sequences. The latter process is the cornerstone for the scnRNA model. It has been tested and corroborated by various methods (Mochizuki and Gorovsky 2004a; Aronica et al. 2008), culminating in the satisfactory solution presented in this study. Theoretically, the enrichment of IES homologous scnRNAs relative to MDS homologous scnRNAs can be achieved in two ways: selective amplification of IES homologous scnRNAs or selective degradation of MDS homologous scnRNAs. As the total amount of scnRNAs does not change dramatically during early to mid-conjugation, selective degradation of MDS homologous scnRNAs—which is a small fraction of the total pool—is obviously more plausible. The investigators further point out that most scnRNAs bear the mark of Dcl1p processing, thus mechanistically ruling out secondary amplification by Slicer activities. However, it remains possible that ncRNA transcripts from the micronucleus are amplified by the RNA-directed RNA polymerase complex (RDRC), which has been to shown to tightly couple with Dicer activities and contribute substantially to small RNA generation in S. pombe as well as Tetrahymena (Lee and Collins 2007).
Interesting parallels can be drawn between scnRNAs in Tetrahymena and piRNAs in metazoa (Ghildiyal and Zamore 2009; Malone and Hannon 2009). Both scnRNAs and piRNAs exclusively associate with piwi clade proteins (Mochizuki et al. 2002), while siRNAs are bound by Argonaute clade proteins. Both scnRNAs and piRNAs (28–30 nt) are also significantly longer than siRNA (22–24 nt) (Mochizuki et al. 2002). In addition, 2′O-methylation catalyzed by the conserved Hen1p methyltransferases is more abundantly present in scnRNAs and piRNAs (Kurth and Mochizuki 2009). In contrast to the widespread distribution of siRNAs in somatic cells, piRNAs are mostly detected in germline cells, particularly at meiosis (Malone and Hannon 2009; Thomson and Lin 2009); similarly, scnRNAs are conjugation-specific and abundantly produced from the meiotic micronuclei (Mochizuki et al. 2002; Mochizuki and Gorovsky 2004a). Furthermore, both the scnRNA and piRNA pathways have been shown to work as adaptive genome defense systems for the germline against the invasion of foreign DNA species (Yao et al. 2003; Liu et al. 2005; Malone and Hannon 2009).
There are also significant differences in the biogenesis of scnRNAs and piRNAs. scnRNA accumulation is dependent on Dcl1p (Malone et al. 2005; Mochizuki and Gorovsky 2005), while piRNA biogenesis is probably Dicer-independent (Malone and Hannon 2009). In the piRNA pathway, primary piRNAs cleave complementary RNA transcripts. This process, facilitated by the slicer activity of additional piwi family proteins, generates complementary secondary piRNAs, resulting in the ping-pong cycle amplification (Malone and Hannon 2009). In Tetrahymena, in association with Twi1p, scnRNAs also bind and presumably cleave complementary ncRNA transcripts in the parental macronucleus during early conjugation and in the developing macronucleus during late conjugation (Aronica et al. 2008). However, there is no secondary amplification of scnRNAs. Indeed, scanning of the parental macronucleus leads to the diminishing of MDS homologous scnRNAs (see below). Nonetheless, the difference in the biogenesis of scnRNAs and piRNAs may reflect more on the versatility of the RNAi pathways than the intrinsic differences in their biology, as RNAi pathways can be primed by divergent input materials, and small RNA populations can be generated and maintained by various mechanisms (Ghildiyal and Zamore 2009).
RNAi-guided heterochromatin formation: scan if you can
A central question in the scnRNA model is the mechanism underlying the selective degradation of MDS homologous scnRNAs. A strong clue is provided by the phenotypic analysis of strains deleting EMA1, which encodes a putative RNA helicase that mediates the interaction of the scnRNA/Twi1p complex and chromatin, most likely through base-pairing with nascent ncRNA transcripts (Aronica et al. 2008). In ΔEMA1 cells, DNA elimination is abolished, and intriguingly, so is the selective degradation of MDS homologous scnRNAs (Aronica et al. 2008). This evidence strongly supports the concept of sequence comparison between the micronuclear and macronuclear genome, mediated by the scnRNA–ncRNA base-pairing. Furthermore, key components involved in the heterochromatin formation, including Ezl1p, are also required for the selective degradation (Liu et al. 2007). As strains defective in the scanning process accumulate MDS homologous scnRNAs, they should, in theory, overdelete MDS in the developing macronucleus. However, this phenotype has not been observed in any of the mutants studied so far, which either completely abolish or underdelete IES. This suggests that the scanning process in the parental macronucleus and the DNA elimination process in the developing macronucleus share the same underlying mechanism. Indeed, RNAi-dependent H3K27 methylation is observed in both the parental macronucleus and developing macronucleus (Liu et al. 2007), supporting RNAi-guided heterochromatin formation as the common denominator. Nonetheless, how RNAi-guided heterochromatin formation leads to the degradation of the scnRNAs involved remains elusive.
Heterochromatin formation in Tetrahymena is dependent on H3K27 methylation and PcG proteins (Liu et al. 2007). Indeed, homologs to PcG proteins can be found in many of the recently sequenced genomes of unicellular eukaryotes. The high degree of evolutionary conservation among PcG proteins contrasts strongly with their highly divergent target DNA sequences. The cis-element for recruiting PcG proteins in Drosophila melanogaster, PcG response element (PRE), is remarkably different even in other closely related Drosophila species (Ringrose and Paro 2007). This scenario closely resembles that of pericentromeric heterochromatin formation, in which conserved chromatin proteins are targeted to fast-evolving DNA sequences. As RNAi-guided heterochromatin formation emerges as a solution for the pericentromeric heterochromatin targeting paradox, the case for ncRNA-mediated recruitment of PcG proteins is also building. Indeed, a survey of large intergenic ncRNA in mammalian cells reveals that ∼20% are bound by Polycomb-repressive complex 2 (PRC2) (Khalil et al. 2009), suggesting that this cis-element-independent recruitment mechanism is widespread. Given the overall similarity between the scnRNA pathway in Tetrahymena and the piRNA pathway in metazoa, and emerging evidence linking sRNA/ncRNA and PcG repression, it is tempting to propose that piRNA may also be involved in the targeting of Polycomb-repressive complexes to establish heterochromatin regions.
Coding or noncoding, that is the question
The new study by Mochizuki and colleagues (Schoeberl et al. 2012) has uncovered a surprisingly strong bias for IESs in the early scnRNA pool, before the scanning process can have any effect. More importantly, global run-on sequencing (GRO-seq) reveals that ncRNAs from the micronucleus, precursors to scnRNAs, are also preferentially generated from IES regions. Indeed, the close similarity between the genomic distribution profiles of ncRNAs and scnRNAs suggests a direct conversion not distorted by secondary amplification. The nuclear run-on result is interpreted as revealing the biased occupancy of elongating RNA polymerase II (RNAP II). This is consistent with previous observations that RNAP II is present in the meiotic micronucleus during meiosis and that its distribution is uneven along the micronuclear chromosomes (Mochizuki and Gorovsky 2004b). These results argue strongly that IES regions in the micronucleus are earmarked for ncRNA transcription and scnRNA generation and therefore are “programmed” for heterochromatin formation and DNA elimination in the developing macronucleus. The subsequent scanning process may only serve to modify and optimize the process.
This raises the question concerning the identity of the epigenetic marks for IES regions in the micronucleus. Since the micronucleus is transcriptionally silent before meiosis and there is no IES in the transcriptionally active macronucleus, RNA is very unlikely to play any roles here. There is no cytosine methylation in Tetrahymena, while adenine methylation is most likely excluded from the micronucleus (Karrer 2012). That leaves only histone modifications associated with heterochromatin, which is paradoxically connected with ncRNA transcription. The germline micronucleus is transcriptionally silent in asexually dividing cells, enriched in H3K27 methylation but devoid of H3K4 methylation (Strahl et al. 1999; Liu et al. 2007). Intriguingly, H3K27 methylation levels increase dramatically in the transcriptionally active meiotic micronucleus, while there is still no detectable H3K4 methylation (Liu et al. 2007). In Tetrahymena strains mutated at H3K27 (K27Q, K27R, and K27A), conjugation progress invariably is aborted after the meiosis stages (Liu et al. 2007), suggesting that histone modifications at or around K27 play a critical but yet undefined role. This scenario is reminiscent of the extensively studied piRNA biogenesis in the Drosophila ovary, which also generates abundant ncRNA transcripts from specific genomic loci. Most of the ncRNA transcripts come from bidirectional transcription of clusters enriched in transposable elements (Malone and Hannon 2009), as is the case for IES regions in the Tetrahymena micronucleus. Intriguingly, bidirectional transcription is facilitated by a chromodomain-containing heterochromatic protein, Rhino, which is required for piRNA biogenesis in Drosophila (Klattenhoff et al. 2009).
Meiosis-specific transcription is observed in a wide range of eukaryotic systems, which predominantly generates ncRNA transcripts often subsequently processed into small RNA species (Malone and Hannon 2009). The process may be part of the genome defense system that silences transposons and other parasitic DNA elements. Note that ncRNAs here are not defined by their protein-coding potential, as many ncRNAs derived from transposons actually can encode functional proteins. Instead, ncRNAs are defined by their biogenesis, particularly the chromatin environment in which they are transcribed. The molecular mechanism underlying ncRNA transcription remains largely elusive. In Arabidopsis, variants of RNAP II are specialized in ncRNA transcription (Haag and Pikaard 2011). In Tetrahymena as well as metazoa, it is the bona fide RNAP II that is involved in the transcription of ncRNA (Mochizuki and Gorovsky 2004b). Indeed, even though ncRNA transcription occurs most efficiently in the meiotic micronucleus, it is also present at low levels in the parental macronucleus and developing macronucleus (Aronica et al. 2008), coincident with highly elevated expression levels of RNAP II components during conjugation. Still, it remains possible that a yet-to-be-characterized accessary component of RNAP II may be involved in promoting ncRNA transcription.
Communication between germline and soma: Lamarckism or Darwinism
The updated scnRNA model presents two routes through which epigenetic information can be passed from the germline micronucleus to the somatic macronucleus during sexual reproduction (Fig. 1). The first is mediated by histone modifications that are nondiffusible in nature. The second is mediated by scnRNAs that are diffusible. Indeed, these two routes can also be construed as the result of deconstruction–reconstruction of the positive feedback loop that connects ncRNA, small RNA, and heterochromatin formation. They can reinforce each other and together form a robust and, more importantly, adaptive system for genome defense. The interplay between these two routes may be a general phenomenon, as it can also be discerned in piRNA biogenesis and transposon silencing in Drosophila (Malone and Hannon 2009).
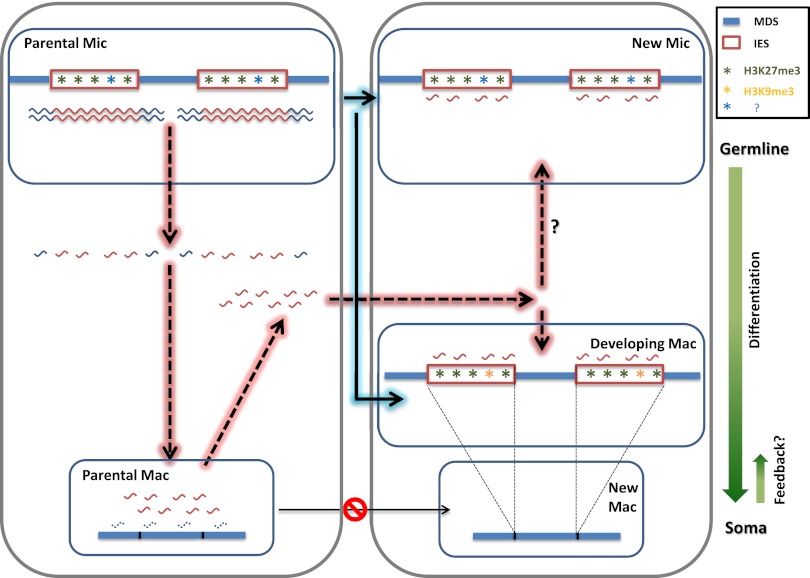
Figure 1.
Flow of epigenetic information between the germline micronucleus and the somatic macronucleus during the sexual reproduction of Tetrahymena. (Blue solid arrows) Transfer of epigenetic information carried by histone modifications. Histone modification patterns in the germline parental micronucleus can be passed to the new germline micronucleus and new somatic macronucleus. (Red dashed arrows) Transfer of epigenetic information carried by small RNAs. In the parental micronucleus, ncRNAs are bidirectionally transcribed, preferentially from the IES regions earmarked by histone modifications. ncRNAs are processed by Dcl1p to generate scnRNAs. scnRNAs, in complex with Twi1p, are translocated into the parental macronucleus, where MDS homologous scnRNAs are selectively degraded. An optimized pool of scnRNAs is then trafficked to the new macronucleus to guide the heterochromatinization and eventual elimination of IES regions, a process that may also be affected by histone modifications inherited from the parental micronucleus. scnRNAs may also be translocated into the new micronucleus to reshape histone modification patterns.
The adaptive nature of the system is best illustrated by the flow of information from the parental macronucleus to the developing macronucleus of the sexual progeny, with scnRNAs as the information carrier. The maternal effect can be perceived as a weak form of soma-to-germline feedback, which regulates the germline–soma differentiation. This occurs readily in Tetrahymena and other ciliates due to several facilitating factors: (1) The germline micronucleus and the somatic macronucleus reside in the same cytoplasmic compartment; (2) transcription of the parental macronucleus and the developing macronucleus are temporally juxtaposed, separated only by a few hours; (3) active trafficking of scnRNAs between the cytoplasm and different nuclear compartments (Mochizuki et al. 2002; Noto et al. 2010). In their study, Mochizuki and colleagues (Schoeberl et al. 2012) have suggested provocatively that the strong form of soma-to-germline feedback may also be present in Tetrahymena conjugation, with scnRNAs as the information carrier. The investigators propose that the histone modifications in the new micronucleus may be reshaped by scnRNAs, just like what happens in the developing new macronuclei. In support, abnormal H3K27 methylation is observed in the new micronucleus in RNAi-deficient mutants (Liu et al. 2007). Reprogramming of micronuclear chromatin states is most likely to occur during the post-zygotic divisions, in which DNA replication potentially exposes the heterochromatin in the micronucleus to the optimized pool of scnRNAs. However, Twi1p is localized only in the cytoplasm and macronucleus (Mochizuki et al. 2002; Noto et al. 2010). Dcl1p is also not detectable in the post-zygotic micronucleus (Malone et al. 2005; Mochizuki and Gorovsky 2005). All of this demands the presence of another set of RNAi machinery for scnRNA-guided heterochromatin formation in the new micronucleus.
RNAi is catalytic in nature, which means signals carried by small RNAs can be readily amplified to enhance their transmission. Indeed, soma-to-germline transfer of information mediated by small RNAs and, more generally, systemic RNAi is gaining experimental support in plants (Chitwood and Timmermans 2010). There is also emerging evidence for trans-generational transfer of epigenetic information in other systems, including metazoa (Ashe et al. 2012; Gu et al. 2012; Lee et al. 2012; Shirayama et al. 2012). This is a problem that needs to be fully explored due to its potentially huge implications in biology. It is often argued that had Lamarck known what Darwin knew, he would have drawn the same conclusion. What conclusions would Lamarck and Darwin have drawn had they known about all of the noncoding messages being passed between the germline and soma?
Acknowledgments
We apologize to colleagues whose original research papers were not directly cited due to the limit of the perspective format. S.G. and Y.L. are supported by National Institute of Health.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.199992.112.
References
- Aronica L, Bednenko J, Noto T, DeSouza LV, Siu KW, Loidl J, Pearlman RE, Gorovsky MA, Mochizuki K 2008. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes Dev 22: 2228–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, et al. 2012. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150: 88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI 2005. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819 [DOI] [PubMed] [Google Scholar]
- Chalker DL, Yao MC 2001. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes Dev 15: 1287–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI 2008. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451: 734–737 [DOI] [PubMed] [Google Scholar]
- Chitwood DH, Timmermans MC 2010. Small RNAs are on the move. Nature 467: 415–419 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD 2009. Small silencing RNAs: An expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Elgin SC 2007. Transcription and RNA interference in the formation of heterochromatin. Nature 447: 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu SG, Pak J, Guang S, Maniar JM, Kennedy S, Fire A 2012. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat Genet 44: 157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Pikaard CS 2011. Multisubunit RNA polymerases IV and V: Purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol 12: 483–492 [DOI] [PubMed] [Google Scholar]
- Karrer KM 2012. Nuclear dualism. Methods Cell Biol 109: 29–52 [DOI] [PubMed] [Google Scholar]
- Ketting RF 2011. The many faces of RNAi. Dev Cell 20: 148–161 [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al. 2009. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138: 1137–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc A, Zaratiegui M, Nora E, Martienssen R 2008. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol 18: 490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth HM, Mochizuki K 2009. 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA 15: 675–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K 2007. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat Struct Mol Biol 14: 604–610 [DOI] [PubMed] [Google Scholar]
- Lee HC, Gu W, Shirayama M, Youngman E, Conte D Jr, Mello CC 2012. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 150: 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Paroo Z 2010. Biochemical principles of small RNA pathways. Annu Rev Biochem 79: 295–319 [DOI] [PubMed] [Google Scholar]
- Liu Y, Mochizuki K, Gorovsky MA 2004. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc Natl Acad Sci 101: 1679–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Song X, Gorovsky MA, Karrer KM 2005. Elimination of foreign DNA during somatic differentiation in Tetrahymena thermophila shows position effect and is dosage dependent. Eukaryot Cell 4: 421–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD 2007. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev 21: 1530–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ 2009. Small RNAs as guardians of the genome. Cell 136: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Anderson AM, Motl JA, Rexer CH, Chalker DL 2005. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol Cell Biol 25: 9151–9164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D 2009. Small RNAs in transcriptional gene silencing and genome defence. Nature 457: 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA 2004a. Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes Dev 18: 2068–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA 2004b. RNA polymerase II localizes in Tetrahymena thermophila meiotic micronuclei when micronuclear transcription associated with genome rearrangement occurs. Eukaryot Cell 3: 1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA 2005. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev 19: 77–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA 2002. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110: 689–699 [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D 2004. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802 [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113 [DOI] [PubMed] [Google Scholar]
- Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SI 2004. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet 36: 1174–1180 [DOI] [PubMed] [Google Scholar]
- Noto T, Kurth HM, Kataoka K, Aronica L, DeSouza LV, Siu KW, Pearlman RE, Gorovsky MA, Mochizuki K 2010. The Tetrahymena argonaute-binding protein Giw1p directs a mature argonaute–siRNA complex to the nucleus. Cell 140: 692–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R 2007. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 134: 223–232 [DOI] [PubMed] [Google Scholar]
- Schoeberl UE, Kurth HM, Noto T, Mochizuki K 2012. Biased transcription and selective degradation of small RNAs shape the pattern of DNA elimination in Tetrahymena. Genes Dev 26: 1729–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D Jr, Mello CC 2012. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150: 65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Ohba R, Cook RG, Allis CD 1999. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci 96: 14967–14972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI 2007. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128: 491–504 [DOI] [PubMed] [Google Scholar]
- Taverna SD, Coyne RS, Allis CD 2002. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell 110: 701–711 [DOI] [PubMed] [Google Scholar]
- Thomson T, Lin H 2009. The biogenesis and function of PIWI proteins and piRNAs: Progress and prospect. Annu Rev Cell Dev Biol 25: 355–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao MC, Chao JL 2005. RNA-guided DNA deletion in Tetrahymena: An RNAi-based mechanism for programmed genome rearrangements. Annu Rev Genet 39: 537–559 [DOI] [PubMed] [Google Scholar]
- Yao MC, Fuller P, Xi X 2003. Programmed DNA deletion as an RNA-guided system of genome defense. Science 300: 1581–1584 [DOI] [PubMed] [Google Scholar]