Proper centrosomal duplication is essential to maintaining genetic stability. This novel study shows that a major cause of Meier-Gorlin syndrome, a microcephalic dwarfism disorder, is the failure of Orc1 to regulate centrosome duplication. The authors identify CyclinE–CDK2 kinase inhibition as essential for Orc1 control of centriole and centrosome copy number and suggest how Orc1 mutations contribute to the phenotype of Meier-Gorlin syndrome patients.
Keywords: Meier-Gorlin syndrome, centriole, centrosome, origin recognition complex, cyclin-dependent protein kinase
Abstract
Like DNA replication, centrosomes are licensed to duplicate once per cell division cycle to ensure genetic stability. In addition to regulating DNA replication, the Orc1 subunit of the human origin recognition complex controls centriole and centrosome copy number. Here we report that Orc1 harbors a PACT centrosome-targeting domain and a separate domain that differentially inhibits the protein kinase activities of Cyclin E–CDK2 and Cyclin A–CDK2. A cyclin-binding motif (Cy motif) is required for Orc1 to bind Cyclin A and inhibit Cyclin A–CDK2 kinase activity but has no effect on Cyclin E–CDK2 kinase activity. In contrast, Orc1 inhibition of Cyclin E–CDK2 kinase activity occurs by a different mechanism that is affected by Orc1 mutations identified in Meier-Gorlin syndrome patients. The cyclin/CDK2 kinase inhibitory domain of Orc1, when tethered to the PACT domain, localizes to centrosomes and blocks centrosome reduplication. Meier-Gorlin syndrome mutations that disrupt Cyclin E–CDK2 kinase inhibition also allow centrosome reduplication. Thus, Orc1 contains distinct domains that control centrosome copy number and DNA replication. We suggest that the Orc1 mutations present in some Meier-Gorlin syndrome patients contribute to the pronounced microcephaly and dwarfism observed in these individuals by altering centrosome duplication in addition to DNA replication defects.
Centrioles are self-replicating structures within centrosomes; are required for cell asymmetry, nuclear localization within a cell, and chromosome organization during mitosis in proliferating cells; and form the core of basal bodies in ciliated cells (Nigg and Stearns 2011). Surprisingly, several DNA replication proteins localize to centrosomes, including the origin recognition complex (ORC), some subunits of the minichromosome maintenance (MCM) helicase complex, and geminin (Prasanth et al. 2004; Tachibana and Nigg 2006; Stuermer et al. 2007; Ferguson and Maller 2008; Hemerly et al. 2009; Ferguson et al. 2010; Knockleby and Lee 2010). In the nucleus, these proteins are required for prereplicative complex (pre-RC) assembly to license during the G1 phase origins of DNA replication for chromosome duplication, a process that ensures that DNA replication only initiates at each origin once per cell division cycle (Stillman 2005). Like chromosome duplication, centriole and centrosome duplication is licensed in proliferating cells as the cells exit mitosis, and their duplication is dependent on Cyclin E–CDK2 protein kinase activity (Hinchcliffe et al. 1999; Lacey et al. 1999; Wong and Stearns 2003; Tsou and Stearns 2006; Nigg and Stearns 2011). Of relevance here is the observation that human Orc1, the largest subunit of ORC, contributes to the control of Cyclin E–CDK2-dependent centriole and centrosome copy number during the normal cell division cycle or following DNA damage-induced centrosome reduplication (Hemerly et al. 2009).
Recently, mutations in Meier-Gorlin syndrome have been found in pre-RC proteins, including ORC subunits Orc1, Orc4, and Orc6, as well as Cdc6 and Cdt1 (Bicknell et al. 2011a,b; Guernsey et al. 2011; de Munnik et al. 2012). Patients have primordial dwarfism, microcephaly, and developmental abnormalities in the ear and patella (Klingseisen and Jackson 2011). Other developmental disorders that cause primordial dwarfism and microcephaly, including Seckel syndrome, have mutations in the centrosome proteins CPAP (CENP-J), PCNT, and CEP152, although defects in a few noncentrosome proteins such as ATM and IGF1 are also associated with microcephalic primordial dwarfism (Rauch et al. 2008; Miyoshi et al. 2009; Thornton and Woods 2009; Kitagawa et al. 2011; Klingseisen and Jackson 2011; Sir et al. 2011; Tang et al. 2011). A survey of the mutations and phenotypes in 45 Meier-Gorlin syndrome patients demonstrated that compared with patients with mutations in Orc4, Cdc6, and Cdt1, all of the patients with mutations in Orc1 were of significantly shorter stature and had increased microcephaly and a significantly proportionally smaller head circumference and brain (de Munnik et al. 2012). All but one of the patients with Orc1 mutations had normal intellect, and the one individual had a mild intellectual disability.
We dissected the domains of Orc1 required to control centriole and centrosome copy number and uncovered overlapping but separate Cyclin A–CDK2 and Cyclin E–CKD2 kinase inhibitory domains in Orc1 as well as a centrosome localization signal (CLS). Mutations in Orc1 present in patients with Meier-Gorlin syndrome alter the Cyclin E–CDK2 kinase inhibitory activity and cause centrosome reduplication in human cells. We suggest that this activity contributes to the more severe dwarfism and microcephaly that is present in patients with Orc1 mutations.
Results
A centrosome-targeting signal in Orc1
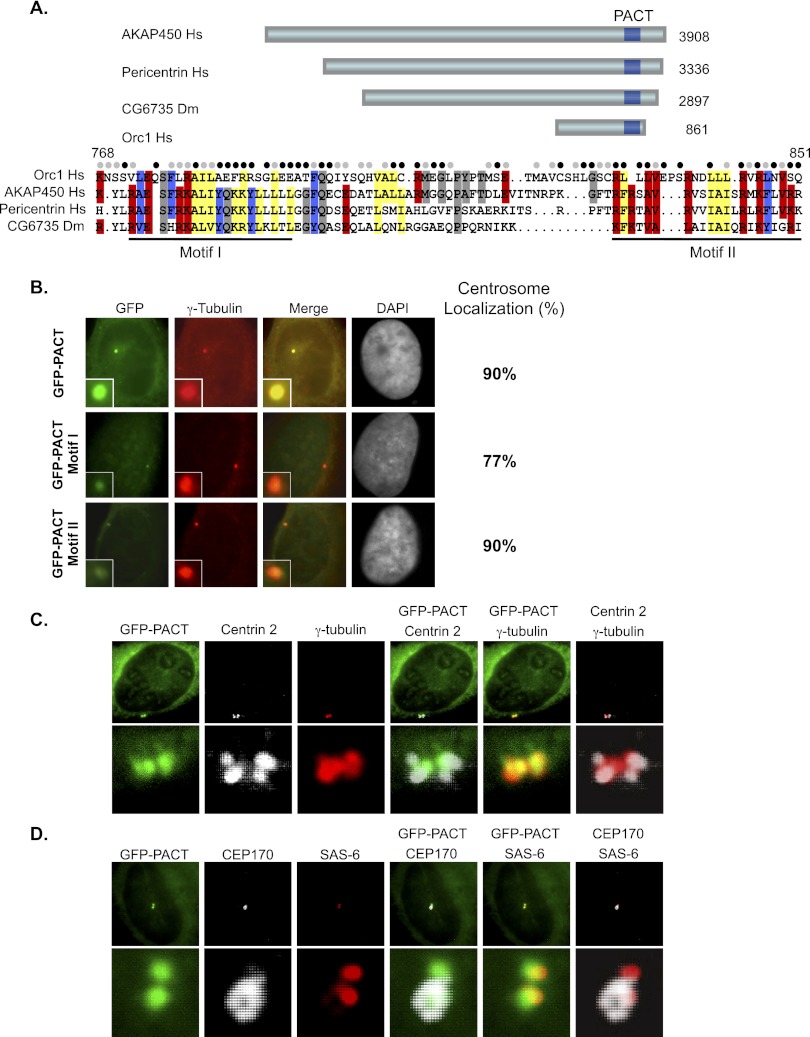
An in silico approach to search for potential centrosome localization domains in human Orc1 revealed a sequence near the C terminus (768–851 amino acids) of the protein that is similar to the centrosome-targeting PACT domains found in pericentrin and AKAP450 (Gillingham and Munro 2000). Like other PACT regions, the Orc1 PACT domain was divided into two conserved regions, named motif I and motif II (Fig. 1A). When fused to GFP and introduced into U2OS cells by transfection of a plasmid, the Orc1 PACT domain localized the GFP signal to centrosomes in 90% of cells (Fig. 1B). Motif I alone caused reduced centrosome localization and caused degradation of the fusion protein, since addition of the proteasome inhibitor MG132 increased the amount of protein (Fig. 1B; Supplemental Fig. S1A). Motif II alone allowed centrosome localization, but the level of fluorescence was reduced compared with the full-length PACT domain, even though the protein was stably expressed (Fig. 1B; Supplemental Fig. S1A). The Orc1 PACT domain targeted GFP to the pericentriolar matrix (stained with antibodies against γ-tubulin and pericentrin) that surrounds the Centrin-2-containing centrioles (Fig. 1C; Supplemental Fig. S1B). Moreover, the Orc1 PACT-GFP protein localized nearer the mother centrioles versus the newly formed daughter centrioles, as evident by comparing GFP-PACT with Centrin-2, SAS-6, and CEP170 staining (Fig. 1C,D).
Figure 1.
Orc1 contains a conserved CLS. (A) Schematic of human Orc1 depicts the presence of a conserved PACT domain toward the C terminus compared with other centrosomal proteins (AKAP450, Homo sapiens; pericentrin, H. sapiens; and CG6735, Drosophila melanogaster). PACT regions are boxed in dark blue. Alignment of PACT sequences is shown below the schematic. The conserved residues of the human Orc1 PACT region are indicated in different colors. The filled circles above the alignment show the conserved residues in Orc1 from different vertebrate classes (pisces, Denio rerio; amphibians, Xenopus laevis; aves, Taeniopygia guttata; and mammals, Mus musculus). (Black circles) Identical residues; (gray circles) similar residues. The Orc1 PACT region is further subdivided into two conserved motifs, as indicated by underlined regions. (B) Transfected U2OS cells expressing human Orc1 PACT (768–851 amino acids), PACT motif I (768–818 amino acids), and PACT motif II (828–851 amino acids) fused with GFP. The cells were costained with γ-tubulin, a centrosome marker, and percentage localization of GFP to centrosomes was evaluated from ∼100 cells in three independent experiments. (C,D, top panels) The GFP-PACT transfected U2OS cells were costained with γ-tubulin (centrosome marker) and Centrin-2 (centriole marker). Staining of GFP-PACT transfected U2OS cells with CEP170 (grandmother centriole marker) and SAS-6 (daughter centrioles or procentrioles marker). The bottom panels are at higher magnification.
Cyclin A–CDK2 and Cyclin E–CDK2 inhibitory domains
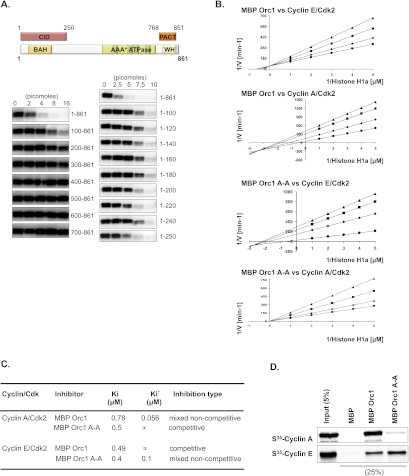
Orc1 controlled Cyclin E–CDK2 kinase-dependent centriole and centrosome duplication in G1-phase cells and inhibited both Cyclin A–CDK2 and Cyclin E–CDK2 kinase activities (Hemerly et al. 2009). To identify how Orc1 controls CDK2 activity, a series of proteins in which the maltose-binding protein (MBP) was fused to the N terminus of either full-length or deletion derivatives of human Orc1 was purified and tested for their ability to inhibit CDK activity using histone H1a as a CDK2 substrate (Fig. 2A). The N-terminal 1- to 250-amino-acid domain was necessary and sufficient to inhibit both Cyclin E–CDK2 (Fig. 2A) and Cyclin A–CDK2 (data not shown) kinase activities. We named this domain the CDK inhibitory domain (CID) (Fig. 2A). However, the mechanism of inhibition was different for the two kinases. Lineweaver-Burk analysis (Segel 1975) suggests that Orc1 inhibits Cyclin E–CDK2 competitively with a Ki of 0.49 μM, but the mode changes to mixed noncompetitive inhibition for Cyclin A–CDK2 with a Ki of 0.78 μM and a Ki′ of 0.056 μM (Fig. 2B,C; Supplemental Fig. S2). The difference in mechanism of inhibition is attributed to the fact that Orc1 interacts directly with Cyclin A in a manner dependent on a cyclin-binding motif (Cy motif; 235KRL237) but binds to Cyclin E in a Cy motif-independent manner (Fig. 2D). The interaction between Orc1 and Cyclin A–CDK2 and Cyclin E–CDK2 was repeated with purified proteins and thus is direct (Supplemental Fig. S3). Cy motif mutation (K235A, L237A; Orc1-A-A) in MBP-Orc1 protein abolished its interaction with Cyclin A and blocked the ability of Orc1 to inhibit Cyclin A–CDK2 via the noncompetitive inhibition mode, while retaining its weaker competitive inhibition mode with a Ki of 0.5 μM (Fig. 2C). The Cy motif mutation of MBP-Orc1 protein did not affect the Ki for inhibition of Cyclin E–CDK2 (0.49 μM vs. 0.4 μM), but the mutant protein also inhibited the Cyclin E–CDK2 kinase noncompetitively with a Ki′ of 0.1 μM, concomitant with a slight increase in binding to Cyclin E (Fig. 2B–D). Thus, inhibition of the different cyclin–CDKs was kinetically quite distinct (see Supplemental Fig. S2A,B).
Figure 2.
Inhibitory mechanism of Orc1 on the CDKs. (A) The inhibition of Cyclin E–CDK2-dependent phosphorylation of histone H1a substrate by MBP-Orc1 and its fragments. The top panel shows a schematic of human Orc1 structure with the CID and PACT. Different amounts of MBP-fused N-terminal deletions (bottom left panel) as well as N-terminal fragments (bottom right panel) of Orc1 were used in the kinase assay. The concentrations of histone H1a and Cyclin E–CDK2 used in the assay were 0.5 μM and 10 nM, respectively. (B) Lineweaver-Burk plots of MBP-Orc1 or MBP-Orc1 A-A (Cy motif mutation) inhibition of histone H1a phosphorylation by Cyclin E–CDK2 or Cyclin A–CDK2. The concentration of Orc1 proteins varied for each fixed concentration of histone H1a substrate. The Orc1 concentrations were 0 μM (•), 0.5 μM (♦), 0.75 μM (▪), and 1 μM (▴). The concentration of histone H1a varied from 0.2 μM to 1 μM. (C) The inhibition constants and mode of inhibition of MBP-Orc1 and MBP-Orc1 A-A on Cyclin E–CDK2 or Cyclin A–CDK2, based on the models in Supplemental Figure S2. (D) In vitro interaction between Orc1 and cyclins in an MBP pull-down assay. MBP-fused wild-type Orc1 or the Cy motif A-A mutant (MBP-Orc1 K235A, L237A) was bound to amylose resin and further incubated with in vitro translated S35-labeled Cyclin E or Cyclin A. Beads were isolated, and bound proteins were separated by gel electrophoresis. Recombinant MBP was used as a control in the assay.
The Orc1 cyclin–CDK2 inhibitory domain and the PACT domain control centriole and centrosome copy number
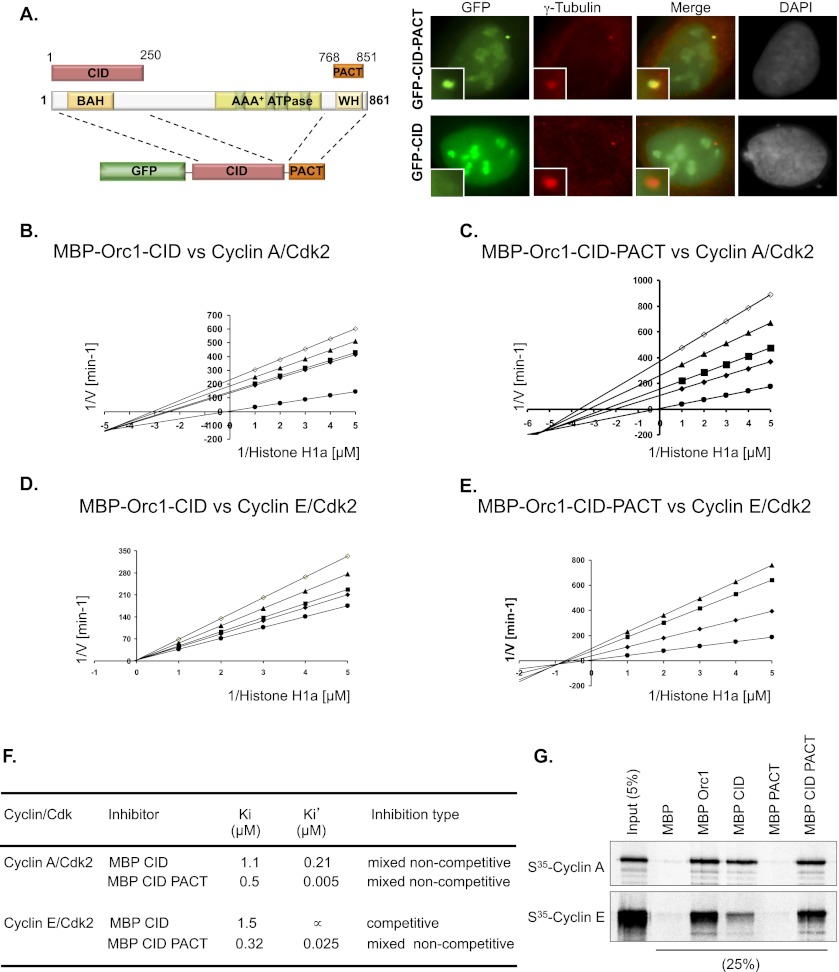
Having identified the PACT and CID domains in Orc1, we determined their effect on centrosome copy number control. In U2OS cells, the transfected GFP-CID did not localize to centrosomes (Fig. 3A, ∼3%, n = 100, in triplicate). Fusion of the Orc1 PACT domain to its CID caused the chimeric GFP-CID-PACT protein to localize to centrosomes (Fig. 3A, 90%, n = 100, in triplicate). Like the Orc1 PACT, GFP-CID-PACT protein localized near the mother centrioles compared with the newly formed procentrioles (daughter centrioles), as evident by localization relative to Centrin-2, SAS-6, and CEP170 (Supplemental Fig. S4A–C). Both Orc1-CID and Orc1-CID-PACT were expressed at similar levels. Thus, the Orc1 PACT domain localized the CID of Orc1 to centrosomes.
Figure 3.
Kinetic analysis of the CID of human Orc1. (A) The CID (1–250 amino acids) of Orc1 was linked to its PACT domain (768–851 amino acids) and tagged with GFP. The left panel shows the schematic of the chimeric Orc1 construct in the pEGFP-C1 plasmid. Transiently transfected U2OS cells overexpressing GFP-Orc1-CID or GFP-Orc1-CID-PACT were stained for γ-tubulin. Insets are higher magnification. (B–E) Lineweaver-Burk plot of kinase inhibition of MBP-Orc1-CID or MBP-Orc1-CID-PACT on cyclin–CDK2 (see label in each panel). The concentrations of Orc1 were 0 μM (•), 0.5 μM (♦), 0.75 μM (▪), 1 μM (▴), and 1.5 μM (○). The concentration of histone H1a varied from 0.2 μM to 1 μM. The secondary plot of slopes or intercepts versus Orc1 concentration was obtained for each of the Lineweaver-Burk plots. (F) The inhibition constants and mode of inhibition of MBP-Orc1 and MBP-Orc1 A-A on Cyclin E–CDK2 or Cyclin A–CDK2. (G) Direct interaction between MBP-Orc1-CID, MBP-Orc1-PACT, or MBP-Orc1-CID-PACT and Cyclin A or Cyclin E. The MBP-fused Orc1 fragments were bound to amylose resin and further incubated with in vitro translated S35-labeled Cyclin E or Cyclin A. Beads were isolated, and bound proteins were separated by gel electrophoresis. Recombinant MBP was used as a control in the assay.
To understand the kinetics of inhibition by the CID and PACT domain of Orc1 on cyclin–CDK2 activity, we purified MBP-Orc1-CID and MBP-Orc1-CID-PACT. The deduced inhibitory constants (Ki and Ki′) obtained from Lineweaver-Burk plots for the Orc1 fragments showed different modes of inhibition on the two different CDK2 kinases. In the case of Cyclin A–CDK2, both of the Orc1 fragments were potent in inhibiting the kinase activity (Fig. 3B,C; Supplemental Fig. S5). Lineweaver-Burk plots showed that both of the Orc1 fragments had mixed noncompetitive inhibition on Cyclin A–CDK2. MBP-Orc1-CID inhibited histone H1a phosphorylation by Cyclin A–CDK2 with a Ki of 1.1 μM and a Ki′ of 0.21 μM, while MBP-Orc1-CID-PACT had a Ki of 0.5 μM and a Ki′ of 5 nM (Fig. 3F). Thus, the PACT domain increased the mixed noncompetitive inhibition of the Orc1 CID on Cyclin A–CDK2.
In the case of Cyclin E–CDK2, the MBP-Orc1-CID was a less potent inhibitor compared with MBP-Orc1-CID-PACT on histone H1a phosphorylation by Cyclin E–CDK2 (Fig. 3D,E; Supplemental Fig. S5). The MBP-Orc1-CID inhibited histone H1a phosphorylation by Cyclin E–CDK2 via competitive inhibition with a Ki of 1.5 μM, while the MBP-Orc1-CID-PACT displayed mixed noncompetitive inhibition with a Ki of 0.32 μM and a Ki′ of 25 nM (Fig. 3F). The MBP-Orc1-CID displayed a competitive mode of inhibition of Cyclin E–CDK2, and the inhibition was stimulated and converted to mixed noncompetitive inhibition by the C-terminal PACT fragment of Orc1 in MBP-Orc1-CID-PACT (Fig. 3F). Consistent with this observation, MBP-Orc1-CID-PACT bound better to Cyclin E compared with MBP-Orc1-CID (Fig. 3G). On the other hand, the CID fragment of Orc1 by itself was a mixed noncompetitive inhibitor of Cyclin A–CDK2 kinase activity and bound Cyclin A as efficiently as MBP-Orc1-CID-PACT (Fig. 3F,G). Thus, like full-length Orc1, the CID and the CID-PACT domains inhibited the two cyclin–CDK complexes differently. We therefore tested how these Orc1 fragments controlled centrosome copy number.
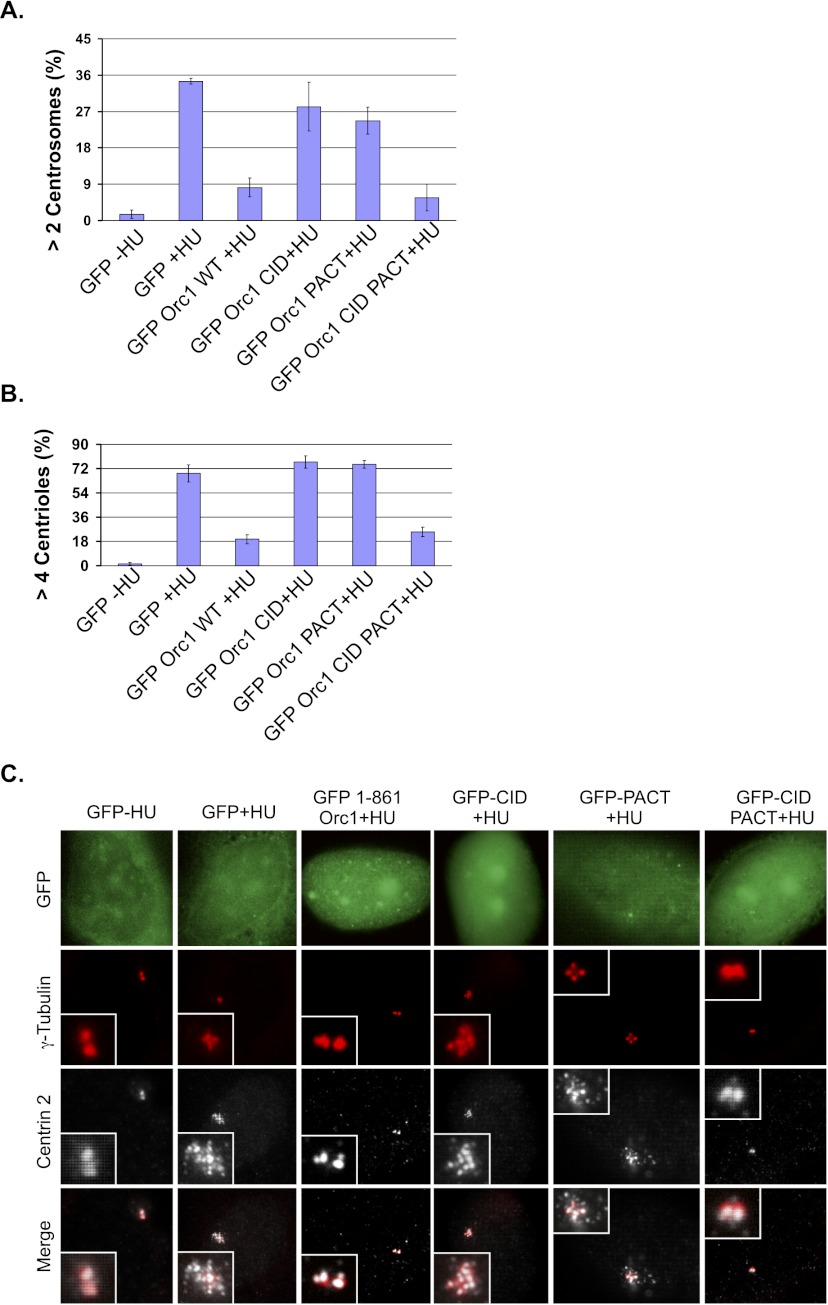
In certain cancer cells, such as U2OS osteosarcoma cells, hydroxyurea (HU) treatment causes centrioles and centrosomes to reduplicate in a single cell division cycle, and Orc1 blocks the HU-induced reduplication (Hemerly et al. 2009). Therefore, we examined whether the PACT and CID fragments, which lack the AAA+ domain of Orc1 required for DNA replication, could suppress centriole and centrosome reduplication induced by HU. Expression of GFP-Orc1-CID-PACT in U2OS cells blocked HU-induced centrosome (γ-tubulin fluorescence) and centriole (Centrin-2 fluorescence) reduplication similar to the full-length GFP-Orc1, while the individual Orc1 domains (GFP-Orc1-CID and GFP-Orc1-PACT) lacked the ability to block reduplication (Fig. 4A–C). Thus, both the cyclin–CDK2 inhibitory domain and the centrosome localization domain were necessary and sufficient for controlling centriole and centrosome copy number.
Figure 4.
Cooperation between Orc1 domains required for centrosome copy number control. (A,B) Quantification of centrosome or centriole numbers in U2OS cells that have been transfected with the indicated constructs and treated with 16 mM HU (+HU) or no HU (−HU). Cells were harvested at 68 h after HU treatment. Amplified centrosome and centriole numbers were scored by γ-tubulin (>2) and Centrin-2 (>4) immunostaining, respectively, in GFP-positive cells. Data are shown as the averages for three experiments ± standard deviations. (C) Overexpressed Orc1 constructs in HU-treated U2OS cells were stained with γ-tubulin for centrosomes and Centrin-2 for centrioles. GFP signal is visualized directly.
Meier-Gorlin mutations differentially alter Orc1 inhibition of Cyclin E–CDK2 versus Cyclin A–CDK2 kinase activities
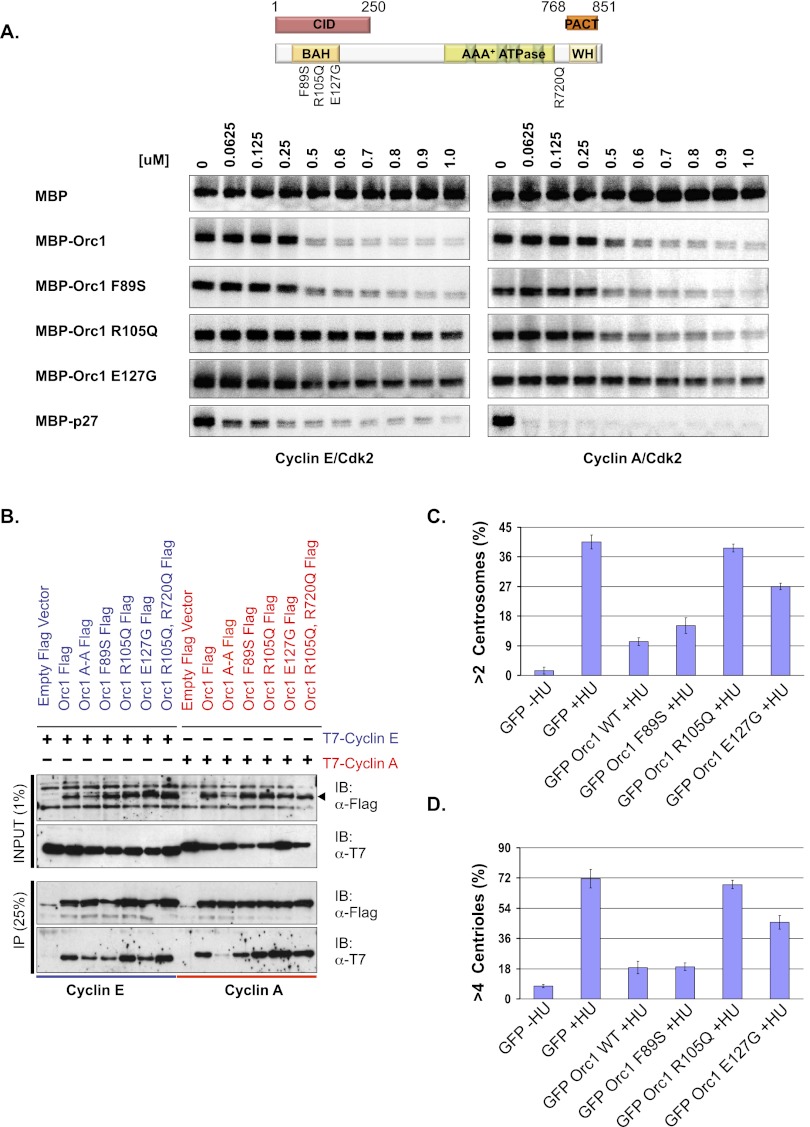
Recently, three studies reported that mutations in pre-RC proteins, including human Orc1, are associated with Meier-Gorlin syndrome, a form of microcephalic primordial dwarfism (Bicknell et al. 2011a,b; Guernsey et al. 2011; de Munnik et al. 2012). Of the mutations reported in human Orc1 in Meier-Gorlin syndrome, the three mutations (F89S, R105Q, and E127G) occurred within the CID that contains a bromo-associated homology (BAH) domain (Fig. 5A, top panel; Zhang et al. 2002; Kuo et al. 2012). Most Meier-Gorlin syndrome patients (seven out of 11 recently characterized) have an Orc1 with a mutation that changed an arginine at amino acid 105 to a glutamine (R105Q) (de Munnik et al. 2012). The presence of Orc1 mutations in the CID, and the fact that other microcephalic primordial dwarfism syndrome mutations map to other genes encoding centrosome-associated proteins, suggested further investigation of their effect on centriole and centrosome biology.
Figure 5.
Mutation blocks Orc1 function in centrosome duplication. (A) The Orc1 mutations identified in Meier-Gorlin syndrome patients are indicated in the schematic. (Top panel) The CID and PACT regions of Orc1 are boxed. The bottom panel shows kinase inhibition of Cyclin A–CDK2 or Cyclin E–CDK2 with MBP-fused Orc1 wild type or its mutant proteins. The indicated molar amounts of MBP-Orc1 or its mutant (F89S, R105Q, or E127) proteins were titrated to inhibit the phosphorylation of histone H1a by Cyclin E–CDK2 or Cyclin A–CDK2. The kinase assay used 0.5 μM histone H1a and 10 nM cyclin–CDK2 in each of the reactions. MBP and MBP-p27 were used as negative and positive controls, respectively. (B) In vivo interaction between mutant Orc1 and Cyclin A or Cyclin E. Flag-tagged wild-type Orc1 or its mutants (Orc1A-A, Orc1F89S, Orc1R105Q, Orc1E127G, and Orc1R105Q, R720Q) or empty Flag vector alone were cotransfected with either T7-Cyclin A or T7-Cyclin E. Immunoprecipitation with anti-Flag antibody from cell lysates from HEK293 cells transiently coexpressing the indicated constructs and immunoblotting with the indicated antibodies. (C,D) Centrosome reduplication assay in HU-arrested U2OS cells. GFP-Orc1 and its mutants (F89S, R105Q, and E127G) were expressed in HU-arrested U2OS cells for 68 h and then stained with γ-tubulin and Centrin-2. Bar graph indicating the percentages of cells transfected with GFP-Orc1 and its mutants (see the text) that accumulate extra γ-tubulin-stained (>2) and Centrin-2-stained (>4) dots following 68 h of treatment with HU. Data are shown as the averages for three experiments ± standard deviations.
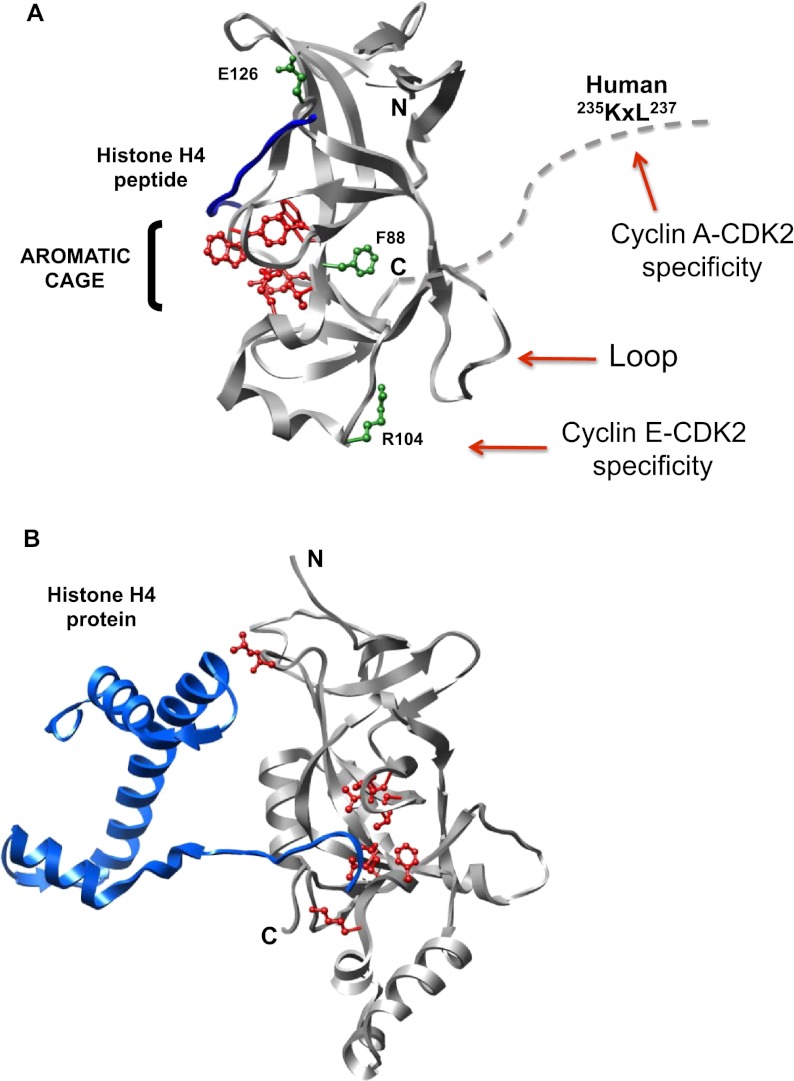
MBP-fused human Orc1 proteins with the Meier-Gorlin mutations were purified, and kinase inhibition assays were performed. Titration of MBP-Orc1F89S recombinant protein showed a potency of inhibition similar to the wild-type MBP-Orc1 on both Cyclin E–CDK2 and Cyclin A–CDK2 kinase activities (Fig. 5A). MBP-p27KIP1 was used as a control CDK inhibitor. The MBP-Orc1E127G protein lost most of its inhibitory activity against both cyclin–CDK2 activities (Fig. 5A, bottom panels). Most interestingly, the MBP-Orc1R105Q protein retained normal inhibition of Cyclin A–CDK2 activity, but could not inhibit Cyclin E–CDK2 activity (Fig. 5A). Thus, the R105Q mutant was the opposite of the Orc1 Cy motif mutation—it inhibited Cyclin A–CDK2 but not Cyclin E–CDK2 activity. Investigation of the interaction between Orc1 and cyclins in vivo suggested that loss of kinase inhibition was not due to loss of interaction between Orc1 and the cyclin proteins, since Flag-Orc1R105Q still interacted with T7-Cyclin E or T7-Cyclin A (Fig. 5B). Only the Orc1 Cy motif mutation (Orc1 A-A) blocked binding to Cyclin A but did not affect Cyclin E binding. Modeling of the human Orc1 sequence based on the crystal structure of the Saccharomyces cerevisiae BAH domain of Orc1 (Zhang et al. 2002) and the recent structure of the murine Orc1 BAH domain (Kuo et al. 2012) suggests that the residues Arg 105 and Glu 127 are exposed on the surface of the human Orc1 CID structure, while Phe 89 is buried inside the structure (Fig. 7, below). These observations suggested that the F89S mutation might have a protein stability defect in vivo but not in vitro. In contrast, the surface R105Q and E127G mutations affected CDK kinase inhibition but not binding to the two cyclins. In the case of the R105Q mutation, it differentially affected Cyclin E–CDK2 versus Cyclin A–CDK2 activity. We tested each of the mutants in the context of full-length Orc1 for binding to a dimethylated histone H4K20me2 peptide, including an Orc1W88A mutant that was shown to be completely defective in binding the H4K20me2 peptide (Kuo et al. 2012). Of note, the R105Q mutation had a minimal effect on histone peptide binding, whereas, as expected, the aromatic cage mutant was completely defective in binding (Fig. 7A, below; Supplemental Fig. S6).
Figure 7.
Molecular models of mouse and yeast BAH domains bound to histone. (A) Ribbon diagram of the mouse Orc1 BAH domain (9–170 amino acids) bound to H4(14–25 amino acids)K20me2 peptide are represented in gray and blue, respectively (Kuo et al. 2012). The residues involved in the murine Orc1 BAH domain that form an aromatic cage that binds the methylated histone H4 peptide are depicted in red ball and stick forms. The corresponding residues of human Meier-Gorlin syndrome patient Orc1 mutations (F89S, R105Q, and E127G) in the murine Orc1 BAH domain are shown in green stick and ball forms (F88S, R104Q, and E126G). The human patient mutant Orc1R105Q abolishes the kinase inhibitory activity of Orc1 on Cyclin E–CDK2, and the corresponding residue in murine Orc1 BAH domain providing Cyclin E–CDK2 specificity is indicated. The C-terminal portion of the murine Orc1 BAH domain was further extended with dotted lines to show the proximity of the human equivalent, the RxL motif (or Cy motif). Mutation of the Cy motif in human Orc1 abolishes its ability to inhibit Cyclin A–CDK2 kinase. (B) Ribbon diagram of the S. cerevisiae Sir3 BAH domain bound to a nucleosome (Armache et al. 2011). Only the histone H4 protein from the nucleosome is shown for comparison with the murine Orc1 BAH domain bound to the H4K20me2 peptide. The yeast BAH domain residues involved in contacting histone H4 protein are shown in red ball and stick forms. The yeast BAH domain and histone H4 proteins are shown in gray and blue ribbons, respectively.
Meier-Gorlin mutations in Orc1 affect centriole and centrosome copy number control
The Meier-Gorlin syndrome mutations in Orc1 offered the opportunity to dissect the effect of Orc1 cyclin–CDK2 kinase inhibitory activity on centrosome and centriole copy number control, particularly since the R105Q mutant disrupted Orc1 inhibition of Cyclin E–CDK2 kinase but not Cyclin A–CDK2 kinase. Wild-type GFP-Orc1 and various GFP-Orc1 mutants present in Meier-Gorlin syndrome patients were expressed in U2OS cells that were then treated with HU, and the cells were scored for the number of centrosomes and centrioles. Strikingly, unlike wild-type Orc1, Orc1R105Q could not prevent reduplication of centrosomes and centrioles, whereas GFP-Orc1E127G was partially defective (Fig. 5C,D). The GFP-Orc1F89S blocked centrosome reduplication almost like the wild-type Orc1, a result that correlated with its ability to inhibit Cyclin E–CDK2 activity in vitro.
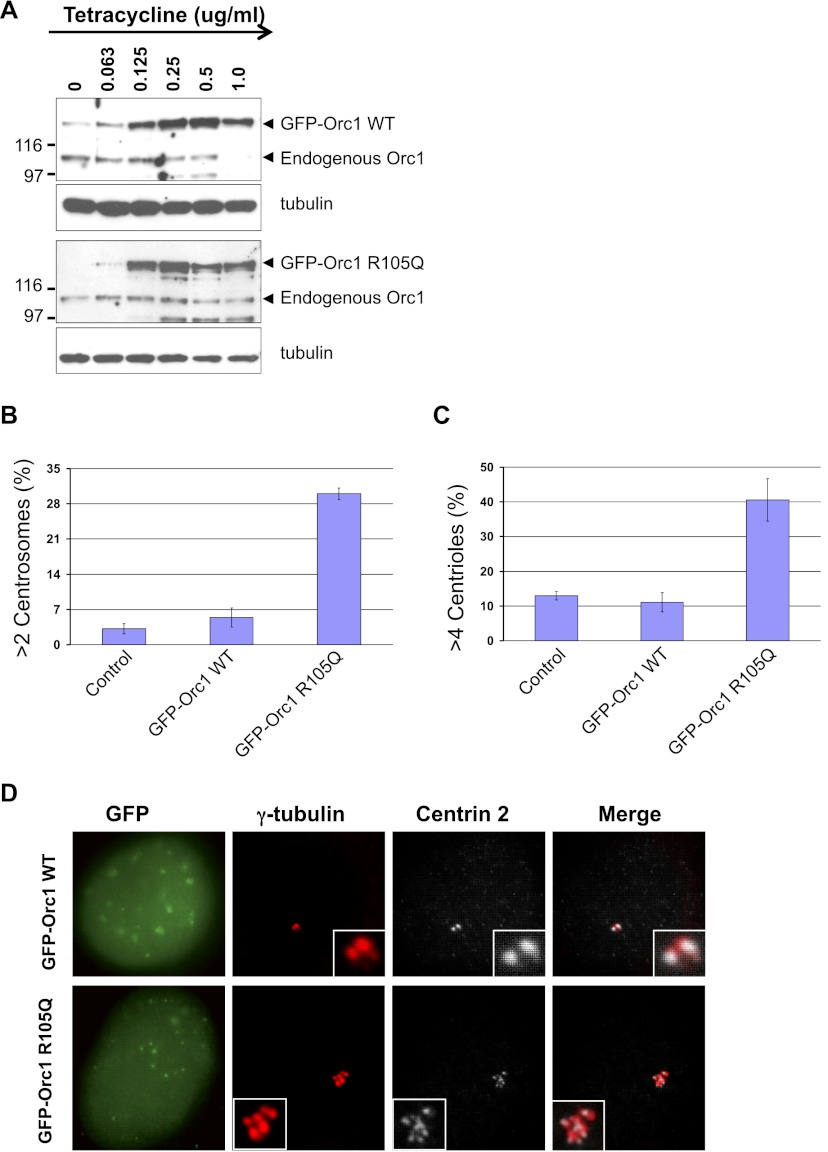
Wild-type and R105Q mutant Orc1 transgenes driven from a hybrid cytomegalovirus (CMV) immediate early promoter that was repressed by a tetracycline-regulated repressor protein were stably integrated into a U2OS cell line that harbors a single FRT integration site (Malecki et al. 2006), giving rise to isogenic cell lines expressing either GFP-Orc1WT or GFP-Orc1R105Q. The GFP-Orc1WT protein was induced by addition of tetracycline, and depending on the amount of tetracycline added, the endogenous wild-type Orc1 protein levels declined (Fig. 6A; Supplemental Fig. S7). In the case of the GFP-Orc1R105Q mutant, the transgene protein was expressed at levels similar to the GFP-Orc1WT protein, but the endogenous Orc1 did not completely disappear (Fig. 6A). After 3 d of induction with tetracycline (1 μg/mL), centriole and centrosome numbers were counted. The wild-type Orc1 did not appreciably change the numbers of either centrioles or centrosomes, but the Orc1R105Q mutant caused reduplication of both centrioles and centrosomes (Fig. 6B–D). The R105Q cells also accumulated increased nuclear volume (data not shown) and eventually greatly slowed proliferation of the cells (Supplemental Fig. S8). Thus, the predominant Orc1 Meier-Gorlin syndrome mutation caused a defect in regulation of centriole and centrosome copy numbers.
Figure 6.
Stable expression of Orc1 containing a Meier-Gorlin syndrome mutation causes amplification of centrioles and centrosomes. (A) Western blot of whole-cell extracts of U2OS Flp-in stable cell lines expressing either GFP-Orc1WT wild-type or GFP-Orc1R105Q mutant protein. The transgenes were induced with the indicated amounts of tetracycline. Monoclonal antibody against human Orc1 was used to detect endogenous as well as GFP-Orc1 proteins. α-Tubulin served as a control for equal loading of each sample. (B,C) Tetracycline-induced stable cell lines expressing GFP-fused wild-type or R105Q mutant Orc1 proteins were monitored for multiple centrosomes or centrioles. The U2OS-TreX cell line was used as a control. The cells were induced for 72 h (3 d) with 1 μg/mL tetracycline. Amplified centrosome and centriole numbers were scored by γ-tubulin (>2) and Centrin-2 (>4) immunostaining, respectively. Data are shown as the average from three experiments ± standard deviations. (D) Tetracycline-induced U2OS stable cell lines expressing GFP-Orc1WT wild-type or GFP-Orc1R105Q mutant proteins were stained with γ-tubulin for centrosomes and Centrin-2 for centrioles. The U2OS-TreX cell line is used as a control. GFP signal was visualized directly.
Discussion
Previous studies suggest that mammalian ORC is associated with nucleosomes that contained methylated histones, including H4K20me, and that such modified histones may be a receptor for ORC binding to chromatin for subsequent DNA replication (Bartke et al. 2010; Vermeulen et al. 2010; Brustel et al. 2011). A recent crystal structure of residues 9–170 amino acids of murine Orc1 showed that the BAH domain specifically bound to a dimethylated peptide derived from histone H4 (H4K20me2) (Kuo et al. 2012). This is the same region of the BAH domain in the yeast Orc1-related Sir3 protein that binds to histone H4 present in a nucleosome (Fig. 7B; Zhang et al. 2002; Armache et al. 2011). The histone H4 dimethylated peptide binds to an aromatic cage on one surface of the Orc1 BAH structure incorporating Tyr 63, Trp 87, Tyr 114, and Trp 119 (murine numbering) (red residues in Fig. 7A), and mutations in this cage disrupt binding of the histone H4K20me2 peptide (Kuo et al. 2012). On the other hand, the mutations in the murine Orc1 BAH domain (F88S and E126G) equivalent to the human Meier-Gorlin mutations F89S and E127G reduced but did not eliminate H4K20me2 peptide binding. The murine equivalent of the R105Q mutant was not tested.
The human Meier-Gorlin syndrome mutation F89 amino acid is buried in the middle of the BAH structure (Fig. 7A) and may influence protein folding in vivo, but in vitro, the F89S mutation had a small effect on both Orc1 cyclin–CDK2 kinase inhibition and inhibition of centrosome reduplication and only had a fourfold effect on the Kd for H4K20me2 peptide binding. In contrast, the R105Q mutation completely abolished the ability of Orc1 to inhibit Cyclin E–CDK2 kinase activity and induced centrosome reduplication in human cells. Interestingly, the R105 amino acid (R104 in the murine structure) localizes to a surface on the Orc1 BAH domain that is on the other side of the three-dimensional (3D) protein structure compared with the histone H4-binding site (Fig. 7A). The R105 amino acid is also predicted to be on the same surface as the C terminus of the BAH domain, likely placing it on the same side of the BAH domain as the Cy motif at amino acids 235–237 (Fig. 7A). Thus, the two different sequences in Orc1 that confer specificity of inhibition of Cyclin E–CDK2 and Cyclin A–CDK2 kinase activities (R105 and the Cy motif, respectively) appear to be localized on the same surface of the BAH domain, away from the histone interaction surface. The R105Q mutation is present in seven of the 11 Orc1 patients recently analyzed (de Munnik et al. 2012) and has the largest effect on Cyclin E–CDK2 and on centrosome and centriole copy number control. The E127 amino acid (E126 in mouse Orc1) (Fig. 7A) that is mutated in only two out of 11 recently characterized Meier-Gorlin syndrome patients, which partially abolishes cyclin–CDK inhibition and is defective in control of centriole and centrosome copy number, is at the top of a turn that then extends into a β sheet and then into an ∼20-amino-acid loop that exists on the same surface as the R105 amino acid (Fig. 7A; Kuo et al. 2012). Thus, we suggest that the surface containing the R105 residue, the loop of the BAH domain, and the extension to the Cy motif that is C-terminal to the BAH domain provide in Orc1 the CID that interacts with Cyclins A and E is bound to CDK2. We further suggest that this surface of Orc1 can differentially bind to Cyclins A and E, conferring specificity to the inhibition of the two cyclin–CDK kinase activities via R105 for Cyclin E or via the Cy motif for Cyclin A. The E127 mutation that affects the inhibitory activity of Orc1 on both Cyclin E–CDK2 and Cyclin A–CDK2 may disrupt the location or shape of the putative cyclin–CDK binding loop.
Some Meier-Gorlin mutations alter pre-RC assembly in human cells and also reduce cell proliferation (Bicknell et al. 2011b). Zebrafish with either reduced Orc1 levels or depletion of Suv420h1 and Suv420h2 that result in reduced levels of H4K20me2 and H4K20me3 have small body size (Bicknell et al. 2011b; Kuo et al. 2012). Furthermore, the Orc1 BAH domain influences the recruitment of ORC to origins of DNA replication in the nucleus (Noguchi et al. 2006). Thus, there is suggestive evidence that the microcephaly and dwarfism observed in Meier-Gorlin syndrome patients might be caused by a defect in Orc1 binding to methylated histones and its effect on DNA replication. A defect in DNA replication is also consistent with the observation that mutations in genes encoding other pre-RC proteins, such as Orc4, Orc6, and Cdt1, can cause microcephaly and dwarfism, but in these latter cases, the dwarfism and microcephaly are relatively mild compared with patients with Orc1 mutations (de Munnik et al. 2012). None of these studies demonstrate that the Meier-Gorlin mutations confer primordial microcephalic dwarfism solely via defects in DNA replication. Therefore, we suggest that the Meier-Gorlin syndrome mutations in Orc1 alter the Cyclin E–CDK2 kinase inhibition and thereby affect centrosome copy number control in cells during development. We further suggest that the resulting centrosome defects contribute to the more severe microcephaly and dwarfism found in Orc1 Meier-Gorlin syndrome mutations. Our analysis of the Orc1R105Q mutant that is present in a majority of patients suggested that Orc1 control of centrosome and centriole copy number in cells acts via its ability to inhibit Cyclin E–CDK2 and not Cyclin A–CDK2 kinase activity. This conclusion is consistent with our previous demonstration that centrosome and centriole reduplication requires Cyclin E but not Cyclin A (Hemerly et al. 2009). Our conclusion that Orc1 mutations might exert their effect via control of centrosome biology is consistent with the finding that defects in other centrosome proteins can cause primordial microcephalic dwarfism. The Meier-Gorlin mutations could alter centrosome activity during cell proliferation or result in basal body defects that cause altered signaling between cells. Moreover, ORC subunits regulate dendrite and spine development in post-mitotic neurons, a process that involves centrosomes, and this may contribute to the increased microcephaly in people with Orc1 mutations (Huang et al. 2005).
We demonstrated that Orc1 has its own centrosome PACT domain localization signal that is sufficient for centrosome localization. Since Orc1 is also a nuclear protein, association with other proteins or post-translational modification must regulate its localization to the two different sites in cells. Although the centrosome-interacting partner of a PACT domain is not known, we showed that this domain targets a functional CID to the centrosome. Human cell Orc1 is present in cells during mitosis and G1 phase and then is degraded by ubiquitin-mediated proteolysis (Kreitz et al. 2001; Mendez et al. 2002; Siddiqui and Stillman 2007), so it is present in cells precisely when Cyclin E–CDK2 activity is important for centrosome duplication. Another DNA replication protein, Mcm5, a subunit of the MCM2–7 helicase complex that, like ORC, is required for pre-RC assembly prior to DNA replication, was shown to localize to centrosomes in a manner dependent on a CLS present in Cyclin A (Ferguson et al. 2010). This study also observed that Orc1 interacted with Cyclin A in a CLS-dependent manner, but then went on to suggest that Orc1 localization to centrosomes required the Cyclin A CLS, a suggestion not consistent with the current data.
In CHO cells, CG-NAP (alternatively called AKAP450 or AKPA350), which has a PACT domain, can recruit Cyclin E–CDK2 to centrosomes, and overexpression of CG-NAP or a fragment of CG-NAP that binds to Cyclin E–CDK2 causes centrosome reduplication (Nishimura et al. 2005). One possible explanation for these results is that an excess amount of Cyclin E–CDK2 activity at centrosomes causes reduplication of centrioles. We propose that Orc1 recruitment to the PACT domain receptor in centrosomes brings a functional Cyclin E–CKD2 inhibitor to the region of the centrosome that has CG-NAP–Cyclin E–CDK2 complexes, thereby controlling the centrosome-associated activity of Cyclin E–CDK2 during this critical period in G1 phase. Later on, when cells enter S phase and Cyclin E itself has been degraded, there is no longer a need for control of Cyclin E–CDK2-mediated reduplication. Thus, it would normally not matter that Orc1 is also destroyed as cells enter S phase.
The observation that Orc1 controls both Cyclin E–CDK2 and Cyclin A–CDK2 kinase activities may be important for the nuclear function of Orc1 during licensing and DNA replication. Cyclin E–CDK2 and Cdc6, an Orc1-related pre-RC assembly protein, cooperate to assemble pre-RCs in mammalian cells during G1 phase (Stoeber et al. 1998; Coverley et al. 2002). On the other hand, Cyclin A–CDK2 is required to activate the pre-RC as cells enter S phase (Coverley et al. 2002). We suggest that the Orc1 CID will play a role in the regulation of Cyclin E–CDK2-dependent pre-RC assembly and that later, at the G1-to-S-phase transition, Cyclin A–CDK2 association with Orc1 may promote Orc1 destruction, thereby providing one mechanism for limiting DNA replication to once per cell division cycle. The localization of an intrinsic Cyclin E–CDK2 inhibitory domain within a key component of the pre-RC that is also targeted to centrosomes provides a mechanism to coordinate DNA replication with centriole and centrosome duplication.
Materials and methods
Plasmid construction and mutagenesis
Human Orc1 and its mutants were cloned into pMALp-c2E and pEGFP-C1 vectors for protein production in bacterial cells and transient overexpression in mammalian cells, respectively. The Orc1 clones were also generated with a C-terminal Flag tag in a pLPC vector (gift from Masashi Narita, Scott Lowe Laboratory, Memorial Sloan Kettering Cancer Center) for mammalian expression. The mutant plasmids were generated following the site-directed mutagenesis protocol (Stratagene). The cloning of N-terminal T7-tagged Cyclins A and E was described previously (Hemerly et al. 2009). All of the plasmid constructs were verified by sequencing. The oligonucleotide sequences used to generate the plasmids are available on request.
Production of purified MBP-fused Orc1 proteins
Wild-type Orc1 and its mutants were cloned in the bacterial expression vector pMALc2E. Transformed Escherichia coli BL21 (DE3) cells bearing the respective Orc1 plasmids were grown on selective medium (100 μg/mL ampicillin) at 37°C until the optical density of the cells reached an OD600 nM of 0.6–0.8, and then the cells were induced with 0.5 mM IPTG. Induction of cells was performed for 8 h at 16°C. For purification of MBP-fused Orc1 proteins, induced E. coli cell pellets were suspended in lysis buffer A (25 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.02% NP-40, 5 mM benzamidine-HCl, 1 mM phenylmethylsulfonyl fluoride, Protease cocktail inhibitor tablets [Roche], 10% glycerol) plus 100 μg/mL lysozyme. Cells were lysed by sonication, and the cell lysate was clarified by centrifugation. The clarified lysate was incubated with prewashed amylose resin with end-to-end shaking for 3 h at 4°C. The amylose resin with bound proteins was washed with three column volumes of buffer A plus 0.05% NP-40 + 400 mM NaCl and further with five column volumes of buffer A alone. Fusion protein was eluted with buffer A containing 10 mM maltose. Fractions containing pure MBP-fused Orc1 proteins were pooled, concentrated, and dialyzed, and protein concentration was estimated using a standard Bradford protein assay. Note that the MBP-Orc1-CID-PACT protein with mutations in the Orc1 Cy motif (K235A, L237A) was insoluble and could not be studied.
MBP pull-down assay
MBP-Orc1 or its mutant fragments were bound to amylose resin (New England Biolabs), and the beads were washed four times with binding buffer (25 mM Tris-Cl at pH 7.5, 100 mM KCl, 0.04% Nonidet P-40, 0.1 mM EDTA, 5 mM magnesium acetate, 1 mM DTT). S35-labeled Cyclin A and Cyclin E were produced using Quick TNT-coupled reticulocyte lysate system (Promega) according to the manufacturer's protocol in the presence of [35S]methionine (PerkinElmer). For each reaction, 30 μL of beads (containing ∼4 μg of MBP-Orc1 wild type, MBP-Orc1 mutants, or MBP) and 10 μL of [35S]-labeled cyclin proteins, prepared as described above, were incubated in 400 μL of binding buffer for 4 h at 4°C. Resin with bound proteins was washed five times with binding buffer, and bound proteins were eluted in SDS sample buffer and analyzed by SDS-PAGE followed by PhosphorImaging analysis.
Immunofluorescence, immunoprecipitation, and antibodies
An indirect immunofluorescence assay was performed on U2OS cells by a procedure described earlier (Hemerly et al. 2009). The following primary and secondary antibodies were used for indirect immunofluorescence: The primary antibodies were rabbit pAb anti-γ-tubulin (#T5192, Sigma), mouse monoclonal anti-γ-tubulin antibody (#T6557, Sigma), rabbit pAb anti-pericentrin (Abcam), rabbit pAb anti-CEP170 (Abcam), mouse monoclonal anti-SAS6 (Santa Cruz Biotechnology), and rabbit pAb anti-Centrin-2 (this study). The secondary antibodies were Dy649 goat anti-mouse (Jackson ImmunoResearch Laboratory), Alexa Fluor 647 goat anti-rabbit (Invitrogen), and Texas Red dye-conjugated AffiniPure donkey anti-rabbit and anti-mouse (Jackson ImmunoResearch Laboratory).
For generation of a Centrin-2 antibody, human Centrin-2 cDNA was cloned in a pGEX6P1 vector (Amersham), and the protein was purified as a GST fusion. The purified GST-Centrin-2 protein was cleaved with PreScission protease (Amersham) and further incubated with glutathione resin to obtain Centrin-2 protein without the GST protein. The purified human Centrin-2 protein was used as an antigen to raise antibodies in rabbits following an immunization protocol. The polyclonal Centrin-2 antibody was tested on HEK293 cell extracts and recognized a band of ∼23 kDa by Western blotting. This was confirmed to be Centrin-2 protein by Western blot analysis on Centrin-2 siRNA-treated extracts that showed the signal was eliminated.
Immunoprecipitation from HEK293 cells was performed using the procedure described previously (Hemerly et al. 2009).
The staining of cells for γ-tubulin was used as a marker for centrosomes, whereas the centrioles within the centrosome were visualized using Centrin-2 antibodies (Stearns et al. 1991; Salisbury et al. 2002). The pair of centrioles within the centrosome is surrounded by an amorphous, proteinaceous material called the pericentriolar material (PCM), and an antibody directed to pericentrin protein was used as marker for the PCM (Doxsey et al. 1994). CEP170 localized to the subdistal appendage of the mother centriole; it stains one centriole during G1 and S and is present at both centrioles in each centrosome during the early G2 phase of the cell cycle (Guarguaglini et al. 2005). The formation of new daughter centrioles or procentrioles was distinguished from the mother centrioles by SAS6 staining, as it localizes asymmetrically next to the mother centriole, where the daughter is synthesized (Strnad et al. 2007).
HU-induced centrosome overduplication assay
The centrosome or centriole reduplication assay was performed on U2OS cells by a procedure described earlier (Hemerly et al. 2009). Cells were stained with either mouse monoclonal anti-γ-tubulin antibody (#T6557, Sigma) or polyclonal anti-γ-tubulin (#T5192, Sigma) for centrosome visualization and polyclonal Centrin-2 antibodies for visualizing centrioles. Overexpressed proteins with the GFP tag were visualized directly. Quantitative analyses of centrosome and centriole numbers were performed separately on 100–200 GFP-positive cells, depending on the expressed proteins. Each experiment for counting centrosomes or centrioles was done in triplicate.
Characterization of Orc1 mutants in U2OS cells
Both wild-type GFP-Orc1 and the GFP-Orc1 R105Q mutant were cloned into the plasmid pcDNA 5/FRT/TO (Life Technologies) so that it is expressed from a tetracycline-regulated CMV-based promoter. The plasmids were integrated into FRT-U2OS-TRex osteosarcoma cells (Malecki et al. 2006) via FRT-mediated recombination, and the integrated transgenes were selected with hygromycin. The expression of Orc1 was assessed following treatment of cells with various concentrations of tetracycline. Centriole and centrosome numbers were determined 3 d after addition of 1 μg/mL tetracycline.
For measurement of cell proliferation, wild-type GFP-Orc1, GFP-Orc1 R105Q mutant, and control U2OS-TRex cell lines were seeded at a density of 1 × 105 cells per well in six-well plates in triplicates. On day 0, tetracycline was added at concentration of 0.5 μg/mL, and the medium was changed every day with the same concentration of tetracycline. The cells were stained with Trypan Blue (Gibco) and counted with a Countess automated cell counter (Invitrogen). The proliferation of cells was followed for 5 d.
Purification of baculovirus-expressed cyclin–CDK2 proteins
Baculoviruses encoding (His6)Cyclin A–CDK2(HA) or (His6)Cyclin E–CDK2(HA) were generated by transfecting Hi5 cells and then amplified. The kinases were purified from Hi5 insect cells coinfected with Cyclin E plus CDK2 or Cyclin A plus CDK2. The insect cells were resuspended and lysed with sonication in lysis buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2 mM MgCl2, 5 mM benzamidine-HCl, 1 mM PMSF, protease cocktail inhibitor tablet, 10% glycerol, and phosphatase inhibitor (10 mM sodium fluoride, 1 mM sodium vanadate, 1 mM β-glycerophosphate). The lysate was clarified by centrifugation, and the supernatant was incubated with prewashed Ni-NTA resin (Qiagen) for 2 h at 4°C. The resin with bound proteins was washed with lysis buffer containing 10 mM imidazole and then eluted with 250 mM imidazole. The fractions containing purified Cyclin E–CDK2 or Cyclin A–CDK2 were pooled and concentrated by centrifugal concentrator (Amicon). The kinase complex was further purified on gel filtration chromatography using Superdex-S100 columns. Fractions containing pure cyclin bound to CDK2 were pooled, dialyzed, and concentrated, and the protein concentration was estimated. Pure proteins were aliquoted, snap-frozen in liquid nitrogen, and stored at −70°C.
Kinase assay
The phosphorylation of histone H1a in the kinase assay was carried out in the presence of 10 nM Cyclin A–CDK2 or Cyclin E–CDK2 in a 15-μL reaction volume—containing 0.1 mg/mL BSA, 0.2 mM cold ATP, and 5 μCi γ-32P ATP (6000 Ci/mmol; PerkinElmer) in reaction buffer (50 mM Tris-HCl at pH 7.5, 10 mM MgCl2, 1 mM DTT)—for 20 min at 37°C. For the kinase inhibition assay with MBP-Orc1 derivatives, the concentrations of MBP-Orc1 and histone H1a varied as indicated in the figures. Reactions were quenched with SDS sample buffer, boiled, and run on 11% SDS–polyacrylamide gels. The gels were stained with Coomassie Brilliant Blue and then destained. The histone bands were excised from the gel and then monitored for incorporation of γ-32P ATP using a scintillation counter. Double reciprocal plots (Lineweaver-Burk plots) (Segel 1975) for MBP-Orc1 and its mutant derivatives' inhibition of CDK activity with histone H1a as a substrate were obtained using regression analysis (least squares method). Microsoft Excel was used to plot and analyze kinetic data (Microsoft). Dissociation constants (Ki) of the inhibitors were calculated from secondary plots of inhibitor concentrations versus the intercept or the slope of the corresponding Lineweaver-Burk plots.
Histone peptide pull-down assay
Biotinylated methylated histone H4 peptide {GKGGAKRHR [K-(Me)2] VLRDNIQGI} was custom-synthesized by Thermo Scientific. In the pull-down assay, 1 μg of the biotinylated peptide was incubated with Streptavidin beads for 2 h at 4°C. The peptide-bound Streptavidin beads were washed with pull-down buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.05% NP-40) and further incubated with purified MBP-Orc1 proteins. Binding with proteins was performed in 400 μL of pull-down buffer for 5 h at 4°C and further washed three times in the same buffer. Bound proteins were mixed with sample loading buffer, run on 8% SDS-PAGE gels, and transferred for Western blotting. The immunoblot was developed using polyclonal anti-MBP rabbit primary and Alexa Fluor 680-conjugated secondary antibodies, followed by quantitation using an Odyssey infrared imaging system (LI-COR Biosciences). Different amounts of MBP-Orc1 proteins were used in the titration experiment for binding and are indicated in the figures. MBP protein and streptavidin beads alone serve as a negative control in the pull-down assays.
Acknowledgments
We thank members of the Stillman laboratory for valuable discussions, and P. Wendel for technical assistance. Stephen Hearn provided valuable assistance with microscopy. We thank Dr. Dinshaw Patel and Dr. Jikui Song for communicating the coordinates of the murine Orc1 BAH domain just prior to its publication. We also thank David Spector, Leemor Joshua-Tor, and David Wah for comments on the manuscript. This research was supported by a grant from the National Cancer Institute (CA13106).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.197178.112.
References
- Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE 2011. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science 334: 977–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T 2010. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143: 470–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell LS, Bongers EM, Leitch A, Brown S, Schoots J, Harley ME, Aftimos S, Al-Aama JY, Bober M, Brown PA, et al. 2011a. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat Genet 43: 356–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell LS, Walker S, Klingseisen A, Stiff T, Leitch A, Kerzendorfer C, Martin CA, Yeyati P, Al Sanna N, Bober M, et al. 2011b. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat Genet 43: 350–355 [DOI] [PubMed] [Google Scholar]
- Brustel J, Tardat M, Kirsh O, Grimaud C, Julien E 2011. Coupling mitosis to DNA replication: The emerging role of the histone H4-lysine 20 methyltransferase PR-Set7. Trends Cell Biol 21: 452–460 [DOI] [PubMed] [Google Scholar]
- Coverley D, Laman H, Laskey RA 2002. Distinct roles for cyclins E and A during DNA replication complex assembly and activation. Nat Cell Biol 4: 523–528 [DOI] [PubMed] [Google Scholar]
- de Munnik SA, Bicknell LS, Aftimos S, Al-Aama JY, van Bever Y, Bober MB, Clayton-Smith J, Edrees AY, Feingold M, Fryer A, et al. 2012. Meier-Gorlin syndrome genotype–phenotype studies: 35 individuals with pre-replication complex gene mutations and 10 without molecular diagnosis. Eur J Hum Genet 20: 598–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey SJ, Stein P, Evans L, Calarco PD, Kirschner M 1994. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell 76: 639–650 [DOI] [PubMed] [Google Scholar]
- Ferguson RL, Maller JL 2008. Cyclin E-dependent localization of MCM5 regulates centrosome duplication. J Cell Sci 121: 3224–3232 [DOI] [PubMed] [Google Scholar]
- Ferguson RL, Pascreau G, Maller JL 2010. The cyclin A centrosomal localization sequence recruits MCM5 and Orc1 to regulate centrosome reduplication. J Cell Sci 123: 2743–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Munro S 2000. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep 1: 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarguaglini G, Duncan PI, Stierhof YD, Holmstrom T, Duensing S, Nigg EA 2005. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell 16: 1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guernsey DL, Matsuoka M, Jiang H, Evans S, Macgillivray C, Nightingale M, Perry S, Ferguson M, LeBlanc M, Paquette J, et al. 2011. Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nat Genet 43: 360–364 [DOI] [PubMed] [Google Scholar]
- Hemerly AS, Prasanth SG, Siddiqui K, Stillman B 2009. Orc1 controls centriole and centrosome copy number in human cells. Science 323: 789–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe EH, Li C, Thompson EA, Maller JL, Sluder G 1999. Requirement of Cdk2–cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science 283: 851–854 [DOI] [PubMed] [Google Scholar]
- Huang Z, Zang K, Reichardt LF 2005. The origin recognition core complex regulates dendrite and spine development in postmitotic neurons. J Cell Biol 170: 527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa D, Kohlmaier G, Keller D, Strnad P, Balestra FR, Fluckiger I, Gonczy P 2011. Spindle positioning in human cells relies on proper centriole formation and on the microcephaly proteins CPAP and STIL. J Cell Sci 124: 3884–3893 [DOI] [PubMed] [Google Scholar]
- Klingseisen A, Jackson AP 2011. Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev 25: 2011–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knockleby J, Lee H 2010. Same partners, different dance: Involvement of DNA replication proteins in centrosome regulation. Cell Cycle 9: 4487–4491 [DOI] [PubMed] [Google Scholar]
- Kreitz S, Ritzi M, Baack M, Knippers R 2001. The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. J Biol Chem 276: 6337–6342 [DOI] [PubMed] [Google Scholar]
- Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, Chen JK, Patel DJ, Gozani O 2012. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature 484: 115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey KR, Jackson PK, Stearns T 1999. Cyclin-dependent kinase control of centrosome duplication. Proc Natl Acad Sci 96: 2817–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, Blacklow SC 2006. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol 26: 4642–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J, Zou-Yang XH, Kim SY, Hidaka M, Tansey WP, Stillman B 2002. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol Cell 9: 481–491 [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Kasahara K, Miyazaki I, Shimizu S, Taniguchi M, Matsuzaki S, Tohyama M, Asanuma M 2009. Pericentrin, a centrosomal protein related to microcephalic primordial dwarfism, is required for olfactory cilia assembly in mice. FASEB J 23: 3289–3297 [DOI] [PubMed] [Google Scholar]
- Nigg EA, Stearns T 2011. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol 13: 1154–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Takahashi M, Kim HS, Mukai H, Ono Y 2005. Centrosome-targeting region of CG-NAP causes centrosome amplification by recruiting cyclin E–cdk2 complex. Genes Cells 10: 75–86 [DOI] [PubMed] [Google Scholar]
- Noguchi K, Vassilev A, Ghosh S, Yates JL, DePamphilis ML 2006. The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo. EMBO J 25: 5372–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B 2004. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J 23: 2651–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Thiel CT, Schindler D, Wick U, Crow YJ, Ekici AB, van Essen AJ, Goecke TO, Al-Gazali L, Chrzanowska KH, et al. 2008. Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science 319: 816–819 [DOI] [PubMed] [Google Scholar]
- Salisbury JL, Suino KM, Busby R, Springett M 2002. Centrin-2 is required for centriole duplication in mammalian cells. Curr Biol 12: 1287–1292 [DOI] [PubMed] [Google Scholar]
- Segel IH. 1975. Enzyme kinetics: Behavior and analysis of rapid equilibrium and steady state enzyme systems. Wiley, New York. [Google Scholar]
- Siddiqui K, Stillman B 2007. ATP-dependent assembly of the human origin recognition complex. J Biol Chem 282: 32370–32383 [DOI] [PubMed] [Google Scholar]
- Sir JH, Barr AR, Nicholas AK, Carvalho OP, Khurshid M, Sossick A, Reichelt S, D'Santos C, Woods CG, Gergely F 2011. A primary microcephaly protein complex forms a ring around parental centrioles. Nat Genet 43: 1147–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Evans L, Kirschner M 1991. γ-Tubulin is a highly conserved component of the centrosome. Cell 65: 825–836 [DOI] [PubMed] [Google Scholar]
- Stillman B 2005. Origin recognition and the chromosome cycle. FEBS Lett 579: 877–884 [DOI] [PubMed] [Google Scholar]
- Stoeber K, Mills AD, Kubota Y, Krude T, Romanowski P, Marheineke K, Laskey RA, Williams GH 1998. Cdc6 protein causes premature entry into S phase in a mammalian cell-free system. EMBO J 17: 7219–7229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P 2007. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell 13: 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuermer A, Hoehn K, Faul T, Auth T, Brand N, Kneissl M, Putter V, Grummt F 2007. Mouse pre-replicative complex proteins colocalise and interact with the centrosome. Eur J Cell Biol 86: 37–50 [DOI] [PubMed] [Google Scholar]
- Tachibana KE, Nigg EA 2006. Geminin regulates multiple steps of the chromosome inheritance cycle. Cell Cycle 5: 151–154 [DOI] [PubMed] [Google Scholar]
- Tang CJ, Lin SY, Hsu WB, Lin YN, Wu CT, Lin YC, Chang CW, Wu KS, Tang TK 2011. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J 30: 4790–4804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton GK, Woods CG 2009. Primary microcephaly: Do all roads lead to Rome? Trends Genet 25: 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MF, Stearns T 2006. Controlling centrosome number: Licenses and blocks. Curr Opin Cell Biol 18: 74–78 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, et al. 2010. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142: 967–980 [DOI] [PubMed] [Google Scholar]
- Wong C, Stearns T 2003. Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat Cell Biol 5: 539–544 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Hayashi MK, Merkel O, Stillman B, Xu RM 2002. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J 21: 4600–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]