Kawakami et al. report a new gene, dsh1+, in S. pombe that is required for RNAi-mediated heterochromatin formation at centromeric repeats. They also show that Dsh1 localizes with Dcr1 at the nuclear periphery to form a nuclear compartment for siRNA amplification, thus providing new information regarding where heterochromatin formation occurs.
Keywords: RNAi, siRNA, heterochromatin, fission yeast
Abstract
In fission yeast, siRNA is generated from pericentromeric noncoding RNA by the RNAi machinery. siRNA synthesis and heterochromatin formation are interdependent, forming a self-reinforcing loop on chromatin. In this system, siRNA is amplified by the RNA-dependent RNA polymerase complex (RDRC) and the endoribonuclease Dcr1, which synthesizes dsRNA and processes the dsRNA, respectively. The amplification is essential for stable heterochromatin formation. Here, a novel gene, dsh1+ (defect of the gene silencing at centromeric heterochromatin), is identified as an essential component of RNAi-directed heterochromatin assembly. Loss of dsh1+ abolishes normal RNAi function and heterochromatic gene silencing at pericentromeres. Dsh1 interacts with Dcr1 and RDRC and couples the reactions of both proteins to the effective production of siRNA in vivo. Dsh1 binds to heterochromatin in the absence of RDRC, while RDRC requires Dsh1 for its chromatin-binding activity, suggesting that Dsh1 recruits RDRC to chromatin. Immunofluorescence analysis shows that Dsh1 forms foci at the nuclear periphery, and some Dsh1 foci colocalize with Dcr1 and RDRC. Dsh1 is required for the colocalization of Dcr1 and RDRC. Moreover, loss of the nuclear periphery localization of Dsh1 abolishes Dsh1 function. Taken together, these results suggest that Dsh1 assembles the RNAi machinery on heterochromatin and forms a perinuclear compartment for amplification of heterochromatic siRNA.
Heterochromatin is a silent chromatin structure that represses gene expression and recombination to maintain genome integrity. Constitutive heterochromatin comprises nucleosomes methylated at Lys 9 of histone H3 (H3K9me) as well as HP1 (heterochromatin protein 1), which recognizes and binds to H3K9me and occurs at repeated sequences, particularly centromeres and subtelomeres. In fission yeast, constitutive heterochromatin localizes at centromeres, telomeres, and the mating type region. Fission yeast centromeres comprise an outer repeat (otr) region (containing dg and dh repeats) and an innermost repeat (imr) region, which flank the central core (cnt) domain, the site of kinetochore assembly (Chikashige et al. 1989). At pericentromeres, heterochromatin is formed on the otr and a portion of the imr. Sequences corresponding to dg and dh are also found in the mating type region and subtelomeres (Grewal and Klar 1997; Mandell et al. 2005). These repeats are rich in H3K9me and the HP1 homologs Swi6 and Chp2 (Cam et al. 2005). H3K9me is catalyzed by the histone methyltransferase Clr4 and is required for the localization of Swi6 to heterochromatin (Rea et al. 2000; Nakayama et al. 2001).
The endogenous RNAi machinery, including Dicer, Argonaute, and RNA-dependent RNA polymerase (encoded by dcr1, ago1, and rdp1, respectively), plays an important role in H3K9me at pericentromeric repeats (Volpe et al. 2002). Although heterochromatin is a silent chromatin structure that blocks transcription, RNA polymerase II (Pol II) transcribes these repeats (Djupedal et al. 2005; Kato et al. 2005; Chen et al. 2008). In fission yeast, siRNAs derived from heterochromatic repeats are present within the cell (Reinhart and Bartel 2002) and are loaded into the RNA-induced transcriptional silencing (RITS) complex, which is composed of Ago1, Tas3, and the chromodomain-containing protein Chp1 (Verdel et al. 2004). RITS is thought to bind to heterochromatic noncoding RNA (ncRNA) using siRNA as a guiding molecule. It then recruits the RNA-dependent RNA polymerase complex (RDRC), which consists of Rdp1, a poly(A) polymerase (Cid12), and a putative helicase (Hrr1), most likely via physical interactions (Motamedi et al. 2004). The RDRC enhances the generation of siRNA by synthesizing dsRNAs from centromeric transcripts as substrates for Dcr1 (Sugiyama et al. 2005; Colmenares et al. 2007). RITS also recruits Clr4 via Stc1 such that the heterochromatin spreads onto the dg and dh repeats (Zhang et al. 2008). The LIM domain protein Stc1 associates with RITS on centromeric transcripts and recruits the Clr4-containing complex (CLRC), thereby coupling RNAi to chromatin modification (Bayne et al. 2010). The RNAi-mediated heterochromatin system also functions at centromere-like repeats in the mating locus and subtelomeres (Cam et al. 2005).
In the RNAi-mediated heterochromatin assembly system, siRNA generation and heterochromatin formation are interdependent, forming a “self-reinforcing loop” system (Noma et al. 2004; Sugiyama et al. 2005). The self-reinforcing as well as the cis-acting nature of siRNA (Noma et al. 2004) during RNAi-mediated heterochromatin formation suggests that the whole process is somehow coupled on chromatin. This is supported by chromatin immunoprecipitation (ChIP) experiments, which show the physical association of Ago1 and Rdp1 with chromatin (Volpe et al. 2002; Cam et al. 2005). Recently, DNA adenine methyltransferase identification methods were used to show that Dcr1 also associates with heterochromatin (Woolcock et al. 2011). Coupling of ncRNA transcription and siRNA generation was further suggested by the isolation of a Pol II mutant that does not affect ncRNA transcription but compromises siRNA synthesis (Kato et al. 2005). Recently, heterochromatic ncRNA was shown to associate with chromatin via cotranscriptional DNA:RNA hybrid formation, which provides a target for the RITS complex (Nakama et al. 2012). In addition, the interaction between the RNAi machinery in vivo hints at the mechanism involved in the coupling: RITS physically interacts with RDRC (Motamedi et al. 2004), and RDRC interacts with Dicer (Colmenares et al. 2007). These interactions are thought to be important for efficient production of siRNA from heterochromatic ncRNA, but the molecular nature and functional relationships between them and siRNA synthesis are not clear.
In many organisms, including fission yeast, heterochromatin localizes to the nuclear periphery. This type of chromosome organization is thought to be important for gene regulation (Towbin et al. 2009). However, the functional importance of this localization in terms of heterochromatin structure/function is not understood. In fission yeast, heterochromatin can be visualized by immunostaining for Swi6, and a previous study identified two to five discrete Swi6 foci at the nuclear periphery in interphase cells (Ekwall et al. 1996). Consistent with the notion that the RNAi machinery functions on or close to heterochromatin, some components of the RITS and RDRC form dot-like structures at the nuclear periphery (Motamedi et al. 2004; Noma et al. 2004). A relationship between the nuclear envelope and RNAi-mediated heterochromatin assembly is further suggested by the fact that nuclear periphery localization of Dcr1, which is independent of heterochromatin or RNAi, is important for RNAi-directed heterochromatin formation in fission yeast (Emmerth et al. 2010). However, the molecular mechanism that connects heterochromatin and RNAi with the nuclear envelope is still unclear.
Therefore, in the present study, we performed a genetic screening in fission yeast and identified a novel gene, dsh1+ (defect of the gene silencing at centromeric heterochromatin), which is essential for RNAi-dependent heterochromatin assembly. We found that Dsh1 interacted with both RDRC and Dcr1. Functional analysis of dsh1+ revealed that Dsh1 recruits RDRC to heterochromatin and stabilizes the RDRC–Dcr1 interaction to facilitate amplification of siRNA from heterochromatic ncRNA. Our analysis also revealed that Dsh1 localizes close to the nuclear periphery independently of RNAi. Thus, Dsh1 assembles the RNAi machinery required for efficient siRNA generation on heterochromatin at the nuclear periphery.
Results
Isolation of dsh1+ as the gene responsible for heterochromatic gene silencing
We first performed a forward genetic screening in the fission yeast Schizosaccharomyces pombe to identify novel factors that contribute to heterochromatin organization at pericentromeres. We used a parental strain in which the native ade6+ and ura4+ genes were deleted and two marker genes, ade6+ and ura4+, were inserted into otr1R and imr1L, respectively (otr1R∷ade6+ and imr1L∷ura4+) (Fig. 1A). Suppression of the ade6+ and ura4+ marker genes by heterochromatin (referred to as “gene silencing”) (Allshire et al. 1995) resulted in the formation of red colonies on a plate containing low concentrations of adenine (Low Ade) that were resistant to the drug 5′-fluoroorotic acid (5′-FOA), which is toxic to cells expressing ura4+. Once heterochromatin is disrupted by a certain mutation, the expression of these two marker genes is increased such that mutant cells form white colonies on Low Ade plates and are killed on 5′-FOA plates (Allshire et al. 1995). Therefore, we screened clones that formed white colonies and were sensitive to 5′-FOA as putative heterochromatin mutants (see the Materials and Methods). Finally, nine clones showing various defects in the silencing of both marker genes (Supplemental Fig. S1A) were isolated. Genetic mapping and DNA sequencing revealed that one of the mutants, “No. 270,” harbored a frameshift mutation at the beginning of the SPBC582.04c ORF (Supplemental Fig. S1B). SPBC582.04c had never been characterized, and its function was unknown. The gene responsible for the phenotype shown by mutant No. 270 was named dsh1+.
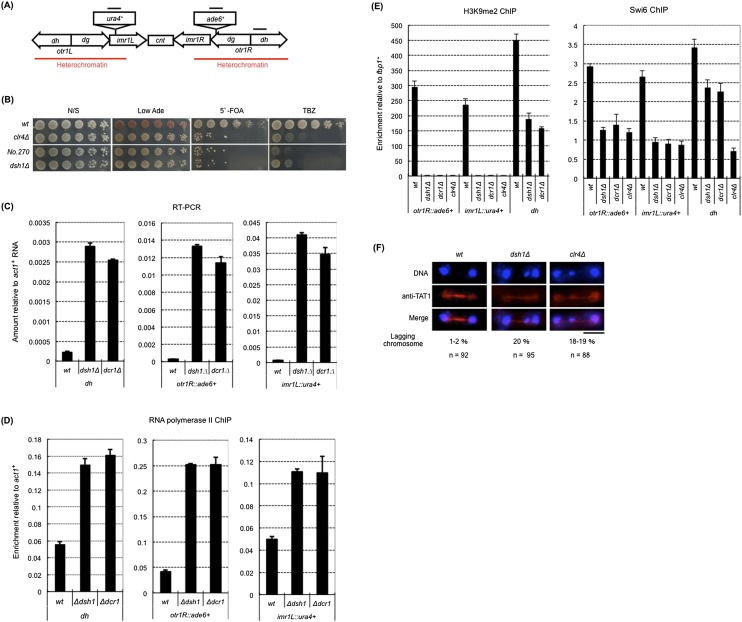
Figure 1.
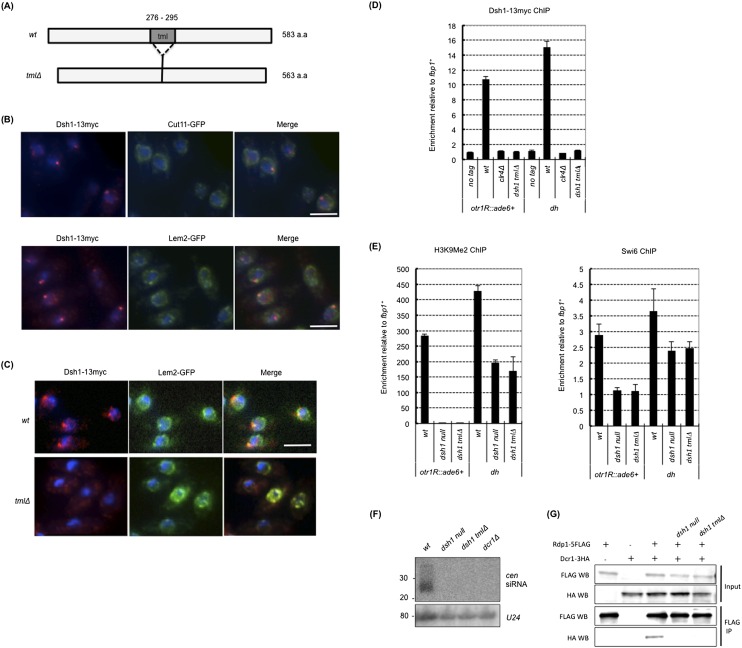
The dsh1 mutant shows defects in heterochromatin structure at the centromere. (A) Schematic representation of centromere 1 in the strain used for all experiments described in this figure. The positions of the inserted marker genes (imr1L∷ura4+ and otr1R∷ade6+) are shown. Black bars indicate the positions of the PCR products in ChIP and RT–PCR. The heterochromatic region is also indicated. Note that the centromere-specific histone H3 variant Cnp1 localized between heterochromatin (cnt and inner part of imr). (B) Tenfold serial dilution plating assays used to examine gene silencing of otr1R∷ade6+ and imr1L∷ura4+ and sensitivity to the microtubule-destabilizing drug TBZ. The indicated strains were spotted onto N/S (YES), Low Ade, YES+5′-FOA, and YES+TBZ plates. (C) RT– PCR analysis of otr1R∷ade6+, imr1L∷ura4+, and dh RNA levels in the indicated strains. Expression relative to act1+ mRNA is shown on the Y-axis. (D,E) ChIP analysis of Pol II (D) and H3K9me2 and Swi6 (E) at otr1R∷ade6+, imr1L∷ura4+, and dh in the indicated strains. Enrichment relative to act1+ (D) or fbp1+ (E) is shown on the Y-axis. (F) Indirect immunofluorescence analysis for visualizing chromosome segregation during late anaphase. Cells growing exponentially were stained with DAPI and an antibody against TAT1 to visualize DNA and tubulin, respectively. The percentage of cells with lagging chromosomes at late anaphase is shown. (n) Number of counted cells. Black bar, 5 μm.
A null mutant of dsh1+ showed the same phenotype as that of mutant No. 270 on Low Ade and 5′-FOA plates (Fig. 1B), which confirmed that the dsh1+ gene was responsible for the silencing defect in mutant No. 270. Consistent with this, the levels of ade6+ and ura4+ mRNAs and the occupancy of Pol II were increased in dsh1Δ cells to a level comparable with that observed in dcr1Δ cells (Fig. 1C,D). These data indicated that dsh1+ is an essential gene for heterochromatic gene silencing at pericentromeric heterochromatin.
dsh1+ is essential for heterochromatin organization at centromeres
To gain further insight into the function of dsh1+ during heterochromatin organization, we analyzed the occupancy of H3K9me and Swi6 at the centromeric heterochromatin using ChIP analysis. The levels of H3K9me2 and Swi6 were significantly reduced at the native heterochromatin repeats (dh) and almost abolished at the inserted marker genes (otr1R∷ade6+ and imr1L∷ura4+) in dsh1Δ cells, as in dcr1Δ cells (Fig. 1E).
The loss of centromeric heterochromatin causes an increase in the number of cells with lagging chromosomes during mitosis and causes hypersensitivity to thiabendazole (TBZ), an inhibitor of spindle assembly (Ekwall et al. 1999). This is because heterochromatin is required for localization of cohesin to the pericentromere (Bernard et al. 2001; Nonaka et al. 2002). We found that, like clr4 Δ cells, dsh1Δ cells harbored increased numbers of lagging chromosomes and were hypersensitive to TBZ (Fig. 1B,F). Thus, we concluded that dsh1+ is essential for centromeric heterochromatin assembly at the level of histone modification and for the normal function of the centromere.
dsh1+ is essential for the RNAi machinery
Because assembly of pericentromeric heterochromatin requires RNAi, and the phenotype of dsh1Δ was similar to that of other RNAi mutants such as dcr1Δ or rdp1Δ (Fig. 1C–E; Sugiyama et al. 2005), we speculated that dsh1+ was a new component of the RNAi machinery. To confirm this hypothesis, we analyzed siRNAs derived from dg and dh repeats. As in dcr1Δ cells, a large amount of ncRNA from dh repeats accumulated in dsh1Δ cells (Fig. 1C), but no siRNA was detected (Fig. 2A). The RITS complex binds to both RNAi-directed heterochromatin and the inserted marker gene (otr1R∷ura4+) in a RNAi-dependent manner. (Noma et al. 2004; Verdel et al. 2004; Cam et al. 2005). Consistent with a defect of RNAi, Ago1, a subunit of the RITS complex, failed to localize to the marker gene and showed decreased localization to the centromeric repeats in dsh1Δ cells (Fig. 2B).
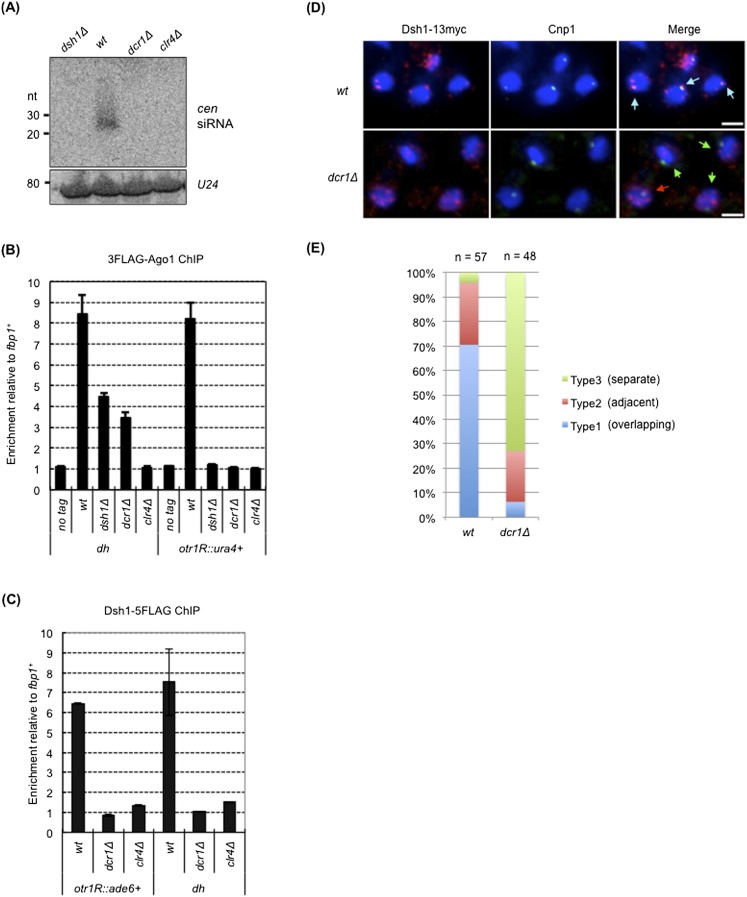
Figure 2.
dsh1+ is an essential component of the RNAi machinery. (A) siRNA analysis by Northern blotting using radiolabeled oligonucleotide probes against dg and dh centromeric repeats. A probe against U24 RNA was also used as an internal control. (B) ChIP analysis of 3×Flag-Ago1 at otr1R∷ura4+ and dh in the indicated strains. Enrichment relative to fbp1+ is shown on the Y-axis. (C) ChIP analysis of Dsh1-5Flag at otr1R∷ade6+ and dh. dcr1Δ and clr4Δ mutants were used as controls for RNAi mutants and heterochromatin mutants, respectively. Enrichment relative to fbp1+ is shown on the Y-axis. (D) Indirect immunofluorescence analysis of Dsh1 localization in a strain expressing Dsh1-13myc. Cells growing exponentially were stained with DAPI, an antibody against myc, and an antibody against Cnp1 to visualize DNA, Dsh1-13myc, and the position of the centromeres, respectively. White bars, 2 μm. (E) The fraction of cells showing overlapping, adjacent, and separate signals of Dsh1-13myc and Cnp1 (indicated by blue, red, and green arrows, respectively, in the right panels in D) in the cells showing both signals are indicated by a bar graph.
We next analyzed the state of gene silencing and the heterochromatin structure at the mating type region, where RNAi- and Atf1/Pcr1-mediated heterochromatin formation systems function in parallel. Each pathway maintains heterochromatin by inducing methylation of H3K9 redundantly. Therefore, at the mating type region, a defect in either system does not affect the heterochromatin, whereas simultaneous disruption of both systems abolished the heterochromatin (Jia et al. 2004). We found that a single deletion of dsh1 did not affect the gene silencing or the levels of H3K9me2 and Swi6 at ura4+ inserted into the K region (kint2∷ura4+), whereas double deletion of dsh1 and atf1 caused a defect in the gene silencing and reduced the levels of H3K9me2 and Swi6, similar to that observed for the clr4Δ single mutant and the dcr1Δatf1Δ mutant (Supplemental Fig. S2). On the other hand, deletion of dsh1+ and dcr1+ did not cause a defect in the silencing or levels of H3K9me2 and Swi6 (Supplemental Fig S2B,C). These results confirmed that dsh1+ is a component of the RNAi machinery.
Dsh1 binds to heterochromatin and localizes to the nuclear periphery
To further analyze the function of Dsh1 in RNAi, we constructed strains expressing Dsh1 with a 5Flag, 13myc epitope tag, or GFP tag. Note that these tags did not affect the gene silencing function of Dsh1 (Supplemental Fig. S3). Using a strain harboring dsh1 tagged with 5Flag, we performed ChIP analysis of Dsh1. Compared with the euchromatic locus (fbp1+), Dsh1 was enriched at the centromeric repeats and at the marker gene inserted into the heterochromatin (dh and otr1R∷ade6+) (Fig. 2C). Binding of Dsh1 to heterochromatin was abolished in clr4Δ or dcr1Δ cells (Fig. 2C), indicating that heterochromatin and/or the RNAi system is required for localization of Dsh1 to heterochromatin. Interestingly, we detected reduced but significant association of Dsh1 with heterochromatin in swi6Δ cells (Supplemental Fig. S4), which is similar to Rdp1 (Sugiyama et al. 2005).
We next analyzed the subcellular localization of Dsh1 by indirect immunofluorescence analysis using cells expressing Dsh1-13myc. We also analyzed the localization of the centromere-specific histone H3 variant Cnp1 using an anti-Cnp1 antibody, which enabled us to compare the localization of Dsh1 with that of the centromeres. Dsh1 localized in the nucleus and formed one to four dot-like structures close to the nuclear periphery (Fig. 2D [top panel], 3B [right panel]). In contrast, Cnp1 formed a single dot-like structure at the nuclear periphery, showing that all three centromeres clustered proximal to the spindle pole body. One of the Dsh1 dots colocalized with the Cnp1 dot or was localized very close to it (Fig. 2D [top panel], E). This was consistent with the results of ChIP analysis, which showed that Dsh1 bound to pericentromeric heterochromatin, which surrounds the Cnp1-bound chromatin domain (Fig. 1A). In dcr1Δ cells, the Dsh1 dots were more dispersed but still remained close to the nuclear periphery, although the colocalization (or close localization) of the Dsh1 dots with the Cnp1 dot was lost (Fig. 2D [bottom panel], E). This supports the conclusions drawn from the results of ChIP analysis: Dsh1 localizes to the centromeres in an RNAi-dependent manner (Fig. 2C). This also showed that heterochromatin association was not a primary cause of nuclear periphery localization of Dsh1 because Dsh1 did not associate with heterochromatin but was retained at the nuclear periphery in dcr1Δ cells. These data strongly suggest that Dsh1 functions on heterochromatin to synthesize siRNA via the RNAi machinery.
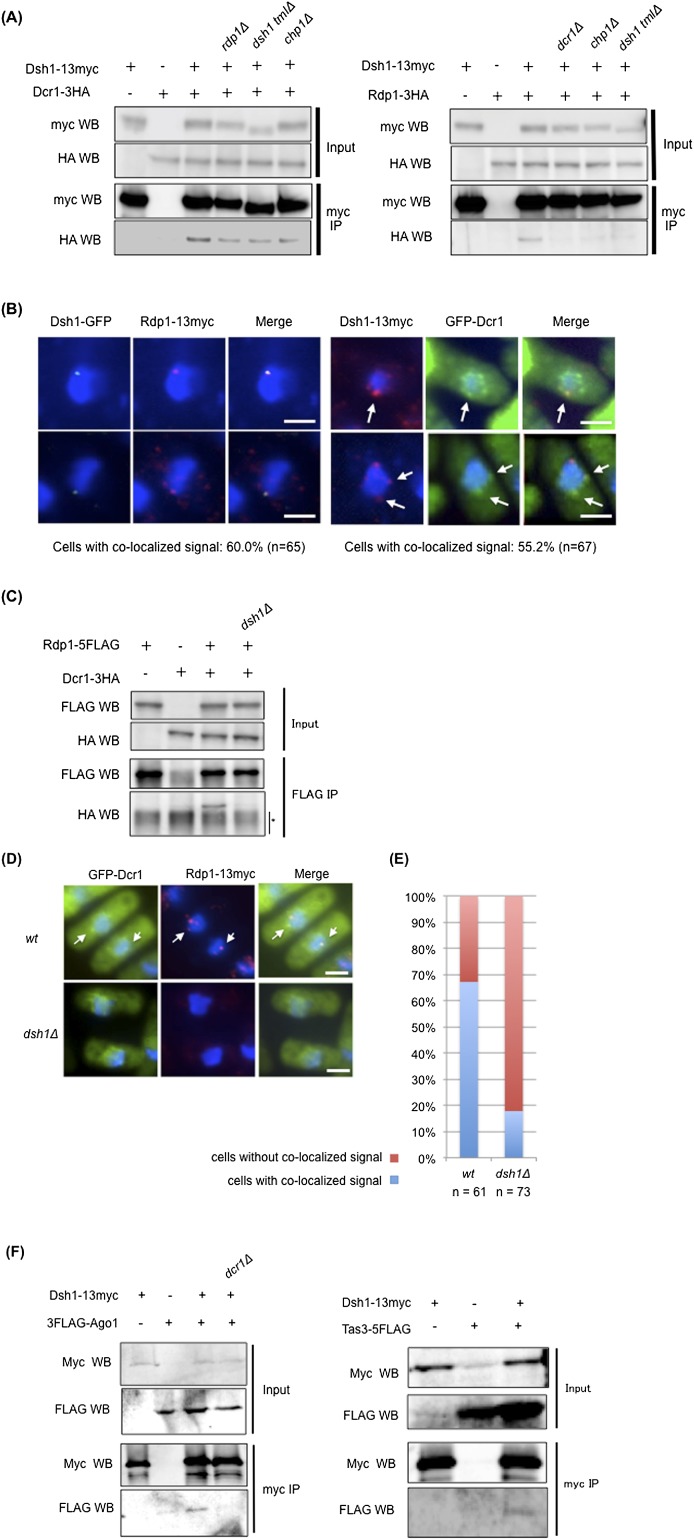
Figure 3.
Dsh1 facilitates the formation of the Dcr1–Rdp1 complex. (A) Coimmunoprecipitation assay analyzing the Dsh1–Dcr1 (left panel) and Dsh–Rdp1 (right panel) interactions. Extracts prepared from the indicated strains were incubated with an antibody against myc. Immunoprecipitated fractions were analyzed by Western blotting with antibodies against myc or HA. dsh1-tmlΔ is described in Figure 5A. (B) Indirect immunofluorescence analysis visualizing the colocalization of Dsh1-GFP and Rdp1-13myc (left) or Dsh1-13myc and GFP-Dcr1 (right) using antibodies against myc and GFP. DNA was also stained with DAPI. Arrows in the right panels indicate the colocalized dots. (Left panel) Note that we usually detected only one Dsh1-GFP dot in the nucleus because signals of the single GFP tag are weaker than those of the 13myc tag. A percentage of the cells showing the colocalized signal is indicated below each panel. (C) Coimmunoprecipitation assay for Rdp1-5×Flag and Dcr1-3HA in the indicated strains. Immunoprecipitates prepared using an anti-Flag antibody were analyzed by Western blotting with antibodies against Flag or HA. (*) Nonspecific background. (D) Indirect immunofluorescence analysis visualizing the colocalization of Rdp1-13myc and GFP-Dcr1 using antibodies against myc and GFP. DNA was also stained with DAPI. White arrows show colocalization of Dcr1 and Rdp1 in D. (E) Quantification of the cells showing colocalization of Rdp1-13myc and GFP-Dcr1 in D. The ratio of the cells with the colocalized signals to the cells showing signals from both proteins was calculated. White bars: B,D, 2 μm. (F) Coimmunoprecipitation assay analyzing the Dsh1–Ago1 (left panel) and Dsh1–Tas3 (right panel) interactions. Extracts prepared from the indicated strains were incubated with an antibody against myc. Immunoprecipitated fractions were analyzed by Western blotting with antibodies against myc or Flag.
Dsh1 facilitates formation of the Dcr1–Rdp1 complex via physical interactions
One of the key steps in RNAi-mediated heterochromatin assembly is recruitment of the RITS complex to heterochromatic ncRNA, which was clearly shown by Bühler et al. (2006), who demonstrated that artificial tethering of RITS to ura4+ RNA by the λN RNA-binding protein promotes ura4-siRNA synthesis and heterochromatin formation at the ura4+ locus. We analyzed the effect of deleting dsh1 on the RITS-tethering system and found that deletion of dsh1 compromised the silencing induced by RITS tethering in the same manner as the deletion of ago1 (Supplemental Fig. S5). This suggests that Dsh1 functions downstream from RITS binding to target RNA.
To gain insight into the molecular function of Dsh1 within the RNAi machinery, we examined physical interactions between Dsh1 and other components of RNAi, such as Dcr1 or Rdp1, using coimmunoprecipitation assays (Fig. 3A; Supplemental Fig. S6). Rdp1 and Dcr1 coprecipitated with Dsh1 (Fig. 3A). Treatment of the extract with DNase I, RNase III, or RNase T1, which degrade DNA, dsRNA, and ssRNA, respectively, did not affect the interaction (Supplemental Fig. S6), indicating that the interactions were not mediated by nucleic acids. These results showed that Dsh1 physically interacted with Dcr1 and Rdp1 within the cell. The interaction between Dsh1 and Dcr1 was detected in chp1Δ or rdp1Δ cells, albeit with slightly reduced efficiency (Fig. 3A, left panel). Since siRNA synthesis is almost abolished in both of these mutants (Motamedi et al. 2004; Halic and Moazed 2010), this result indicates that the Dsh1–Dcr1 interaction does not require siRNA or integrity of the RITS complex or RDRC. In contrast, only a marginal interaction between Dsh1 and Rdp1 was detected in chp1Δ or dcr1Δ cells (Fig. 3A, right panel), suggesting that siRNA synthesis and/or the integrity of the RITS complex is required for the Dsh1–Rdp1 interaction.
We next analyzed the colocalization of Dsh1 with Dcr1 or Rdp1 using indirect immunofluorescence analysis. Rdp1 formed nuclear dots at the perinuclear region, and cytoplasmic dots were detected in some cells (Fig. 3B, bottom left panels), as previously reported (Motamedi et al. 2004). In 60% of the cells, at least one of the Rdp1 dots colocalized with the nuclear dot representing Dsh1 (Fig. 3B, left panels). It is difficult to detect signals generated by endogenous Dcr1 using immunofluorescence techniques, but peripheral nuclear signals were detected after overexpression of GFP-Dcr1 (Emmerth et al. 2010). Under these conditions, some of the nuclear dots representing Dsh1 colocalized with those representing Dcr1 in 55% of the cells (Fig. 3B, right panels).
Because a previous study indicated that Dcr1 and Rdp1 form a complex within the cell and that this interaction facilitates dsRNA generation (Colmenares et al. 2007), we hypothesized that Dsh1 functionally bridges Dcr1 and Rdp1 at the nuclear periphery. To test this, we first verified whether the interaction between Dcr1 and Rdp1 requires Dsh1 using coimmunoprecipitation assays. In wild-type cells, Dcr1 and Rdp1 formed a complex and were coprecipitated as previously reported (Colmenares et al. 2007); however, loss of dsh1 completely abolished this interaction (Fig. 3C). We also examined the Dcr1–Rdp1 interaction using indirect immunofluorescence (Fig. 3D,E) in cells overexpressing GFP-Dcr1. Both proteins formed several foci at the nuclear periphery in wild-type cells, and colocalization of Dcr1 and Rdp1 was observed (Fig. 3D [top panels], E). It was difficult to detect clear evidence of colocalization in dsh1Δ cells because the nuclear foci of Rdp1 were diminished, although Dcr1 foci remained visible at the nuclear periphery (Fig. 3D [bottom panels], E). Together, these data showed that Dsh1 facilitates stable association between Dcr1 and Rdp1 at the nuclear periphery.
We also examined the interaction between Dsh1 and the RITS complex. We found that Dsh1 was coprecipitated with components of the RITS complex, Ago1 and Tas3 (Fig. 3F), although the interaction seems to be relatively weak. We failed to detect any interactions with Chp1 under the same conditions used to detect interactions between Ago1 or Tas3 (Supplemental Fig. S7), probably because Chp1 is readily degraded in the extracts. Like the Dsh1–Rdp1 interaction, the interaction between Dsh1 and Ago1 required Dcr1 (Fig. 3F, left panel). Previous reports indicated that the RITS complex interacted with RDRC and that the interaction requires Dcr1 and Clr4 (Motamedi et al. 2004). We detected coimmunoprecipitation of Rdp1 with Chp1, a component of the RITS complex, and the coimmunoprecipitation was not observed in the dsh1Δ cells (Supplemental Fig. S8). Therefore, Dsh1 is also required for interaction between the RITS complex and RDRC.
Dsh1 and RDRC together enhance siRNA generation by Dicer
Because Dsh1 interacts with Dcr1 and is essential for siRNA generation (Figs. 2A, 3A), we examined whether Dsh1 was also essential for the endoribonuclease activity of Dcr1 in vivo by monitoring siRNA generation from long ura4+ hairpin RNA. When long hairpin ura4 RNA is generated by transcription of ura4+ sequence-inverted repeats, cells produce siRNA derived from the double-stranded portion of the ura4+ hairpin RNA (Iida et al. 2008; Simmer et al. 2010). In this system, siRNA generation depends completely on Dcr1, whereas a substantial amount of siRNA is formed in rdp1Δ cells (Fig. 4A; Iida et al. 2008; Simmer et al. 2010); this indicates that Dcr1 can process hairpin RNA to generate siRNA without Rdp1 and that Rdp1 is likely to enhance the reaction by producing dsRNA as a better substrate for Dcr1. In this system, we found that small amounts of ura4+ siRNAs were still synthesized in dsh1Δ cells, similar to that observed in rdp1Δ cells (Fig. 4A). Importantly, the same amount of siRNA was generated in the rdp1Δdsh1Δ double mutant as in each of the single mutants, showing that Rdp1 and Dsh1 function in the same pathway to enhance Dcr1-dependent siRNA synthesis. These results indicate that Dsh1 is dispensable for the enzymatic activity of Dcr1 but has an enhancing role in Dcr1-dependent siRNA generation in cooperation with Rdp1.
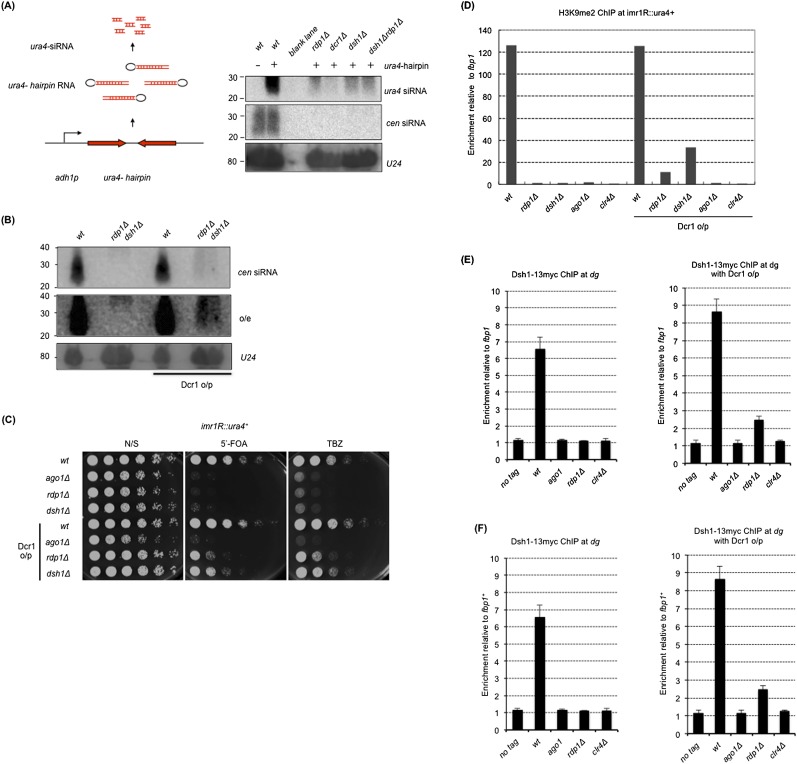
Figure 4.
Dsh1 promotes siRNA amplification by recruiting Rdp1 to heterochromatin. (A, left) Schematic representation of the construct inducing ura4+-hairpin RNA. siRNA analysis by Northern blotting with radiolabeled oligonucleotide probes against ura4+-hairpin RNA. (Right) Probes against centromeric repeats (dg and dh) and U24 RNA were used as controls. (B) siRNA analysis by Northern blotting with oligonucleotide probes against dg repeats in the indicated strains. (Middle panel) Overexposure (o/e) of the blot shown in the top panel is also shown. (Bottom panel) A probe against U24 RNA was used as an internal control. (C) Tenfold serial dilution assays examining the effect of modest Dcr1 o/p on gene silencing. Because Dcr1 o/p is toxic to cells, GFP-Dcr1 was only modestly overproduced under the control of the nmt1(3×) promoter under repressive conditions (i.e., cultured in YES medium) as previously described (Emmerth et al. 2010). Indicated strains were spotted on N/S (YES), YES+5′-FOA, and YES+TBZ plates. (D) ChIP analysis of H3K9me at imr1R∷ura4+ in the indicated strains with or without Dcr1 o/p. Cells growing exponentially in YES medium were used. Enrichment relative to fbp1+ is shown on the Y-axis. (E,F) ChIP analysis of Rdp1-13myc (E) and Dsh1-13myc (F) in the indicated strains with (right panels) or without (left panels) Dcr1 o/p. Enrichment at dg relative to fbp1+ is shown on the Y-axis.
The long hairpin RNA experiment indicates that Dcr1 can generate siRNA in vivo when dsRNA is supplied. Since both strands of the centromeric repeats are transcribed (Volpe et al. 2002; Irvine et al. 2006; Chen et al. 2008) and part of the transcripts might form hairpin structures (Djupedal et al. 2009), Dcr1 would generate siRNA from ncRNA transcribed from the centromeric repeats without the need for Rdp1. Indeed, deep sequencing analysis of siRNA indicates that small amounts of siRNA are produced in rdp1Δ cells (Djupedal et al. 2009; Halic and Moazed 2010). Considering our assumption that Rdp1 and Dsh1 together enhance synthesis of siRNA by Dcr1, we speculated that overproduction of Dcr1 (Dcr1 o/p) would rescue the defects of siRNA synthesis in rdp1Δ or dsh1Δ cells by generating siRNA from the dsRNA derived from heterochromatic ncRNA independently of Rdp1. As expected, Dcr1 o/p allowed the synthesis of a small amount of siRNA from dg repeats without rdp1+ or dsh1+, which was detected by Northern blotting (Fig. 4B). Importantly, siRNA generated independently of Rdp1 or Dsh1 functions during RNAi-dependent heterochromatin formation because Dcr1 o/p partially rescued the defects in the silencing of imr1R∷ura4+, sensitivity to TBZ (Fig. 4C), and the levels of H3K9me2 at imr1R∷ura4+ (Fig. 4D) in dsh1Δ or rdp1Δ cells. Rescue by Dcr1 o/p was not observed in ago1Δ cells (Fig. 4C,D), which indicates that partial heterochromatin formation by Dcr1 o/p depends on the RITS complex, as does normal RNAi-mediated heterochromatin formation. These results indicate that the activity of Dcr1 under normal conditions is not sufficient to induce heterochromatin formation without Rdp1 or Dsh1; however, this can be partially rescued by Dcr1 o/p. Thus, Dsh1 and Rdp1 collaborate to amplify siRNA generation by Dcr1, which results in the formation of fully active heterochromatin.
Binding of RDRC to heterochromatin requires Dsh1
We speculated that Dsh1 recruits Rdp1 to the otr repeats because Dsh1 bound to heterochromatin (Fig. 2C), interacted with Rdp1 (Fig. 3A), and was required for nuclear localization of Rdp1 (Fig. 3D). To test this, we performed ChIP analysis of Rdp1-13myc and Dsh1-13myc on heterochromatin in rdp1Δ and dsh1Δ cells either with or without Dcr1 o/p, respectively (Fig. 4E,F). Association of Rdp1 with centromeric repeats was abolished in both dsh1Δ cells and clr4Δ cells (Fig. 4E, left panel). Similarly, association of Dsh1 with the repeats was also abolished in rdp1Δ and clr4Δ cells (Fig. 4F, left panel). Note that reduced but significant amounts of H3K9me and Swi6 were retained at centromeric repeats in dsh1Δ and rdp1Δ cells (Fig. 1E; Sadaie et al. 2004), which suggests that H3K9me and/or Swi6 are not sufficient to retain Dsh1 and Rdp1 on chromatin. When Dcr1 was overproduced in each mutant, chromatin binding of Rdp1 was not detected (Fig. 4E, right panel), even in dsh1Δ cells in which siRNA production and heterochromatin formation were partially recovered, suggesting that Rdp1 requires Dsh1 for its heterochromatin binding. On the other hand, we detected significant interaction between Dsh1 and dg repeats in rdp1Δ cells when siRNA and heterochromatin were partially recovered by Dcr1 o/p (Fig. 4F, right panel). However, chromatin binding of Dsh1 was not detected in ago1Δ and clr4Δ cells (Fig. 4F, right panel) in which heterochromatin was not formed by Dcr1 o/p. These results indicate that Dsh1 could associate with heterochromatin in the absence of Rdp1 when RNAi-mediated heterochromatin was partially formed, whereas Rdp1 requires Dsh1 for chromatin association under the same conditions.
The putative transmembrane domain of Dsh1 is essential for perinuclear and heterochromatin localization as well as RNAi-dependent heterochromatin formation
We searched several databases to identify the functional domains within the Dsh1 protein. The only domain identified was a putative transmembrane domain spanning amino acids 276–295 (Fig. 5A; Supplemental Fig. S9A), which we named as the TML (transmenbrane-like) domain. The transmembrane domain is a functional domain that targets proteins to the cell membrane and is present in transmembrane proteins from many organisms. Although the numerical value representing the possibility of a transmembrane domain was not very high (Supplemental Fig. S9A), we expected that Dsh1 was a nuclear membrane protein because of its perinuclear localization (Fig. 3B).
Figure 5.
The TML domain of Dsh1 is essential for perinuclear and heterochromatin localization and function in siRNA-directed heterochromatin formation. (A) Schematic representation of the Dsh1 protein. The position of the TML domain (tml) is indicated by a gray box. A schematic representation of a tml deletion mutant is also shown. (B) Indirect immunofluorescence analysis of the localization of Dsh1-13myc and Cut11-GFP (top panels) or Lem2-GFP (bottom panels). White bars, 5 μm. (C) Indirect immunofluorescence analysis of the localization of Dsh1-13myc or Dsh1-tmlΔ-13myc. Strains expressing Dsh1-13myc (or Dsh1-tmΔ-13myc) and Lem2-GFP were used. Cells growing exponentially were stained with DAPI, an antibody against myc, and an antibody against GFP to visualize DNA, Dsh1-13myc, and Lem2-GFP, respectively. Lem2-GFP was stained to visualize the position of the nuclear periphery and spindle pole body. White bars, 5 μm. (D) ChIP analysis of Dsh1-13myc at otr1R∷ade6+ and dh in indicated strains. Enrichment relative to fbp1+ is shown on the Y-axis. The positions of the PCR products are shown in Figure 1A. (E) ChIP analysis of H3K9me2 (left) and Swi6 (right) in dsh1 mutants. The primers are the same as outlined in Figure 1E. Enrichment relative to fbp1+ is shown on the Y-axis. (F) siRNA analysis in dsh1 mutants. The probes used were the same as outlined in Figure 2A. (G) Coimmunoprecipitation assay for Rdp1-5Flag and Dcr1-3HA in dsh1 mutants. Immunoprecipitates prepared using an antibody against Flag were analyzed by Western blotting with antibodies against Flag and HA.
To analyze the nuclear membrane localization of Dsh1 in detail, we compared the localization of Dsh1 with nuclear membrane proteins Cut11 and Lem2 by immunofluorescence (Fig. 5B). Cut11 is a nuclear membrane protein whose ortholog, Ndc1, is a component of the nuclear pore complex (West et al. 1998; Brohawn et al. 2009), while Lem2 is an inner nuclear membrane protein (Hiraoka et al. 2011). We found that Dsh1-13myc localized at the nuclear periphery but rarely colocalized with the Cut11 foci and tended to localize more proximal to nucleoplasm than Cut11 (Fig. 5B, top panels), which was similar to the reported localization of modestly overexpressed GFP-Dcr1 (Emmerth et al. 2010). In contrast, one of the Lem2 foci colocalized with a Dsh1 focus (Fig. 5B, bottom panels). Since we observed one of the Dsh1 foci colocalized or closely localized with Cnp1 (Fig. 2D,E), and Lem2 was shown to localize to the spindle pole body (Hiraoka et al. 2011), the colocalization of Dsh1 and Lem2 appeared to occur at the spindle pole body, where centromeric heterochromatin clustered.
To examine the role of the TML domain, we generated a strain expressing Dsh1-13myc lacking a TML domain (Dsh1-tmlΔ-13myc) and performed indirect immunofluorescence analysis. The levels of Dsh1-tmlΔ-13myc protein were similar to those in the wild-type strain (Supplemental Fig. S9B). Dsh1-tmlΔ-13myc failed to localize to the perinuclear region and showed a diffuse pattern in the cytoplasm and nucleus (Fig. 5C). Next, to test whether Dsh1 is a membrane protein, we performed biochemical fractionation of Dsh1 using Swi6 and Lem2 as a control of a heterochromatic protein and an inner nuclear membrane protein, respectively (Supplemental Fig. S10). Both Dsh1-13myc and Dsh1-tmlΔ-13myc were insoluble proteins and were not solubilized by extraction with high salt (1 M NaCl) or with high salt and a detergent (1% triton), while the former extraction solubilized Swi6, and the latter extraction solubilized Lem2. This result suggests that Dsh1 tightly interacts with insoluble protein/structure independently on the TML domain, which made biochemical fractionation of Dsh1 difficult. ChIP analysis indicated that Dsh1-tmlΔ-13myc also failed to localize at centromeric heterochromatin (Fig. 5D). These results suggested that the TML domain is essential for the localization of Dsh1 to the nuclear periphery and to chromatin, although it is unclear at this stage whether Dsh1 is a nuclear membrane protein and the TML domain truly functions as transmembrane domain.
We next assessed the importance of the TML domain of Dsh1 for heterochromatin organization and siRNA generation. Silencing assays and ChIP analysis showed that loss of the TML domain completely abolished Dsh1 functions in terms of heterochromatin organization. dsh1-tmlΔ cells showed the same phenotype as dsh1Δ cells; i.e., loss of silencing (Supplemental Fig. S11) accompanied by a loss and a decrease of H3K9me/Swi6 from the marker gene (otr1R∷ade6+) and the pericentromeric repeats (dh), respectively (Fig. 5E). siRNA was not detected in dsh1-tmlΔ cells (Fig. 5F). Accordingly, we also found that loss of the TML domain disrupted the interaction between Dcr1 and Rdp1 (Fig. 5G). Importantly, deletion of the TML domain caused the loss of the Dsh1–Rdp1 interaction, but the Dsh1–Dcr1 interaction was still retained (Fig. 3A). These data suggest that the perinuclear localization mediated by the TML domain plays an essential role in the function of Dsh1.
Discussion
Once H3K9 methylation is induced at centromeric repeats during de novo establishment of heterochromatin, the RNAi machinery establishes the self-reinforcing loop of siRNA generation and H3K9 methylation to spread and maintain heterochromatin. The self-reinforcing loop, which includes an siRNA amplification step catalyzed by RDRC and Dcr1, is assembled on heterochromatin (Sugiyama et al. 2005). The present study identified and characterized a novel gene, dsh1+, as being essential for RNAi-mediated heterochromatin assembly in fission yeast. The results suggest that Dsh1 is a key factor for the amplification of siRNA on heterochromatin and at the nuclear periphery (Fig. 6). The role of Dsh1 in RNAi-directed heterochromatin formation and its implications for compartmentalization of the RNAi machinery at the nuclear periphery are discussed below.
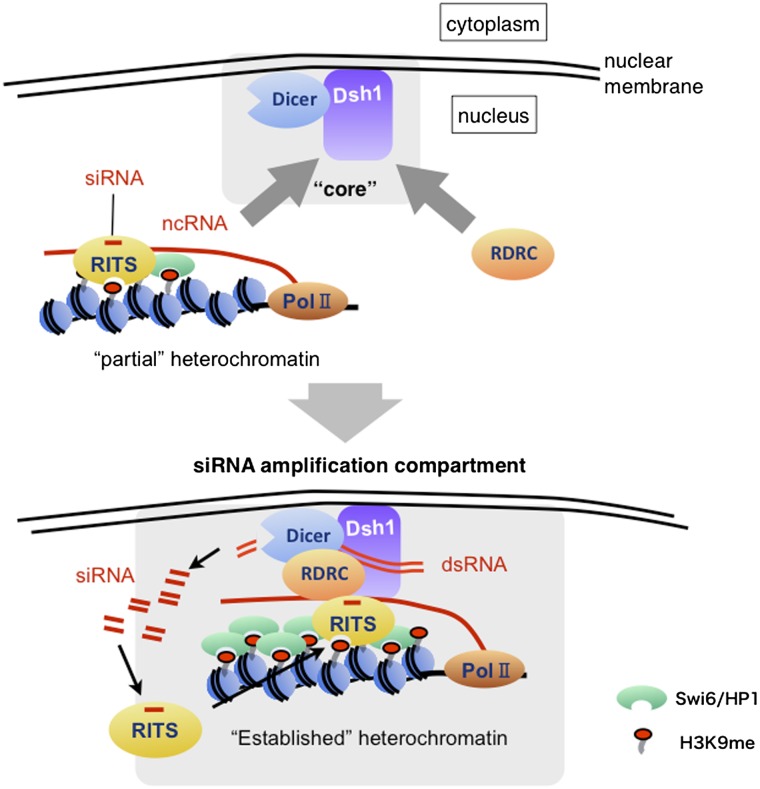
Figure 6.
Model showing the role of Dsh1 in amplifying siRNA for RNAi-directed heterochromatin formation. Dsh1 and Dcr1 localize at the nuclear periphery to form the “core” nuclear compartment for siRNA amplification. During the establishment of heterochromatin, RDRC and pre-established heterochromatin are recruited to the “core” compartment to assemble the siRNA amplification compartment, in which RNAi machinery forms a self-reinforcing loop to amplify heterochromatic siRNA. Dsh1 associates with chromatin by interacting with siRNA-containing RITS. For simplicity, some factors, including CLRC, are omitted. For details, see the Discussion section.
Dsh1 couples Dicer to RDRC for efficient siRNA production
Dsh1 forms a bridge between Dcr1 and RDRC within the cell to ensure efficient siRNA generation. This is supported by several lines of evidence. First, we showed that Dsh1 associated with Dcr1 and Rdp1 and that depletion of Dsh1 in vivo disrupted the Dcr1–RDRC interaction and siRNA generation. Second, RDRC and Dsh1 stimulated the generation of siRNA from long hairpin RNA by Dcr1. Importantly, epistasis analysis of dsh1+ and rdp1+ double mutants indicated that Dsh1 and RDRC function in the same pathway to generate siRNA. Third, Dcr1 o/p partially rescued the defects in siRNA generation and heterochromatin formation induced by depletion of Rdp1 or Dsh1, indicating that the function of Dsh1/RDRC is, at least in part, to enhance siRNA production for heterochromatin formation.
The Dcr1 o/p experiment also suggests that a substantial amount of dsRNA is formed, probably via annealing of convergent transcripts or the self-formation of hairpin-like structures, without either Dsh1 or RDRC; however, normal levels of Dcr1 result in poor utilization of the dsRNA. Hence, we suggest that Dsh1 connects Dcr1 to RDRC to supply the dsRNA produced by RDRC to Dcr1 more efficiently. Previous in vitro experiments using purified Dcr1 and RDRC indicate that coupling of dsRNA synthesis by RDRC with Dcr1 reaction does not stimulate the siRNA synthesis by Dcr1 (Colmenares et al. 2007). The absence of a stimulatory effect during the coupling reaction can be explained by the lack of Dsh1. On the other hand, Dcr1 stimulates dsRNA formation in vitro, which is catalyzed by purified RDRC (Colmenares et al. 2007), suggesting the existence of a direct interaction between Dcr1 and RDRC, although no direct interaction was detected in the absence of Dsh1 in vivo (Fig. 3C). Accordingly, we believe that Dsh1 stabilizes the interaction in vivo to enable the efficient coupling of dsRNA synthesis by RDRC and the subsequent siRNA generation by Dcr1.
Our results showed that siRNA synthesis induced by Dcr1 o/p functioned in heterochromatin formation. However, the siRNA generated by Dcr1 o/p in the absence of Dsh1/RDRC was far less abundant than that produced in wild-type cells and was not sufficient to yield fully active heterochromatin (Fig. 4B–D). One of the reasons for this incomplete heterochromatin formation may be that heterochromatin formed by Dcr1 o/p inhibits the transcription of RNA used as a substrate for Dcr1. In other words, one of the major functions of RDRC/Dsh1 is to supply sufficient amounts of dsRNA from the small amount of RNA transcribed from heterochromatin.
Dsh1 couples RDRC and Dcr1 reactions on heterochromatin
Binding of Dsh1 to centromeric repeats is completely dependent on Dcr1 and Clr4 and partially dependent on Swi6. A substantial amount of siRNA is generated and associates with RITS in swi6Δ cells, whereas siRNA is almost abolished in dcr1Δ or clr4Δ cells (Motamedi et al. 2004; Halic and Moazed 2010). In contrast, H3K9me is retained at centromeric repeats in both swi6Δ and dcr1Δ cells but is abolished in clr4Δ cells (Fig. 1E; Sadaie et al. 2004). Therefore, the amount of siRNA, rather than the level of H3K9me, seems to correlate with binding of Dsh1 to centromeric repeats. Note that in dcr1Δ cells, the RITS complex, which does not associate with siRNA, is still retained on the centromeric repeats, probably through binding of the Chp1 chromodomain to H3K9me (Fig. 2B). When siRNA was synthesized in rdp1Δ cells by Dcr1 o/p, Dsh1 associated with heterochromatin (Fig. 4E). We also found that Dsh1 interacted with the RITS complex in a Dcr1-dependent manner (Fig. 3F). Taken together, we suggest that the RITS complex associated with siRNA is the prime determinant for the association between heterochromatin and Dsh1.
Similar to Dsh1, heterochromatin binding of Rdp1 depends on Dcr1 and Clr4 and partially depends on Swi6 (Sugiyama et al. 2005), supporting the idea that Rdp1 and Dsh1 associate with heterochromatin in the same manner. We showed that RNAi-directed heterochromatin was partially formed by Dcr1 o/p in dsh1Δ and rdp1Δ cells. The RITS complex is expected to localize at the partial heterochromatin under these conditions. Dsh1 bound to the partial heterochromatin in the absence of RDRC, probably via interaction with the siRNA-containing RITS complex, but RDRC did not bind to the partial heterochromatin in the absence of Dsh1, indicating that Dsh1 is required for the association of RDRC with heterochromatin. Therefore, we propose that Dsh1 initially recruits RDRC onto heterochromatin through a physical interaction with RDRC. Alternatively, Dsh1 may recruit RDRC onto heterochromatin by promoting interaction between the RITS complex and RDRC, as Dsh1 is required for the RITS–RDRC interaction (Supplemental Fig. S8). Based on the fact that the RITS complex and RDRC associate with centromeric ncRNAs, it is suggested that the RITS–RDRC interaction may contribute to the initiation of dsRNA synthesis via stabilization of the RDRC–ncRNA interaction (Motamedi et al. 2004). Hence, Dsh1 might contribute to dsRNA synthesis by promoting the RITS–RDRC interaction. In any case, Dsh1 not only connects RDRC and Dcr1, but also connects the RNAi machinery to heterochromatin in concert with the siRNA-containing RITS complex (Fig. 6).
siRNA generation at the nuclear periphery
The formation of perinuclear foci of Dcr1 and its association with chromatin appear to be important for RNAi-directed heterochromatin formation (Emmerth et al. 2010; Woolcock et al. 2011). Dsh1 predominantly localized to the nucleus and formed foci close to the nuclear periphery. Some of the Dsh1 foci colocalized with Dcr1 foci. Thus, we propose that the Dsh1/Dcr1 foci at the nuclear periphery represent a compartment for siRNA amplification, in which the self-reinforcing loop is established on heterochromatin in combination with RDRC and the RITS complex (Fig. 6). Several lines of evidence support this proposal. First, Dsh1 foci were formed at the nuclear periphery and colocalized with centromeres in a Dcr1-dependent manner. Second, nuclear Rdp1 foci colocalized with Dcr1 and Dsh1 foci at that nuclear periphery, and the colocalization of Rdp1 and Dcr1 was dependent on Dsh1; loss of Dsh1 resulted in loss of nuclear periphery localization of Rdp1. Importantly, the Dsh1-dependent colocalization of Dcr1 and Rdp1 at the nuclear periphery corresponded with the Dsh1-dependent stimulation of siRNA synthesis by RDRC. Third, perinuclear localization of Dsh1 was lost after deletion of the TML domain, resulting in the loss of siRNA amplification and association with Rdp1. Importantly, Dsh1 was still able to interact with Dcr1 after deletion of the TML domain, which suggests that the loss of nuclear periphery localization is the main reason for the loss of siRNA amplification.
Protein–protein interaction between Dsh1 and Dcr1 was independent of RNAi, whereas those between Dsh1 and RDRC were dependent on RNAi. In addition, formation of perinuclear Dsh1 and Dcr1 foci was independent of RNAi, whereas that of Rdp1 was not. Therefore, we suspect that Dcr1/Dsh1 foci form the “core” of the siRNA amplification compartment at the nuclear periphery. During the initial stage of heterochromatin establishment, H3K9 methylation and siRNA synthesis begin at the target loci via an as-yet-unknown mechanism, and the RITS complex harboring siRNA is localized on the “partial” heterochromatin through binding to H3K9me and/or ncRNA. Then, both RDRC and the partial heterochromatin are recruited to the “core” compartment via Dsh1 to establish the self-reinforcing loop and finally establish “fully active” heterochromatin at the nuclear periphery (Fig. 6).
Dcr1 forms nuclear foci in close proximity to the nuclear pore complex (Emmerth et al. 2010). Since homologs of Dcr1 in humans and Caenorhabditis elegans localize in the nucleus and associate with the nuclear pore complex (Ando et al. 2011; Beshore et al. 2011), it is likely that a similar interaction between the nuclear pore complex and Dcr1 exists in fission yeast to form the perinuclear foci. We showed that the TML domain, which has a trans-membrane-like amino acid sequence, is essential for the perinuclear localization of Dsh1, which raises the possibility that the TML domain actually does function as a trans-membrane domain and that Dsh1 is actually a nuclear membrane protein. Indeed, Dsh1 tended to colocalize with the inner nuclear membrane Lem 2 (Fig. 3B). However, we could not address this point by biochemical fractionation because of the insolubility of the Dsh1 protein and the Dsh1-tmlΔ mutant protein. Furthermore, swapping the Dsh1 TML domain for the transmembrane domains of other nuclear membrane proteins did not result in transmembrane activity (data not shown). Further studies are necessary to understand the molecular mechanism underlying the perinuclear localization of Dsh1. Although it is uncertain whether Dsh1 is a membrane protein at this stage, our data about Dsh1, together with previous data that showed the importance of nuclear periphery localization of Dcr1 (Emmerth et al. 2010), suggested the importance of the TML domain in the compartmentalization of RNAi machinery at the nuclear periphery for heterochromatic siRNA amplification.
Woolcock et al. (2011) reported that both Dcr1 and Rdp1 associate with some euchromatic genes, ncRNA genes, and long terminal repeat retrotransposons and that both proteins might be involved in cotranscriptional gene silencing. It is possible that Dsh1 is involved in this regulatory system and that some of the Dsh1–Dcr1 foci may represent the compartment for the Dicer-dependent euchromatic regulation.
Compartmentalization appears to be important for both RNAi-based post-transcriptional gene silencing and transcriptional gene silencing in various organisms. In mammals, in which RNA-dependent polymerases do not seem to exist, Dicer and siRNA-containing effector complexes (known as RNA-induced silencing complexes [RISCs]) are coupled within a cytoplasmic granule compartment called the “P-body” to produce siRNA and silence target RNA post-transcriptionally (Jakymiw et al. 2007; Pontes and Pikaard 2008). Plants have siRNA-directed DNA methylation systems that resemble fission yeast systems (Henderson and Jacobsen 2007). Increasing evidence suggests that these RNAi factors function collaboratively within a specialized nuclear structure called the Cajal body to produce siRNA (Pontes and Pikaard 2008). Thus, Dsh1/Dcr1 nuclear foci might represent a functional nuclear compartment similar to a Cajal body. Although Dsh1 is only conserved in fission yeast (the Schizosaccharomyces family), functionally homologous proteins might exist in other systems. Further analysis of Dsh1 will provide insights into the “coupling” of RNAi-dependent regulatory systems within specialized cellular compartments.
Materials and methods
S. pombe strains, media, and genetic procedures
All of the strains used in this study and their genotypes are listed in Supplemental Table S1. The media and genetic procedures used in this study have been previously described (Moreno et al. 1991). Gene replacement for deletion or tagging was performed as previously described (Bahler et al. 1998). All integrations were confirmed by PCR. The tmlΔ mutant of dsh1+ was generated using a PCR-based method with long oligonucleotides harboring small deletions corresponding to the TML domain. The deletion of tml was confirmed by DNA sequencing. The primers used for constructing the tml deletion mutants are listed in Supplemental Table S2. In most experiments, cells were grown in S. pombe medium (YES or EMMS) at 30°C. For modest GFP-Dcr1 o/p under the control of the nmt1 (3×) promoter, cells were cultured in YES at 30°C as previously described (Emmerth et al. 2010).
Tenfold serial dilution plating assay
Cells were grown to saturation in YES medium at 30°C, and 10-fold serial dilutions were spotted onto culture plates containing various media. Plates were incubated for 2 or 3 d at 30°C before imaging. For TBZ sensitivity analysis, cells were spotted on YES+TBZ plates (YES containing 15 μg/mL TBZ). For the otr1R∷ade6+silencing assay, cells were spotted on Low Ade plates (YES lacking adenine). For the imr∷ura4+ or kint2∷ura4+ or RITS-tethered ura+ silencing assays, cells were spotted on −Ura plates (EMMS lacking uracil) and on YES/EMMS +5′-FOA plates (YES or EMMS containing 1 mg/mL 5′-FOA).
Indirect immunofluorescence analysis
Indirect immunofluorescence analysis was performed essentially as previously described (Dohke et al. 2008), except that cells were incubated with primary antibodies for 3.5 h at room temperature and with secondary antibodies for 12 h at 4°C. Anti-TAT1, anti-Cnp1, anti-myc (4A6, Millipore), and anti-GFP (A6455, Invitrogen) were used as primary antibodies. Antibodies against mouse IgG conjugated to Alexa 546 (Invitrogen) and against rabbit IgG conjugated to Alexa 488 (Invitrogen) were used as secondary antibodies.
Other methods
Details of other methods used in this study, including RT–PCR, siRNA analysis, coimmunoprecipitation assay, ChIP assay, and biochemical fractionation, are described in the Supplemental Material.
Acknowledgments
We thank T. Iida, Y. Chikashige, Y. Hiraoka, M. Buhler, D. Moazed, and R. Allshire for providing the fission yeast strains; T. Urano for the H3K9me antibody; and D.B Goto for technical advice regarding small RNA analysis. We thank K. Ishii for discussion and for providing materials for experiments. We also thank our laboratory members for helpful discussion and support. Y.M. was supported by a Grant-in-Aid for Scientific Research (A) from the Japan Society for the Promotion of Science and by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. K.K. is a Research Fellow of the Japan Society for the Promotion of Science.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.190272.112.
References
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev 9: 218–233 [DOI] [PubMed] [Google Scholar]
- Ando Y, Tomaru Y, Morinaga A, Burroughs AM, Kawaji H, Kubosaki A, Kimura R, Tagata M, Ino Y, Hirano H, et al. 2011. Nuclear pore complex protein mediated nuclear localization of dicer protein in human cells. PLoS ONE 6: e23385 doi: 10.1371/journal.pone.0023385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, Pringle JR 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Bayne EH, White SA, Kagansky A, Bijos DA, Sanchez-Pulido L, Hoe KL, Kim DU, Park HO, Ponting CP, Rappsilber J, et al. 2010. Stc1: A critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell 140: 666–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294: 2539–2542 [DOI] [PubMed] [Google Scholar]
- Beshore EL, McEwen TJ, Jud MC, Marshall JK, Schisa JA, Bennett KL 2011. C. elegans Dicer interacts with the P-granule component GLH-1 and both regulate germline RNPs. Dev Biol 350: 370–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Partridge JR, Whittle JR, Schwartz TU 2009. The nuclear pore complex has entered the atomic age. Structure 17: 1156–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler M, Verdel A, Moazed D 2006. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell 125: 873–886 [DOI] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI 2005. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819 [DOI] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI 2008. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451: 734–737 [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Kinoshita N, Nakaseko Y, Matsumoto T, Murakami S, Niwa O, Yanagida M 1989. Composite motifs and repeat symmetry in S. pombe centromeres: Direct analysis by integration of NotI restriction sites. Cell 57: 739–751 [DOI] [PubMed] [Google Scholar]
- Colmenares SU, Buker SM, Buhler M, Dlakić M, Moazed D 2007. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell 27: 449–461 [DOI] [PubMed] [Google Scholar]
- Djupedal I, Portoso M, Spahr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K 2005. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev 19: 2301–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djupedal I, Kos-Braun IC, Mosher RA, Soderholm N, Simmer F, Hardcastle TJ, Fender A, Heidrich N, Kagansky A, Bayne E, et al. 2009. Analysis of small RNA in fission yeast; centromeric siRNAs are potentially generated through a structured RNA. EMBO J 28: 3832–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohke K, Miyazaki S, Tanaka K, Urano T, Grewal SI, Murakami Y 2008. Fission yeast chromatin assembly factor 1 assists in the replication-coupled maintenance of heterochromatin. Genes Cells 13: 1027–1043 [DOI] [PubMed] [Google Scholar]
- Ekwall K, Nimmo ER, Javerzat JP, Borgstrom B, Egel R, Cranston G, Allshire R 1996. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J Cell Sci 109: 2637–2648 [DOI] [PubMed] [Google Scholar]
- Ekwall K, Cranston G, Allshire RC 1999. Fission yeast mutants that alleviate transcriptional silencing in centromeric flanking repeats and disrupt chromosome segregation. Genetics 153: 1153–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerth S, Schober H, Gaidatzis D, Roloff T, Jacobeit K, Buhler M 2010. Nuclear retention of fission yeast dicer is a prerequisite for RNAi-mediated heterochromatin assembly. Dev Cell 18: 102–113 [DOI] [PubMed] [Google Scholar]
- Grewal SI, Klar AJ 1997. A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics 146: 1221–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Moazed D 2010. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell 140: 504–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE 2007. Epigenetic inheritance in plants. Nature 447: 418–424 [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Maekawa H, Asakawa H, Chikashige Y, Kojidani T, Osakada H, Matsuda A, Haraguchi T 2011. Inner nuclear membrane protein Ima1 is dispensable for intranuclear positioning of centromeres. Genes Cells 16: 1000–1011 [DOI] [PubMed] [Google Scholar]
- Iida T, Nakayama J, Moazed D 2008. siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription. Mol Cell 31: 178–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine DV, Zaratiegui M, Tolia NH, Goto DB, Chitwood DH, Vaughn MW, Joshua-Tor L, Martienssen RA 2006. Argonaute slicing is required for heterochromatic silencing and spreading. Science 313: 1134–1137 [DOI] [PubMed] [Google Scholar]
- Jakymiw A, Pauley KM, Li S, Ikeda K, Lian S, Eystathioy T, Satoh M, Fritzler MJ, Chan EK 2007. The role of GW/P-bodies in RNA processing and silencing. J Cell Sci 120: 1317–1323 [DOI] [PubMed] [Google Scholar]
- Jia S, Noma K, Grewal SI 2004. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304: 1971–1976 [DOI] [PubMed] [Google Scholar]
- Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y 2005. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science 309: 467–469 [DOI] [PubMed] [Google Scholar]
- Mandell JG, Bahler J, Volpe TA, Martienssen RA, Cech TR 2005. Global expression changes resulting from loss of telomeric DNA in fission yeast. Genome Biol 6: R1 doi: 10.1186/gb-2004-6-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D 2004. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802 [DOI] [PubMed] [Google Scholar]
- Nakama M, Kawakami K, Kajitani T, Urano T, Murakami Y 2012. DNA–RNA hybrid formation mediates RNAi-directed heterochromatin formation. Genes Cells 17: 218–233 [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113 [DOI] [PubMed] [Google Scholar]
- Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SI 2004. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet 36: 1174–1180 [DOI] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y 2002. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol 4: 89–93 [DOI] [PubMed] [Google Scholar]
- Pontes O, Pikaard CS 2008. siRNA and miRNA processing: New functions for Cajal bodies. Curr Opin Genet Dev 18: 197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599 [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Bartel DP 2002. Small RNAs correspond to centromere heterochromatic repeats. Science 297: 1831. [DOI] [PubMed] [Google Scholar]
- Sadaie M, Iida T, Urano T, Nakayama J 2004. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J 23: 3825–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Buscaino A, Kos-Braun IC, Kagansky A, Boukaba A, Urano T, Kerr AR, Allshire RC 2010. Hairpin RNA induces secondary small interfering RNA synthesis and silencing in trans in fission yeast. EMBO Rep 11: 112–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI 2005. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci 102: 152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, Meister P, Gasser SM 2009. The nuclear envelope—a scaffold for silencing? Curr Opin Genet Dev 19: 180–186 [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837 [DOI] [PubMed] [Google Scholar]
- West RR, Vaisberg EV, Ding R, Nurse P, McIntosh JR 1998. cut11+: A gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in Schizosaccharomyces pombe. Mol Biol Cell 9: 2839–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolcock KJ, Gaidatzis D, Punga T, Buhler M 2011. Dicer associates with chromatin to repress genome activity in Schizosaccharomyces pombe. Nat Struct Mol Biol 18: 94–99 [DOI] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SI 2008. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol 15: 381–388 [DOI] [PubMed] [Google Scholar]