The DNA glycosylase DEMETER is thought to activate maternally expressed imprinted genes in Arabidopsis by removing DNA methylation established by the DNA methyltransferase MET1. Here, Grossniklaus and colleagues challenge this model by identifying an upstream imprinting control region (ICR) upstream that activates MEA expression independently of DNA methylation. This study provides the first example of an ICR without DNA methylation, suggesting that factors other than DME and MET1 are required for imprinting at the MEA locus.
Keywords: Arabidopsis, DEMETER, DNA methylation, genomic imprinting, MEDEA, imprinting control region
Abstract
Genomic imprinting is exclusive to mammals and seed plants and refers to parent-of-origin-dependent, differential transcription. As previously shown in mammals, studies in Arabidopsis have implicated DNA methylation as an important hallmark of imprinting. The current model suggests that maternally expressed imprinted genes, such as MEDEA (MEA), are activated by the DNA glycosylase DEMETER (DME), which removes DNA methylation established by the DNA methyltransferase MET1. We report the systematic functional dissection of the MEA cis-regulatory region, resulting in the identification of a 200-bp fragment that is necessary and sufficient to mediate MEA activation and imprinted expression, thus containing the imprinting control region (ICR). Notably, imprinted MEA expression mediated by this ICR is independent of DME and MET1, consistent with the lack of any significant DNA methylation in this region. This is the first example of an ICR without differential DNA methylation, suggesting that factors other than DME and MET1 are required for imprinting at the MEA locus.
Genomic imprinting is a form of epigenetic gene regulation, which leads to the differential expression of an allele according to its parent of origin. Its discovery dates back to 1970, when Kermicle (1970) described the maternal effect of the R gene, which controls maize kernel coloration. Later, an analogous phenomenon was identified in mice when pronuclear transplantation experiments revealed that both maternal and paternal genomes were required to achieve normal development (McGrath and Solter 1984; Surani et al. 1984). Imprinted genes encode for diverse proteins that function in growth and cellular proliferation, typically in extraembryonic tissues involved in nourishing the newly developing organism; i.e., the placenta in mammals and the endosperm in plants (Grossniklaus 2005; Feil and Berger 2007). The endosperm results from double fertilization in angiosperms: While one sperm cell fertilizes the egg cell, giving rise to the embryo, the second sperm cell fuses with the central cell, leading to the development of the endosperm (Maheshwari 1950). Genomic imprinting in mammals and seed plants evolved independently, but likely in response to similar selective pressures that maintain a fine balance between competing interests of the maternal and paternal genomes in resource allocation (Haig and Westoby 1989; Moore and Reik 1996; Messing and Grossniklaus 1999).
Although some imprinted plant genes are also expressed in the embryo, most show preferential expression in the triploid endosperm, and some of them are essential for seed development (for review, see Raissig et al. 2011). MEDEA (MEA) and FERTILIZATION-INDEPENDENT SEED2 (FIS2) are maternally expressed genes encoding evolutionary conserved Polycomb group (PcG) proteins (Grossniklaus et al. 1998; Luo et al. 1999). Plant PcG proteins form several variants of multiprotein complexes that maintain a silenced state of gene expression over many cell divisions through histone modifications (Pien and Grossniklaus 2007). The MEA–FIE (FERTILIZATION-INDEPENDENT ENDOSPERM) complex, which regulates cell proliferation in the endosperm and embryo, contains the PcG proteins MEA, FIS2, FIE, and MULTICOPY SUPPRESSOR OF IRA 1 (MSI1) (Ohad et al. 1999; Luo et al. 2000; Spillane et al. 2000; Köhler et al. 2003a). Mutations in any of these FIS class genes (mea, fie, fis2, and msi1) lead to maternal-effect seed abortion (for review, see Grossniklaus et al. 2001), which, in the case of MEA and FIS2, is due to their maternal-specific expression (Kinoshita et al. 1999; Vielle-Calzada et al. 1999; Jullien et al. 2006b). To date, PHERES1 (PHE1), which is directly regulated by MEA, represents the only well-studied paternally expressed imprinted gene in plants (Köhler et al. 2003b, 2005). While MEA and FIS2 are required for normal seed development (Grossniklaus et al. 1998; Luo et al. 1999), and PHE1 plays a role in seed abortion in hybrids (Josefsson et al. 2006), two other maternally expressed genes that were reported to be imprinted, FLOWERING WAGENINGEN (FWA) and AGAMOUS-LIKE 36 (AGL36), are not essential for seed development (Kinoshita et al. 2004; Shirzadi et al. 2011). Recently, several studies using allele-specific RNA profiling of the seed transcriptome describe many novel candidate imprinted genes in Arabidopsis (Gehring et al. 2011; Hsieh et al. 2011; Wolff et al. 2011), rice (Luo et al. 2011), and maize (Waters et al. 2011; Zhang et al. 2011). Yet, little is known concerning their role during seed development or their allele-specific regulation.
In contrast, the molecular mechanism underlying the maternal monoallelic expression of MEA, FIS2, and FWA, which results from genomic imprinting (for review, see Grossniklaus 2005), has been studied in some detail. Imprinting of all three loci results from a combination of maternal allele activation and paternal allele silencing. DNA and histone methylation function as epigenetic marks to distinguish maternal and paternal alleles, with DNA methylation playing a critical role in the regulation of all three loci (Vielle-Calzada et al. 1999; Luo et al. 2000; Kinoshita et al. 2004; Jullien et al. 2006b). The current model for imprinting control of FIS2 and FWA involves repressive DNA methylation of both parental alleles by the maintenance DNA methyltransferase MET1 throughout vegetative development. The silencing of the paternal MEA allele, however, depends on repressive histone H3 Lys 27 methylation (H3K27me) mediated by a vegetatively acting PcG complex (Jullien et al. 2006a). During male gametogenesis, paternal allele silencing is maintained by MET1 for FIS2 and FWA but by the PcG protein FIE at the paternal MEA allele, since in MET1-deficient pollen, the paternal MEA allele is not derepressed (Gehring et al. 2006; Jullien et al. 2006a,b). In contrast, during female gametogenesis, the DNA glycosylase DEMETER (DME) removes maternal DNA methylation at all three loci, which results in expression of the maternal allele in the central cell and, subsequently, during seed development (Choi et al. 2002; Kinoshita et al. 2004; Gehring et al. 2006; Jullien et al. 2006b). This demethylation process also involves a histone chaperone, illustrating the interplay of DNA methylation and chromatin level regulation (Ikeda et al. 2011).
In addition to the shared regulation of imprinting at the FIS2 and FWA loci, additional mechanisms appear to operate at the MEA locus: MEA is expressed in both the embryo and endosperm, and paternal MEA allele expression has not been detected during early seed development, suggesting that it is imprinted in both fertilization products at these stages, at least in some accessions (Vielle-Calzada et al. 1999; Luo et al. 2000; Spillane et al. 2007; Raissig et al. 2011). Thus, it is currently unknown how the maternal MEA allele is activated in the embryo in the absence of DME activity, which is thought to be restricted to the central cell (Choi et al. 2002). Nevertheless, maternal MEA allele activation in the central cell by DME has been the main focus of imprinting regulation in Arabidopsis, and possible DME target regions at the MEA locus have been identified: The AtREP2 helitron, CG sites 3 kb and 500 bp upstream of the MEA coding region, and the MEA-intergenic subtelomeric repeat (ISR) (Cao and Jacobsen 2002) downstream from the MEA coding region were shown to be methylated (Xiao et al. 2003). Indeed, DME establishes allele-specific hypomethylation of the maternal MEA allele at the −500-bp region and the MEA-ISR, suggesting that these regions control MEA-imprinted expression via their methylation status (Gehring et al. 2006). However, Arabidopsis accessions lacking the MEA-ISR remain imprinted at the MEA locus (Spillane et al. 2004), and the methylation status of the −500-bp region is not only controlled by DME, but varies depending on the accession; i.e., this region is unmethylated in the Landsberg erecta (Ler) accession, despite MEA being imprinted in Ler (Spillane et al. 2004; Gehring et al. 2006; Schoft et al. 2011). Taken together, this challenges DME as the regulator of imprinted MEA expression and raises the question of the actual cis-regulatory element for MEA imprinting.
Here we report on a minimal 200-base-pair (bp) fragment from the MEA cis-regulatory region that faithfully recapitulates MEA-like expression and functionally complements the mea mutation. Hence, it contains all of the necessary elements for transcriptional activation and imprinting control. We show that activation by DME is not mediated by this 200-bp fragment, thereby uncoupling maternal activation by DME from the imprinting control region (ICR). Genetic analysis of seed abortion indicated that DME and MET1 are only indirectly involved in MEA imprinting regulation. Maternally, dme-induced seed abortion could not be rescued by a functional MEA transgene; paternally, rescue of mea-induced seed abortion by met1 mutant pollen was not linked to a functional paternal MEA allele. As suggested previously (Gehring et al. 2006), allele-specific expression analysis showed that paternal MEA silencing is independent of MET1, consistent with the lack of significant methylation in the MEA-ICR. We propose a new model of MEA imprinting, in which DME and MET1 affect higher-order chromatin structure through targeting of transposon-related sequences but are not directly involved in the regulation of MEA imprinting.
Results
Cis-activating regions and ICRs reside in the 200-bp MEA promoter
In order to identify the minimal cis-regulatory region for imprinted MEA expression, we undertook a systematic deletion analysis of the MEA cis-regulatory sequences (Fig. 1A). The −4-kb MEA upstream sequence, which was shown to confer imprinted expression (Spillane et al. 2004), contains the previously unidentified gene At1g02570. It was recently annotated based on expressed sequence tags found in transcription profiling studies and encodes a protein of unknown function (Schmid et al. 2003; Castelli et al. 2004). As it resides between regions implicated in MEA regulation, we analyzed expression of At1g02570 by RT–PCR before and after fertilization. We found no expression during early seed development when MEA is expressed (Supplemental Fig. S1), suggesting that this gene does not share regulatory cis-elements with MEA.
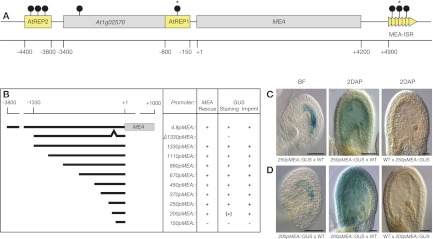
Figure 1.
MEA promoter dissection. (A) The MEA locus contains two helitron transposons, AtREP2 and AtREP1, 5′ of the translational start site and a tandem repeat region, termed MEA-ISR, 3′of the gene. At1g02570 resides in the formerly designated MEA promoter (see also Supplemental Fig. S1). Numbers are relative to the translational start site. (Gray boxes) Genes; (yellow boxes) transposons and repeats; (arrowheads) 182-bp direct repeats; (lollipops) sites of DNA methylation as reported (Xiao et al. 2003; Gehring et al. 2006); (stars) hypomethylation of maternal MEA endosperm alleles at 7–9 d after pollination (DAP). (B) The 4.8pMEA∷MEA transgene contains 3.8 kb of MEA upstream sequence fused to MEA cDNA and was shown to complement the mea-induced seed abortion phenotype (Makarevich et al. 2006). The 4.8pMEA∷GUS transgene was previously described (Spillane et al. 2004) and contains 3.8 kb of MEA upstream sequence plus 1 kb of MEA coding region. The other transgenes consist of 1330-bp to 150-bp MEA promoter sequence fused to MEA genomic DNA (pMEA∷MEA) or the bacterial uidA reporter gene (pMEA∷GUS). In the Δ1330pMEA∷GUS transgene, the region between the −200-bp and −150-bp MEA upstream sequence is deleted. Plus signs [+] indicate positively tested for rescue, staining, or imprinting; minus signs [−] indicate negatively tested for rescue, staining, or imprinting; the plus sign in parenthesis [(+)] indicates deviation from MEA-like GUS staining; and empty fields indicate that the corresponding promoter fusion was not tested. (C,D) Expression of a 250pMEA∷GUS transgene (C) and a 200pMEA∷GUS transgene (D). The transgenes were reciprocally crossed to Ler wild-type plants. Maternal GUS activity is detected with both transgenes before fertilization (BF) and 2 DAP. No paternal GUS activity is detected. For detailed GUS expression analysis, see Supplemental Figure S2. Bar, 50 μm.
Using the previously described 4.8pMEA∷GUS reporter construct, which comprises 3.8 kb of upstream and 1 kb of coding region of the MEA gene (Spillane et al. 2004), successive 5′ deletions were introduced, leading to fragment lengths ranging from 1330 bp to 150 bp of MEA cis-regulatory sequence. We constructed transcriptional fusions to the Escherichia coli uidA gene (pMEA∷GUS), encoding β-glucuronidase (GUS), and to MEA genomic DNA (pMEA∷MEA) for expression and functional analyses, respectively (Fig. 1B). Several independent primary transformants for each transgene were recovered and scored for MEA-like expression (Supplemental Tables S1, S2). Only transgenic lines containing a single copy of the insertion, as determined by Southern blot analysis, were chosen for experiments investigating MEA imprinting regulation (data not shown).
We studied maternal GUS expression from before fertilization until 4 d after pollination (DAP), corresponding to the globular stage of embryo development (Fig. 1C; Supplemental Fig. S2A,C,D). The plant line harboring the 4.8pMEA∷GUS transgene was used as a reference, its GUS-staining pattern reflecting MEA expression (Supplemental Fig. S2A). pMEA∷GUS transgenes with 1330 bp to 250 bp of MEA cis-regulatory sequences resulted in GUS-staining patterns that were indistinguishable from the MEA-like reference pattern: GUS activity was first detected in gynoecia before fertilization, with the entire embryo sac displaying a strong blue staining. After fertilization, GUS activity was found in the embryo, in the free nuclei in the peripheral endosperm, and at the chalazal cyst region of the endosperm. At 4 DAP, weak GUS activity was detected in globular stage embryos and at the chalazal pole in the endosperm. Therefore, the minimum element necessary to confer MEA-like expression resides in the 250-bp MEA upstream sequence.
Reducing the fragment length further to only 200 bp of the upstream sequence resulted in a slightly different GUS-staining pattern, which extended into the surrounding sporophytic endothelium (Fig. 1D; Supplemental Fig. S2F,G). The altered expression observed with the 200pMEA∷GUS indicates that a sporophytic repressor-binding site is located between −250 and −200 bp. We observed no GUS activity in plants with the 150pMEA∷GUS transgene and therefore proposed that the 50-bp fragment, which extends from −200 bp to −150 bp, is required for cis-activation of MEA expression. Indeed, deletion of this 50 bp in the context of the 1330pMEA∷GUS transgene resulted in a loss of expression in all independent primary transformants analyzed (Fig. 1B; Supplemental Table S1). The 50-bp fragment alone did not result in any detectable expression when fused to a min35S∷GUS transgene (data not shown), indicating that this fragment is necessary but not sufficient for cis-activation of MEA.
To test for potential loss of imprinting of the reporter transgenes, we reciprocally crossed plants containing the different pMEA∷GUS transgenes and looked for possible paternal pMEA∷GUS expression. All reporter transgenes showing MEA-like expression were active only when inherited from the mother (Fig. 1C,D; Supplemental Fig. S2A,C,D,F,G), whereas paternally inherited transgenes were silent (Fig. 1C,D; Supplemental Fig. S2B,E,H). Thus, a cis-regulatory fragment as short as 200 bp is able to confer imprinted expression to a GUS reporter gene, suggesting the presence of an ICR within this fragment.
The 200-bp fragment mediates functional MEA expression rescuing seed abortion
In order to functionally test the pMEA fragments, we investigated seed abortion in mea/MEA plants transformed with pMEA∷MEA transgenes. Heterozygous mea/MEA mutant plants show 50% seed abortion, and all seeds carrying a maternally inherited mea mutation abort irrespective of the paternal contribution (Grossniklaus et al. 1998). We scored seed abortion in transgenic mea/MEA plants to look for complementation of the mea-induced 50% seed abortion phenotype. In all primary transformants except the ones carrying the 150pMEA∷MEA transgene, we found rescue of the mea mutant phenotype illustrated by reduced seed abortion frequencies (Supplemental Table S2). Thus, the 200-bp cis-regulatory fragment is necessary and sufficient for functional expression of pMEA∷MEA transgenes, recapitulating the results with the pMEA∷GUS transgenes at the functional level.
Taken together, our systematic analysis has uncovered a 200-bp minimal fragment of the MEA cis-regulatory region that contains the elements necessary and sufficient for transcriptional activation and imprinting control. An additional element between −250 bp and −200 bp is needed to repress sporophytic expression in the ovule. Thus, we used the 250pMEA∷GUS transgene, reflecting MEA-like expression, to investigate MEA imprinting control in combination with allele-specific expression analyses of the endogenous MEA locus.
MEA-ICR sequence elements are found upstream of or downstream from other potentially imprinted loci
We investigated whether sequence elements from the MEA-ICR were also present at other potentially imprinted loci. To this aim, we performed a WU-BLAST analysis (http://www.arabidopsis.org/wublast/index2.jsp) of the entire 250pMEA promoter sequence and of the promoter sequence required for proper MEA-like expression (100-bp element between −250 and −150 from the MEA start codon) against 3 kb of upstream and downstream sequences of all TAIR10 loci (http://www.arabidopsis.org/wublast/index2.jsp). We then compared the output (684 loci) with all potentially imprinted genes that were recently reported (Gehring et al. 2011; Hsieh et al. 2011; McKeown et al. 2011; Wolff et al. 2011). Interestingly, we found that 15 of these recently published imprinted candidate genes do have conserved sequences upstream of or downstream from the respective gene, suggesting that some MEA-ICR sequence elements might be conserved between genes regulated by genomic imprinting (see Supplemental Table S3). A permutation test using 1000 randomized gene samples (n = 684) showed that the probability of finding >14 of the recently described imprinted candidate genes by chance is only P = 0.051.
In addition, we performed a motif analysis of the MEA-ICR and the putative regulatory sequences of the six imprinted candidate genes with the highest similarity scores (i.e., the smallest P-values) using the PLACE database (Higo et al. 1999). Interestingly, we found that GT1-binding sites and DOF-binding elements, both of which are abundant in the MEA-ICR (nine and five sites, respectively), were also present in the putative regulatory sequences of all six imprinted candidate genes analyzed (Supplemental Table S4). Surprisingly, a pollen-associated binding element, which we speculate might be involved in recruiting repressors to the paternal allele in the male gametophyte, was also found in all of these sequences. An overview of the identified motifs, including other expected cis-regulatory elements such as TATABOX5, GATABOX, and a poly-A signal box, is shown in Supplemental Table S4. However, none of these six candidate imprinted genes was analyzed for regulation by MET1 or DME, such that we have no information on their dependence on DNA methylation. Expression of three of the candidates was analyzed in a fie mutant background (At3g19160, At2g18880, and At4g29650) (Wolff et al. 2011), but disruption of PRC2 (Polycomb-repressive complex 2) had no effect on their expression.
Taken together, these bioinformatic analyses showed that some sequence elements of the MEA-ICR are conserved in putative regulatory sequences of other imprinted loci. Yet these motifs constitute only a small part of the conserved region, as most of the similarity is based on the high A+T content of the MEA-ICR (70%). Nevertheless, the imprinted candidate genes with the highest similarity do share common motifs, such as GT1-binding sites and DOF-binding elements, possibly reflecting conserved regulatory mechanisms.
The MEA-ICR mediates activation of maternal MEA expression independent of DME
Allele-specific demethylation of the maternal MEA allele by DME in the central cell was proposed to selectively activate the maternal MEA allele, whereas the paternal MEA allele remains silenced (Gehring et al. 2006). However, the 250pMEA∷GUS transgenes are maternally active and paternally silent even though they lack the −500-bp region targeted by DME-dependent demethylation. To elucidate the impact of DME on MEA-imprinted expression, we analyzed the maternal activity of two pMEA∷GUS transgenes in the dme-4 mutant background (Guitton et al. 2004). We crossed plants homozygous for a single locus of either the 4.8pMEA∷GUS or 250pMEA∷GUS transgene to dme-4/DME plants and analyzed the progeny for maternal GUS activity. All F1 plants are hemizygous for the pMEA∷GUS transgene, and half of them are dme-4/DME or DME/DME, respectively. F1 plants segregating the dme-4 mutation were emasculated and analyzed for their GUS-staining pattern before fertilization.
In DME wild-type plants hemizygous for either 250pMEA∷GUS or 4.8pMEA∷GUS, we observed 50% and 47% GUS staining in unfertilized ovules, respectively, consistent with Mendelian inheritance of the pMEA∷GUS transgenes by one-half of the female gametophytes (Fig. 2A,B). In plants hemizygous for the pMEA∷GUS transgene and heterozygous dme-4/DME, one-fourth of the ovules are predicted to inherit both the wild-type DME allele and the pMEA∷GUS transgene, whereas one-fourth will inherit the mutant dme-4 allele along with the pMEA∷GUS transgene. If DME is a direct activator of maternal MEA allele expression, we would expect to see only 25% GUS-staining ovules in dme-4/DME plants. Indeed, we found a significant reduction (P = 0.0003) from 47% to 34% GUS-staining ovules in dme-4/DME plants with the 4.8pMEA∷GUS transgene (Fig. 2A,B), suggesting that the 4.8pMEA∷GUS transgene was partly subject to DME-dependent repression. In plants hemizygous for 250pMEA∷GUS and dme-4/DME, we obtained 46% GUS-staining ovules (Fig. 2A,B). This is not significantly different (P = 0.9667) from the 50% GUS staining found in the DME wild-type background, suggesting that all of the ovules inheriting the dme-4 mutation expressed 250pMEA∷GUS.
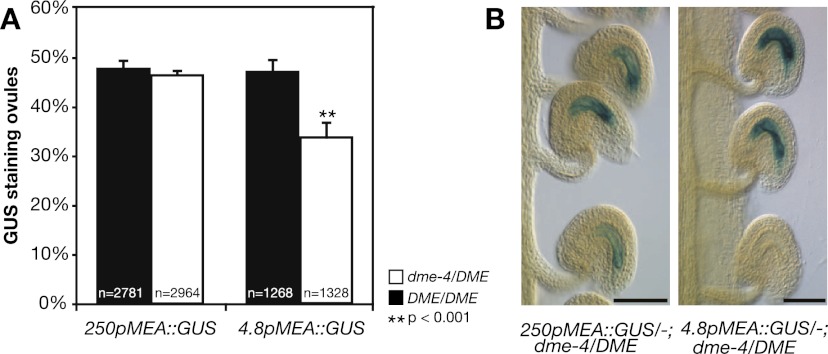
Figure 2.
Maternal MEA activation by DME. (A) Percentage of ovules expressing the 250pMEA∷GUS and 4.8pMEA∷GUS reporter transgenes in DME/DME and dme-4/DME plants before fertilization. At least four independent DME/DME and four independent dme-4/DME segregants were analyzed for each transgene. Error bars indicate SEM. (n) Total number of ovules analyzed for each genotype; (p) level of significance relative to the difference between the two segregants (t-test). (B) Maternal pMEA∷GUS expression of unfertilized ovules in dme-4/DME mutant background. Bar, 50 μm.
Thus, DME activation of maternal transgene expression before fertilization is dependent on the MEA promoter length, which is likely due to the presence of the AtREP2 helitron 4 kb upstream of MEA. This is supported by a previous study, which demonstrated that 4.2pMEA∷GUS and 4.2pMEA∷GFP transgenes containing 450 bp of AtREP2 are only active when a maternal wild-type DME copy is provided (Choi et al. 2002). Our 4.8pMEA∷GUS transgene, containing 3.8 kb of MEA upstream sequence with only 100 bp of AtREP2, is partially dependent on DME activation, whereas maternal activation of the 250pMEA∷GUS transgene is completely independent of DME function. As the 250pMEA∷GUS transgene shows exclusive maternal expression, we conclude that DME is not required for imprinting control beyond the native genomic context; i.e., DME is not targeted to the MEA-ICR for activation of maternal MEA expression.
The MEA-ICR mediates paternal transgene silencing by maternal MEA
The MEA promoter analysis revealed the existence of a MEA-ICR in the 200-bp fragment. Subsequently, we could show that maternal MEA allele activation by DME is not targeted to the MEA-ICR on the maternal allele. Therefore, we sought to test whether the previously suggested mechanism for paternal MEA allele silencing, involving DNA and histone methylation (Gehring et al. 2006; Jullien et al. 2006a,b), is mediated by the MEA-ICR.
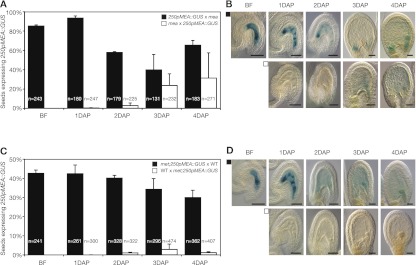
The MEA–FIE complex represses the MEA paternal allele via deposition of repressive H3K27 dimethylation (H3K27me2), which has been found in a region close to the MEA transcriptional start site (Gehring et al. 2006). We asked whether the MEA-ICR still responds to repression by the MEA–FIE complex. Therefore, we analyzed GUS expression in reciprocal crosses of plants homozygous for the 250pMEA∷GUS transgene and homozygous for mea-1. Pollination of female plants homozygous for the 250pMEA∷GUS transgene with mea-1 mutant pollen (Fig. 3A,B) resulted in the same maternal GUS-staining pattern as in females pollinated with wild-type pollen (Supplemental Fig. S2C). Although the 250pMEA∷GUS transgene is imprinted and paternally not expressed after fertilization of a wild-type ovule (Supplemental Fig. S2E), we found paternal GUS expression starting from 3 DAP in maternal mea-1 mutant plants (Fig. 3A,B). The number of seeds expressing paternal 250pMEA∷GUS in the endosperm increased during development and peaked 4 DAP, with 31% of the seeds showing paternal 250pMEA∷GUS expression. Derepression during 3–4 DAP of the paternally inherited 250pMEA∷GUS transgene in maternal mea mutant seeds suggests that the MEA-ICR mediates the repressive function of the MEA–FIE complex.
Figure 3.
Expression analysis of 250pMEA∷GUS in mea-1 and met1-3 mutants. (A) Percentage of seeds expressing 250pMEA∷GUS from before fertilization (BF) until 4 DAP. Reciprocal crosses were made between plants homozygous for 250pMEA∷GUS and mea-1/mea-1 (mea) plants. Error bars indicate SEM of two biological replicates. (n) Total number of seeds. (B) Maternal 250pMEA∷GUS expression was detected throughout seed development (top row), and weak paternal 250pMEA∷GUS expression was first detected after 3 DAP (bottom row). (C) Percentage of seeds expressing 250pMEA∷GUS from before fertilization (BF) until 4 DAP. Reciprocal crosses were made between met1-3/MET plants hemizygous for 250pMEA∷GUS (met; 250pMEA∷GUS) and Ler wild-type plants (WT). Error bars indicate SEM of two biological replicates. (n) Total number of seeds. (D) Maternal 250pMEA∷GUS expression was detected throughout seed development (top row), and no paternal 250pMEA∷GUS expression was detected (bottom row).
MET1 is not involved in paternal transgene silencing
We found that the maternal MEA protein is required for repression of the paternal 250pMEA∷GUS transgene 3–4 DAP, which is transmitted by the pollen in a transcriptionally silent state (Gehring et al. 2006). As the paternal MEA allele provided by met1 pollen showed no expression in wild-type endosperm 7 DAP, it was concluded that the methylation status of the paternal MEA allele is irrelevant for its transcriptional state (Gehring et al. 2006). However, derepression of the paternal MEA allele in a met1 mutant background might only be visible during early seed development, when the MEA–FIE complex is not yet functionally targeting the MEA locus.
We tested the impact of MET1 loss of function during male and female gametogenesis on the expression of the 250pMEA∷GUS transgene. In contrast to previous studies that did not distinguish between indirect effects of met1-3 due to global DNA hypomethylation (Saze et al. 2003; Mathieu et al. 2007) and direct effects of met1-3 due to MEA hypomethylation, we isolated met1-3/MET plants from a segregating population of wild-type plants pollinated with met1-3/MET pollen. In these plants, MET1 activity is missing only in the gametophytes, and thus pre-existing epigenetic misregulation by hypomethylation of genes other than MEA can be excluded. We used only met1-3/MET plants that showed full methylation at the 180-bp centromeric repeat (Martinez-Zapater et al. 1986) as an indication for wild-type methylation levels in those plants. We crossed wild-type pollen to females heterozygous for met1-3 and hemizygous for 250pMEA∷GUS and investigated maternal GUS activity (Fig. 3C,D). We observed GUS staining in almost all prefertilization ovules and developing seeds after fertilization inheriting the reporter construct (maximum 50%) as in wild-type females (Supplemental Fig. S2C). In contrast, when we used pollen from plants heterozygous for met1-3 and hemizygous for the 250pMEA∷GUS transgene to fertilize wild-type females, we found no (1 DAP) or only very few (2, 3, and 4 DAP) seeds with paternal GUS activity (Fig. 3C,D). Thus, a lack of MET1 activity during female or male gametogenesis has no effect on imprinted expression of the 250pMEA∷GUS transgene.
Silencing of the endogenous paternal MEA allele is controlled by maternal MEA
The MEA-ICR in the paternally inherited 250pMEA∷GUS transgene responds to repression by maternal MEA but not by MET1. In order to correlate the control of paternal transgene silencing with the control of endogenous paternal MEA allele silencing, we quantified MEA allele-specific transcripts in mea mutants and combinations of mea mutants with met1-3 mutants.
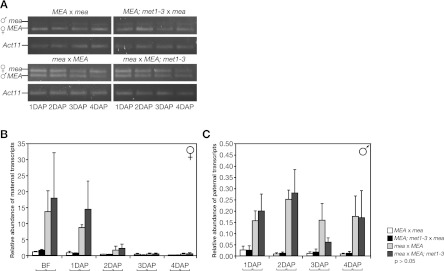
We first investigated the role of maternal MEA on paternal MEA allele silencing during early seed development. Therefore, we reciprocally crossed MEA wild-type plants with mea homozygous plants and quantified MEA allele-specific transcripts from 1–4 DAP (Fig. 4). Maternal transcripts in reciprocal crosses of MEA/MEA and mea/mea plants accumulated to their highest level before fertilization and decreased afterward (Fig. 4B). No paternal transcripts were detectable in a maternal MEA wild-type background, whereas in a maternal mea mutant background, paternal MEA allele silencing was released already at 1 DAP (Fig. 4C). Derepression of the paternal MEA allele continued until 4 DAP and resulted in more or less constant levels of paternal MEA transcripts. Remarkably, the level of derepressed paternal MEA transcripts in maternal mea/mea mutants represented only 19.5% (0.1563 of 0.8008) of the amount of maternal MEA transcripts in the maternal wild-type background (Fig. 4B,C; Supplemental Table S5). However, maternal transcription is no longer autorepressed and highly up-regulated in mea/mea mutant plants (Baroux et al. 2006), so the paternal MEA transcripts represented only 1.8% (0.1563 of 8.6833) of the amount of maternal mea transcripts in mea/mea plants (Supplemental Table S5). Thus, derepression of the paternal MEA allele in a maternal mea mutant background does not result in equivalent expression levels of the two parental alleles. The low level of derepressed paternal MEA expression indicates weak paternal MEA promoter activity, which might explain why paternal 250pMEA∷GUS expression is only detected 3–4 DAP (Fig. 3A,B). Taken together, we observed derepression of a paternally inherited 250pMEA∷GUS transgene and derepression of the endogenous paternal MEA allele in maternal mea mutants. This suggests that the MEA-ICR is the target of the MEA–FIE complex at the endogenous MEA locus.
Figure 4.
Quantification of MEA allele-specific transcription levels. (A) Allele-specific RT–PCR on RNA extracted from hand-pollinated siliques at 1–4 DAP. Reciprocal crosses were made between MEA/MEA (MEA) and mea-2/mea-2 (mea) plants, and between MEA/MEA; met1-3/MET (MEA; met1-3) and mea plants. The RT–PCR products shown are the end products after 40 cycles and show qualitatively whether there is maternal and/or paternal MEA expression but are unsuitable to infer quantitative differences. The paternal (♂) and maternal (♀) RT–PCR products are indicated. Actin 11 (Act11) was used as loading control. (B,C) Quantification of maternal (B) and paternal (C) transcripts by RT-qPCR. Transcript levels were normalized to Act11. No significant differences in transcript levels were found between crosses with and without met1-3 (braces below the X-axis indicate pairwise t-tests). Note the different scales for maternal and paternal transcripts. Error bars indicate SEM of three biological replicates.
Since a lack of maternal MEA–FIE PcG activity only leads to very weak derepression of the endogenous paternal MEA allele, we wondered whether DNA methylation might be involved in keeping it largely silenced. Thus, we asked again whether MET1 has any residual role in paternal MEA allele silencing and crossed mea/mea mother plants with either wild-type or met1-3 mutant pollen and analyzed allele-specific MEA expression levels (Fig. 4A,C). In mea/mea mutant mothers, the paternal MEA allele was derepressed when transmitted by both wild-type and met1-3 pollen, with no significant change in the level of derepression (Fig. 4C). This shows that MEA, presumably as part of the maternal MEA–FIE complex, represses the paternal MEA allele independent of its methylation status maintained by MET1 during male gametogenesis. Thus, even after removal of both known repressing factors, the maternal MEA–FIE complex and MET1, the paternal MEA allele is still expressed at extremely low levels compared with the maternal MEA allele; in other words, it is still imprinted. Furthermore, we detected no paternal MEA transcripts in the reciprocal cross when mea homozygous mutant pollen was crossed to met1-3 heterozygous females. We conclude that paternal MET1 during male gametogenesis and maternal MET1 during early seed development are not required for MEA paternal silencing and thus play no significant role in imprinting at the MEA locus.
The MEA-ICR is unmethylated
Our comparative analysis of MEA transgene and endogene regulation revealed that the MEA-ICR is not targeted by DME and MET1. Thus, contrary to what was previously suggested (Gehring et al. 2006; Jullien et al. 2006a), MEA imprinting regulation is not primarily controlled by differential DNA methylation. Therefore, we speculated that there is either no DNA methylation at all at the MEA-ICR or no differential DNA methylation between active and silent MEA alleles.
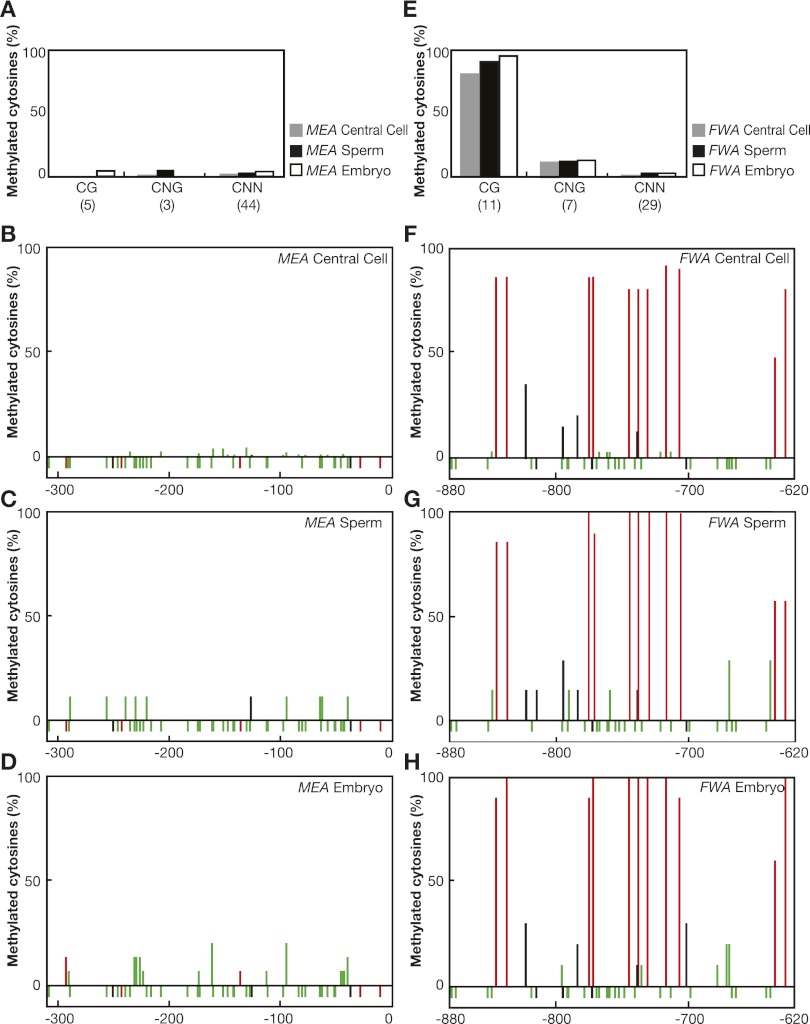
We analyzed MEA promoter methylation in isolated central cells and sperm cells as well as in isolated two-cell stage embryos where the maternal MEA allele is expressed (Vielle-Calzada et al. 1999; Spillane et al. 2007). In parallel, we monitored FWA promoter methylation, which exhibits imprinting control through a differentially methylated SINE-related element in its promoter (Kinoshita et al. 2004, 2007). In sperm cells, we found high levels of FWA promoter methylation in the CG context, consistent with previously reported methylation levels in pollen (Fig. 5E,G; Kinoshita et al. 2004). Surprisingly, we found only a small reduction of CG methylation in the central cell at the FWA locus, suggesting that DNA methylation is fully removed after fertilization only (Fig. 5E,F). Contrary to this, we detected almost no methylation in the 250-bp MEA promoter from sperm cells and central cells in any sequence context (Fig. 5A–C). In addition, we analyzed methylation in two-cell stage embryos early after fertilization. We detected high methylation levels of FWA in the CG contexts in the embryo (Fig. 5E,H), consistent with MET1-dependent silencing of parental FWA alleles. However, we found no methylation in the 250-bp MEA promoter in the embryo, where the maternal MEA allele is expressed (Fig. 5A,D).
Figure 5.
Promoter methylation of MEA and FWA. (A,E) Percentage of cytosine methylation at CG, CNG, and CNN sites in the MEA promoter (A) and the FWA promoter (E). DNA was isolated from central cells, sperm cells, and two-cell stage embryos; bisulfite-treated; sequenced; and analyzed. Numbers in brackets indicate the number of sites present in the investigated promoter region. (B–D,F–H) Percentage of cytosine methylation at each position is indicated with a red (CG), black (CNG), or green (CNN) bar. Unmethylated cytosines are shown below the 0% line. Numbers are relative to the translational start site and indicate the investigated promoter region.
In summary, MET1-dependent FWA silencing in sperm cells, central cells, and the embryo correlates with DNA methylation in the SINE-related repeat region of its promoter. However, the MEA-ICR in the 250-bp MEA promoter carries no DNA methylation in any reproductive cell. This confirms our finding that DME is not targeted to the MEA-ICR for maternal MEA allele activation and that MET1 is not involved in paternal MEA allele silencing. Thus, MEA is regulated differently from FIE and FWA, and presently unknown factors, together with the MEA–FIE complex, must be responsible for the imprinted expression of MEA.
Discussion
The MEA-ICR maps to a 200-bp region and displays no differential DNA methylation
In plants, the primary DNA sequences responsible for genomic imprinting remained elusive. Studies involving transgenes to identify the cis-determinants for imprinted expression in Arabidopsis and maize indicated that plants ICRs are located close to the imprinted loci (Luo et al. 2000; Kinoshita et al. 2004; Gehring et al. 2006; Gutierrez-Marcos et al. 2006; Makarevich et al. 2008). We identified the 200-bp upstream region adjacent to the MEA translational start site as the minimal sequence necessary to confer cis-activation and imprinted expression of a GUS transgene. The proximity of the MEA-ICR and the MEA locus is in contrast to mammalian ICRs, which can be located >100 kb distal from the imprinted loci (Ferguson-Smith and Surani 2001).
Mammalian ICRs are typically a few kilobases in length and exhibit parental allele-specific DNA methylation (Bartolomei 2009). However, the MEA-ICR maps to a 200-bp fragment and is essentially unmethylated, excluding DNA methylation as the epigenetic mark distinguishing maternal and paternal MEA alleles. This is in contrast to the cis-elements involved in imprinting at the FWA and PHE1 loci. Maternal-specific expression of FWA in the endosperm is due to differential methylation of a SINE-related element located in the FWA promoter (Kinoshita et al. 2007). Yet our analysis of DNA methylation in gametes shows that differential methylation at FWA is only established after fertilization. This suggests that the primary germline imprint at the FWA locus is not the DNA methylation mark itself. Imprinting of PHE1 results in preferential paternal expression in the endosperm and correlates with differential methylation of tandemly repeated motifs located 3 kb downstream from the PHE1 gene (Makarevich et al. 2008). Furthermore, differential DNA methylation between the parental alleles has been described for the maize imprinted genes ZmFie1 and ZmFie2 (Gutierrez-Marcos et al. 2006). Interestingly, ZmFie2 is unmethylated in both central cells and sperm cells prior to fertilization, and the differential methylation pattern is only established after fertilization, also indicating that the primary germline imprint is not a DNA methylation mark. In addition, several of the potentially imprinted genes recently identified by transcriptome profiling are unaffected by mutations in one or even all of the known imprinting factors (i.e., DME, MET1, and FIE) (Hsieh et al. 2011; Wolff et al. 2011), suggesting additional, yet-undiscovered imprinting regulators.
MEA is an imprinted gene that is not controlled by differential DNA methylation at the ICR. A related situation may occur in the mouse Prader-Willi/Angelman region showing a complex imprinting control involving several cis-acting elements, one of which is not differentially methylated but is required to establish parental imprints at other sites (Kaufman et al. 2009). Moreover, it was recently shown that in macaques, some ICRs that acquire a germline DNA methylation imprint in mice are not methylated in the germline and acquire a differential methylation mark only post-fertilization (A Ferguson-Smith, pers. comm.). Thus, primary imprints that do not involve germline DNA methylation appear to exist in both plants and mammals. Future studies will show whether common regulatory mechanism indeed exist between nonmethylated ICRs in mammals and plants.
Imprinting control at the MEA-ICR is independent of DME and MET1
Maternal allele expression of MEA and other maternally expressed imprinted genes depends on the removal of MET1-dependent DNA methylation (Choi et al. 2002; Kinoshita et al. 2004; Jullien et al. 2006b). Consistent with the lack of significant DNA methylation at the MEA-ICR, the imprinted 250pMEA∷GUS transgene is maternally activated independent of DME, suggesting that DME is only required in the endogenous context, probably targeting a region different from the MEA-ICR. Although involved in imprinting, DNA methylation in flowering plants primarily silences transposons and repeat elements (Henderson and Jacobsen 2007). Thus, a 590-bp AtREP2 transposon element that is located −4 kb upstream of the MEA start codon represents a likely DME target. Indeed, the previously described 4.2pMEA∷GUS transgene containing 450 bp of the AtREP2 is fully dependent on DME for activation (Choi et al. 2002), whereas the 4.8pMEA∷GUS transgene containing 3.8 kb of MEA upstream sequence with 100 bp of the AtREP2 is only partially dependent on DME (this study). Therefore, we hypothesize that DME is only indirectly involved in the activation of endogenous maternal MEA transcription by demethylation of the AtREP2.
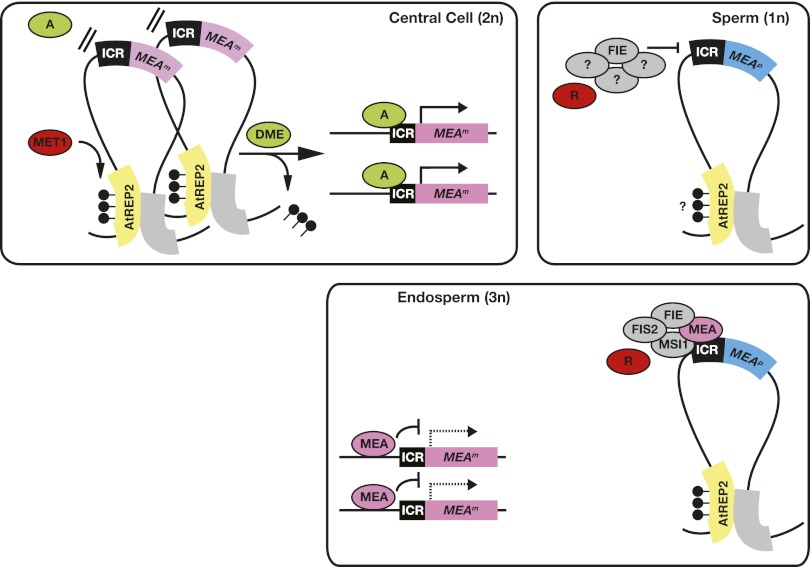
Based on our results, we propose a new model of MEA imprinting regulation (Fig. 6). The methylated AtREP2 would interact with an unidentified region of the MEA locus to establish a silent higher-order chromatin structure; e.g., a repressive chromatin loop. This prevents the MEA promoter from being accessed by an unknown transcriptional activator binding the MEA-ICR. Demethylation of AtREP2 by DME in the central cell resolves the repressive chromatin loop and allows the transcriptional activator to access the MEA-ICR. The repressive chromatin loop is not resolved in the male gametophyte, where DME is not expressed, resulting in exclusive maternal MEA allele expression. Since the paternal MEA allele is not fully activated if both known repressing activities, MET1 and the MEA–FIE complex, are removed, additional paternal repressors involved in imprinting control have to be postulated, possibly including a PcG complex with a histone methyltransferase other than MEA.
Figure 6.
Model of MEA imprinting control through a higher-order chromatin structure. Methylation at AtREP2 is maintained by MET1 in central cells and sperm cells. AtREP2 might interact with another region, thereby forming a repressive chromatin loop preventing the MEA locus from being accessed by a transcriptional activator (A). Specific expression of DME in the central cell removes methylation and resolves the repressive chromatin loop. This allows the transcriptional activator (A) to access the MEA-ICR. As a consequence, in the endosperm, the two maternal MEA alleles (MEAm) are expressed. Paternal MEA allele (MEAp) silencing is maintained by a proposed PcG complex containing FIE during male gametogenesis (Jullien et al. 2006a). After fertilization, MEAp is repressed partially by the maternal MEA–FIE complex and another paternal repressor (R). Since parental alleles in the endosperm are differentially targeted by trans-acting factors, they must have been marked in the germline, as illustrated by the purple and blue color of the maternal and paternal allele, respectively. The nature of this mark is unknown. The model explicitly shows MEAm activation in the central cell for imprinted expression in the endosperm; however, the same model is proposed for the egg cell and embryo. (Lollipops) DNA methylation; (dashed line) autorepressed MEAm transcription.
In mammals, chromosome conformation capture experiments revealed that chromosome looping is involved in imprinting control (Lopes et al. 2003; Kurukuti et al. 2006; Yoon et al. 2007; Engel et al. 2008). More specifically, interactions of differentially methylated regions (DMRs) at the mouse H19/Igf2 locus were shown to partition maternal and paternal chromatin into distinct loops, generating an epigenetic switch to control allele-specific expression (Murrell et al. 2004). Our findings raise the possibility that MEA imprinting control might depend on a similar mechanism involving higher-order chromatin structure controlled by DME and MET1.
This hypothesis is consistent with recent reports that DME is involved in genome-wide demethylation of the maternal genome in the endosperm, especially of transposons and repeat elements (Gehring et al. 2009; Hsieh et al. 2009). Intriguingly, all characterized imprinted genes in plants are hypomethylated on the maternal allele regardless of which allele is expressed. This suggests that DME-dependent demethylation in the endosperm is not specifically targeting imprinted genes, but rather is a nearly universal process that reshapes DNA methylation of the entire maternal genome in the endosperm.
The imprinting factors required for paternal MEA silencing remain unknown
Two epigenetic silencing marks were found at specific sites of the MEA locus: DNA methylation and histone H3K27 di- and trimethylation (H3K27me) (Xiao et al. 2003; Gehring et al. 2006; Jullien et al. 2006a). We report that lack of MET1 during male gametogenesis does not derepress the paternal MEA allele 1–4 DAP. This complements previous studies with met1 mutant pollen that showed no paternal MEA allele expression 7–9 DAP (Gehring et al. 2006).
Whereas DNA methylation is irrelevant for paternal MEA allele silencing, PcG-mediated histone methylation is necessary for paternal MEA allele silencing (Gehring et al. 2006; Jullien et al. 2006a). Maternal MEA is involved in deposition of repressive H3K27me at the paternal MEA allele close to the translational start site (Gehring et al. 2006). We found derepression of a paternally inherited 250pMEA∷GUS transgene in the maternal mea mutant background, suggesting that the MEA-ICR in the 250-bp MEA promoter is targeted by the maternal MEA–FIE complex. However, it is unclear how the MEA–FIE complex gains access to the silent chromatin loop of the paternal allele to maintain silencing after fertilization. Possibly, the repressive machinery, including the MEA–FIE complex and other proposed repressors, has access to cis-regulatory elements in repressive chromatin loops, whereas the activating machinery is efficiently prevented from binding to the MEA-ICR.
We found derepression of the paternal MEA allele in the maternal mea mutant background already at 1 DAP. This contradicts recent findings of delayed paternal derepression, which were explained by the need for passive loss of repressive H3K27me on the paternal MEA allele (Jullien et al. 2006a). Surprisingly, derepressed paternal MEA transcripts in maternal mea mutant plants represent only 14% of maternal MEA transcripts in maternal wild-type plants. This resembles the observed residual transcriptional activity of the silent maternal PHE1 allele (Köhler et al. 2005). Similarly, in mice, paternal alleles of several imprinted genes in the IC2-imprinted domain are not completely silent (Lewis et al. 2004). Even though the silent paternal MEA allele is derepressed in mea mutant plants, parental transcript levels are clearly not equivalent and still show parent-of-origin-dependent differences. Assuming equivalent parental expression levels in the background of compromised imprinting, the main components involved in paternal MEA allele silencing remain to be identified because the paternal MEA allele is still imprinted when MET1 and the MEA–FIE complex are missing. As the MEA-ICR confers paternal MEA silencing beyond the native genomic context, loop formation is not sufficient to explain paternal MEA silencing. Thus, another unknown repressor binding to the MEA-ICR, along with the proposed PcG complex (Jullien et al. 2006a), may be required for paternal MEA repression (Fig. 6).
In summary, our promoter dissection identified the MEA-ICR in the 200-bp MEA upstream sequence. The MEA-ICR carries no significant methylation in sperm cells, central cells, and two-cell stage embryos, which to our knowledge is the first example of an ICR without differential DNA methylation. DME, the key factor necessary for specific activation of maternally expressed imprinted genes in Arabidopsis, is dispensable for activation of maternal MEA allele transcription. Instead, DME and MET1 may be involved in the regulation of a higher-order chromatin structure at the MEA locus, thereby only indirectly controlling the specific marking and activation of the maternal MEA allele by unknown factors. However, a repressive chromatin structure at the paternal MEA locus alone cannot explain paternal MEA silencing, which is mediated through the MEA-ICR beyond the native genomic context by still unknown MEA imprinting factors.
Material and methods
Plant material
The Ler accession was used as the wild type. The mutant alleles used were mea-1, mea-2 (Ler) (Grossniklaus et al. 1998), dme-4 (C24) (Guitton et al. 2004), and met1-3 (Col) (Saze et al. 2003). The 4.8pMEA∷GUS transgenic line was described before (Spillane et al. 2004). The dme-4 (C24) and the met1-3 (Col-0) mutants were introgressed into the Ler background by crossing them at least five times as pollen parents. For genotyping assays, methylation status evaluation, and growth conditions, see the Supplemental Material.
Generation of pMEA∷MEA and pMEA∷GUS constructs
All pMEA∷MEA constructs were cloned into pCAMBIA3300 containing the corresponding MEA promoter sequence and the entire MEA ORF amplified from genomic Ler DNA. All pMEA∷GUS constructs contain the corresponding MEA promoter sequence amplified from genomic Ler DNA and were cloned in-frame to the GUS reporter gene in pCAMBIA 1381Z.
Promoter deletions were done using different primer pairs amplifying differently sized amplicons and were subsequently cloned in the above-mentioned vectors. For a detailed cloning procedure, see the Supplemental Material.
Microscopy and GUS staining
Histochemical analysis of GUS reporter gene expression was essentially done as described in Baroux et al. 2006. Microscopic inspection was carried out under differential contrast (DIC) optics using a Leica DMR microscope (Leica Microsystems). A detailed description can be found in the Supplemental Material.
RT–PCR analyses
Reverse transcription was performed as previously published (Baroux et al. 2006) on 20 gynoecia before fertilization or on 10–15 siliques at 1–4 DAP, depending on the stage indicated in the corresponding figure. In all experiments, transcript levels were normalized to the level of ACTIN11 (Huang et al. 1997). For detailed protocol and primers used, see Supplemental Material.
Bisulfite DNA sequencing of isolated reproductive cells
Central cells were isolated using laser capture microscopy, sperm cells were isolated using a Percoll density gradient (M Schauer and U Grossniklaus, unpubl.), and embryos were isolated as previously described (Autran et al. 2011). DNA isolation and bisulfite conversion were essentially performed as described in the Epigenetics Protocols Database “Bisulphite sequencing of small DNA/cell samples” (PROT35; http://www.epigenome-noe.net/research tools/protocols.php). Subsequently, regions of interest (250-bp MEA promoter and SINE-related tandem repeat in the FWA promoter) were amplified. Purified bisulfite PCR products were cloned into the pGEM-T vector (Promega) and several independent clones were sequenced (for sperm cell and embryo sample), or purified PCR products were directly sequenced with the 454 sequencer according to the standard protocol (central cell samples).
All sequences were analyzed with the BiQ Analyzer software (Bock et al. 2005) for quality control and removal of identical clones in a standardized manner. For a more detailed description, see the Supplemental Material.
Acknowledgments
We thank J. Paszkowski (University of Geneva) and C. Baroux (University of Zürich) for seeds, O. Mittelsten Scheid for the 180-bp repeat probe, M. Schauer for sharing the sperm isolation protocol, K. Kritsas for help with the permutation test, and B. Müller, S. Kessler, and S. Pien for critical comments on the manuscript. This work was supported by the University of Zürich; fellowships of the Forschungskredit der Universität Zürich (to H.J.P.W.), the Roche Research Foundation (to H.S. and M.R.), and the Deutsche Forschungsgemeinschaft (to A.S.); and grants from the Swiss National Science Foundation (to U.G.), Deutsche Forschungsgemeinschaft (to J.W.), and the EPIGENOME Network of Excellence of the European Union (to J.W. and U.G.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.195123.112.
References
- Autran D, Baroux C, Raissig MT, Lenormand T, Wittig M, Grob S, Steimer A, Barann M, Klostermeier UC, Leblanc O, et al. 2011. Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell 145: 707–719 [DOI] [PubMed] [Google Scholar]
- Baroux C, Gagliardini V, Page DR, Grossniklaus U 2006. Dynamic regulatory interactions of Polycomb group genes: MEDEA autoregulation is required for imprinted gene expression in Arabidopsis. Genes Dev 20: 1081–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei MS 2009. Genomic imprinting: Employing and avoiding epigenetic processes. Genes Dev 23: 2124–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T 2005. BiQ Analyzer: Visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics 21: 4067–4068 [DOI] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE 2002. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci 99: 16491–16498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelli V, Aury JM, Jaillon O, Wincker P, Clepet C, Menard M, Cruaud C, Quétier F, Scarpelli C, Schächter V, et al. 2004. Whole-genome sequence comparisons and ‘full-length’ cDNA sequences: A combined approach to evaluate and improve Arabidopsis genome annotation. Genome Res 14: 406–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL 2002. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110: 33–42 [DOI] [PubMed] [Google Scholar]
- Engel N, Raval AK, Thorvaldsen JL, Bartolomei SM 2008. Three-dimensional conformation at the H19/Igf2 locus supports a model of enhancer tracking. Hum Mol Genet 17: 3021–3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Berger F 2007. Convergent evolution of genomic imprinting in plants and mammals. Trends Genet 23: 192–199 [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC, Surani MA 2001. Imprinting and the epigenetic asymmetry between parental genomes. Science 293: 1086–1089 [DOI] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL 2006. DEMETER DNA glycosylase establishes MEDEA Polycomb gene self-imprinting by allele-specific demethylation. Cell 124: 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Bubb KL, Henikoff S 2009. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324: 1447–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Missirian V, Henikoff S 2011. Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS ONE 6: e23687 doi: 10.1371/journal.pone.0023687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U 2005. Genomic imprinting in plants: A predominantly maternal affair. In Annual plant reviews: Plant epigenetics (ed. P Meyer), pp. 174–200, Blackwell, Sheffield, UK. [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB 1998. Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis. Science 280: 446–450 [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Spillane C, Page DR, Kohler C 2001. Genomic imprinting and seed development: Endosperm formation with and without sex. Curr Opin Plant Biol 4: 21–27 [DOI] [PubMed] [Google Scholar]
- Guitton AE, Page DR, Chambrier P, Lionnet C, Faure JE, Grossniklaus U, Berger F 2004. Identification of new members of FERTILISATION INDEPENDENT SEED Polycomb group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131: 2971–2981 [DOI] [PubMed] [Google Scholar]
- Gutierrez-Marcos JF, Costa LM, Dal Pra M, Scholten S, Kranz E, Perez P, Dickinson HG 2006. Epigenetic asymmetry of imprinted genes in plant gametes. Nat Genet 38: 876–878 [DOI] [PubMed] [Google Scholar]
- Haig D, Westoby M 1989. Parent-specific gene expression and the triploid endosperm. Am Nat 134: 147 [Google Scholar]
- Henderson IR, Jacobsen SE 2007. Epigenetic inheritance in plants. Nature 447: 418–424 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T 1999. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D 2009. Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Shin J, Uzawa R, Silva P, Cohen S, Bauer MJ, Hashimoto M, Kirkbride RC, Harada JJ, Zilberman D, et al. 2011. Inaugural article: Regulation of imprinted gene expression in Arabidopsis endosperm. Proc Natl Acad Sci 108: 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, An YQ, McDowell JM, McKinney EC, Meagher RB 1997. The Arabidopsis ACT11 actin gene is strongly expressed in tissues of the emerging inflorescence, pollen, and developing ovules. Plant Mol Biol 33: 125–139 [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Kinoshita Y, Susaki D, Ikeda Y, Iwano M, Takayama S, Higashiyama T, Kakutani T, Kinoshita T 2011. HMG domain containing SSRP1 is required for DNA demethylation and genomic imprinting in Arabidopsis. Dev Cell 21: 589–596 [DOI] [PubMed] [Google Scholar]
- Josefsson C, Dilkes B, Comai L 2006. Parent-dependent loss of gene silencing during interspecies hybridization. Curr Biol 16: 1322–1328 [DOI] [PubMed] [Google Scholar]
- Jullien PE, Katz A, Oliva M, Ohad N, Berger F 2006a. Polycomb group complexes self-regulate imprinting of the Polycomb group gene MEDEA in Arabidopsis. Curr Biol 16: 486–492 [DOI] [PubMed] [Google Scholar]
- Jullien PE, Kinoshita T, Ohad N, Berger F 2006b. Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell 18: 1360–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman Y, Heled M, Perk J, Razin A, Shemer R 2009. Protein-binding elements establish in the oocyte the primary imprint of the Prader-Willi/Angelman syndromes domain. Proc Natl Acad Sci 106: 10242–10247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle JL 1970. Dependence of the R-mottled aleurone phenotype in maize on mode of sexual transmission. Genetics 66: 69–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Yadegari R, Harada JJ, Goldberg RB, Fischer RL 1999. Imprinting of the MEDEA Polycomb gene in the Arabidopsis endosperm. Plant Cell 11: 1945–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T 2004. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303: 521–523 [DOI] [PubMed] [Google Scholar]
- Kinoshita Y, Saze H, Kinoshita T, Miura A, Soppe WJ, Koornneef M, Kakutani T 2007. Control of FWA gene silencing in Arabidopsis thaliana by SINE-related direct repeats. Plant J 49: 38–45 [DOI] [PubMed] [Google Scholar]
- Köhler C, Hennig L, Bouveret R, Gheyselinck J, Grossniklaus U, Gruissem W 2003a. Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J 22: 4804–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U 2003b. The Polycomb group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev 17: 1540–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Page DR, Gagliardini V, Grossniklaus U 2005. The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat Genet 37: 28–30 [DOI] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R 2006. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci 103: 10684–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W 2004. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet 36: 1291–1295 [DOI] [PubMed] [Google Scholar]
- Lopes S, Lewis A, Hajkova P, Dean W, Oswald J, Forne T, Murrell A, Constância M, Bartolomei M, Walter J, et al. 2003. Epigenetic modifications in an imprinting cluster are controlled by a hierarchy of DMRs suggesting long-range chromatin interactions. Hum Mol Genet 12: 295–305 [DOI] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chaudhury AM 1999. Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci 96: 296–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A 2000. Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci 97: 10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Taylor JM, Spriggs A, Zhang H, Wu X, Russell S, Singh M, Koltunow A 2011. A Genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet 7: e1002125 doi: 10.1371/journal.pgen.1002125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari P 1950. An introduction to the embryology of angiosperms. In McGraw Hill publications in the botanical sciences (ed. EW Sinnott), McGraw Hill Book Company, New York. [Google Scholar]
- Makarevich G, Leroy O, Akinci U, Schubert D, Clarenz O, Goodrich J, Grossniklaus U, Köhler C 2006. Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep 7: 947–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevich G, Villar CB, Erilova A, Kohler C 2008. Mechanism of PHERES1 imprinting in Arabidopsis. J Cell Sci 121: 906–912 [DOI] [PubMed] [Google Scholar]
- Martinez-Zapater JM, Estelle MA, Somerville CR 1986. A highly repeated DNA sequence in Arabidopsis thaliana. Mol Gen Genet 204: 417–423 [Google Scholar]
- Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J 2007. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130: 851–862 [DOI] [PubMed] [Google Scholar]
- McGrath J, Solter D 1984. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37: 179–183 [DOI] [PubMed] [Google Scholar]
- McKeown PC, Laouielle-Duprat S, Prins P, Wolff P, Schmid MW, Donoghue MTA, Fort A, Duszynska A, Comte A, Thi Lao N, et al. 2011. Identification of imprinted genes subject to parent-of-origin specific expression in Arabidopsis thaliana seeds. BMC Plant Biol 11: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J, Grossniklaus U 1999. Genomic imprinting in plants. Results Probl Cell Differ 25: 23–40 [DOI] [PubMed] [Google Scholar]
- Moore T, Reik W 1996. Genetic conflict in early development: Parental imprinting in normal and abnormal growth. Reproduction 1: 73–77 [DOI] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Reik W 2004. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet 36: 889–893 [DOI] [PubMed] [Google Scholar]
- Ohad N, Yadegari R, Margossian L, Hannon M, Michaeli D, Harada JJ, Goldberg RB, Fischer RL 1999. Mutations in FIE, a WD Polycomb group gene, allow endosperm development without fertilization. Plant Cell 11: 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien S, Grossniklaus U 2007. Polycomb group and trithorax group proteins in Arabidopsis. Biochim Biophys Acta 1769: 375–382 [DOI] [PubMed] [Google Scholar]
- Raissig MT, Baroux C, Grossniklaus U 2011. Regulation and flexibility of genomic imprinting during seed development. Plant Cell 23: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H, Mittelsten Scheid O, Paszkowski J 2003. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet 34: 65–69 [DOI] [PubMed] [Google Scholar]
- Schmid KJ, Sorensen TR, Stracke R, Torjek O, Altmann T, Mitchell-Olds T, Weisshaar B 2003. Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res 13: 1250–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoft VK, Chumak N, Choi Y, Hannon M, Garcia-Aguilar M, Machlicova A, Slusarz L, Mosiolek M, Park JS, Park GT, et al. 2011. Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc Natl Acad Sci 108: 8024–8047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirzadi R, Andersen ED, Bjerkan KN, Gloeckle BM, Heese M, Ungru A, Winge P, Koncz C, Aalen RB, Schnittger A, et al. 2011. Genome-wide transcript profiling of endosperm without paternal contribution identifies parent-of-origin-dependent regulation of AGAMOUS-LIKE36. PLoS Genet 7: e1001303 doi: 10.1371/journal.pgen.1001303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillane C, MacDougall C, Stock C, Kohler C, Vielle-Calzada JP, Nunes SM, Grossniklaus U, Goodrich J 2000. Interaction of the Arabidopsis Polycomb group proteins FIE and MEA mediates their common phenotypes. Curr Biol 10: 1535–1538 [DOI] [PubMed] [Google Scholar]
- Spillane C, Baroux C, Escobar-Restrepo JM, Page DR, Laoueille S, Grossniklaus U 2004. Transposons and tandem repeats are not involved in the control of genomic imprinting at the MEDEA locus in Arabidopsis. Cold Spring Harb Symp Quant Biol 69: 465–475 [DOI] [PubMed] [Google Scholar]
- Spillane C, Schmid KJ, Laoueille-Duprat S, Pien S, Escobar-Restrepo JM, Baroux C, Gagliardini V, Page DR, Wolfe KH, Grossniklaus U 2007. Positive darwinian selection at the imprinted MEDEA locus in plants. Nature 448: 349–352 [DOI] [PubMed] [Google Scholar]
- Surani MA, Barton SC, Norris ML 1984. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 308: 548–550 [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada JP, Thomas J, Spillane C, Coluccio A, Hoeppner MA, Grossniklaus U 1999. Maintenance of genomic imprinting at the Arabidopsis MEDEA locus requires zygotic DDM1 activity. Genes Dev 13: 2971–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters AJ, Makarevitch I, Eichten SR, Swanson-Wagner RA, Yeh C-T, Xu W, Schnable PS, Vaughn MW, Gehring M, Springer NM 2011. Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm. Plant Cell 23: 4221–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff P, Weinhofer I, Seguin J, Roszak P, Beisel C, Donoghue MTA, Spillane C, Nordborg M, Rehmsmeier M, Köhler C 2011. High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet 7: e1002126 doi: 10.1371/journal.pgen.1002126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Gehring M, Choi Y, Margossian L, Pu H, Harada JJ, Goldberg RB, Pennell RI, Fischer RL 2003. Imprinting of the MEA Polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev Cell 5: 891–901 [DOI] [PubMed] [Google Scholar]
- Yoon YS, Jeong S, Rong Q, Park KY, Chung JH, Pfeifer K 2007. Analysis of the H19ICR insulator. Mol Cell Biol 27: 3499–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Zhao H, Xie S, Chen J, Xu Y, Wang K, Zhao H, Guan H, Hu X, Jiao Y, et al. 2011. Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm. Proc Natl Acad Sci 108: 20042–20047 [DOI] [PMC free article] [PubMed] [Google Scholar]