This study provides in vivo and in vitro evidence for the role of small RNAs in Rho termination and shows that a trans-encoded small RNA can act cotranscriptionally. The ChiX small RNA in Salmonella is capable of inducing Rho-dependent transcription termination within a cistron in a manner dependent on NusG function. This is the first report of a trans-encoded small RNA triggering termination through the Rho/NusG pathway.
Keywords: noncoding small RNA, Rho-dependent termination, rut, NusG, ChiP
Abstract
Gene regulation by bacterial trans-encoded small RNAs (sRNAs) is generally regarded as a post-transcriptional process bearing exclusively on the translation and/or the stability of target messenger RNA (mRNA). The work presented here revealed the existence of a transcriptional component in the regulation of a bicistronic operon—the chiPQ locus—by the ChiX sRNA in Salmonella. By studying the mechanism by which ChiX, upon pairing near the 5′ end of the transcript, represses the distal gene in the operon, we discovered that the action of the sRNA induces Rho-dependent transcription termination within the chiP cistron. Apparently, by inhibiting chiP mRNA translation cotranscriptionally, ChiX uncouples translation from transcription, causing the nascent mRNA to become susceptible to Rho action. A Rho utilization (rut) site was identified in vivo through mutational analysis, and the termination pattern was characterized in vitro with a purified system. Remarkably, Rho activity at this site was found to be completely dependent on the function of the NusG protein both in vivo and in vitro. The recognition that trans-encoded sRNA act cotranscriptionally unveils a hitherto neglected aspect of sRNA function in bacteria.
Small noncoding RNAs make up a large family of ubiquitous regulators controlling a wide range of mechanisms in all living cells. In higher eukaryotes, microRNAs participate in processes related to development, aging, and disease. In prokaryotes, small RNAs (sRNAs) regulate mechanisms that promote adaptation to environmental changes and stress conditions. The largest class of sRNA regulators acts by base-pairing with sequences in the 5′ untranslated region (UTR) or within the first few codons of the target messenger RNA (mRNA) (Vogel 2009; Gottesman and Storz 2011). In many bacterial species, pairing is stimulated by the Hfq protein, which acts as a docking platform to facilitate the encounter of sRNAs and cognate mRNAs (Brennan and Link 2007; Vogel and Luisi 2011). Most commonly, formation of the sRNA:mRNA duplex occludes the ribosome-binding site, inhibiting translation initiation (Storz et al. 2004; Vogel 2009; Gottesman and Storz 2011. No longer protected by ribosomes, the mRNA becomes vulnerable to the activity of the degradosome (Carpousis 2007; Dreyfus 2009). Although not essential for regulation in most systems, mRNA degradation is thought to confer robustness to the process and to make it irreversible (Morita et al. 2006). In competing with the ribosome for mRNA binding, sRNAs might be expected to perturb the coupling of transcription and translation.
In bacteria, translating ribosomes closely follow the transcribing RNA polymerase throughout the elongation cycle (Proshkin et al. 2010). Conditions that uncouple the two processes—e.g., nonsense mutations or severe amino acid starvation—make the transcription elongation complex (EC) prone to pausing and premature termination (Adhya and Gottesman 1978; Platt 1986; Richardson 1991; Proshkin et al. 2010). A central player in this chain of events is the transcription termination factor Rho. A ring-shaped RNA–DNA helicase, Rho is thought to bind to nascent RNA and thread the RNA through its central canal in a 5′-to-3′ direction until it reaches a pausing EC, where it dissociates the RNA from template DNA (Nudler and Gottesman 2002; Richardson 2002; Banerjee et al. 2006; Boudvillain et al. 2010; Peters et al. 2011). A recent variation of this model incorporates the finding that Rho travels with the EC from the beginning of the transcription cycle and can bind RNA polymerase in the absence of RNA (Mooney et al. 2009a; Epshtein et al. 2010). A panoply of auxiliary factors can transiently associate with the EC and affect its processivity and susceptibility to Rho action. One such factor is NusG, a protein conserved in the three domains of life (Werner 2012) that binds Rho through its C-terminal domain and stimulates termination at a subset of sites (Sullivan and Gottesman 1992; Burns et al. 1999). NusG can also bind ribosomal protein S10 (NusE); this interaction, mutually exclusive with the Rho interaction, provides a physical framework for the coupling of transcription and translation (Burmann et al. 2010).
Rho function requires specific RNA sequences named Rho utilization (rut) sites, characterized by a high cytidine/low guanosine content and relatively little secondary structure (Ciampi 2006; Peters et al. 2011). Besides being located at the end of genes or intercistronic regions, rut sites can be found within protein-coding regions. These intragenic sites are normally hidden by translating ribosomes but become exposed and available for Rho binding if translation is inhibited. The resulting intragenic termination is responsible for the long-known polar effects of nonsense mutations in polycistronic operons. Thought to be mostly relevant for cell economy (Richardson 1991), Rho-dependent polarity has seldom been associated with translational regulatory mechanisms. The recent finding that the magnesium-sensing mgtA riboswitch in Salmonella exerts its function by governing the accessibility of a nearby rut site sets a precedent for a novel type of Rho-mediated regulation (Hollands et al. 2012).
The present study was aimed at elucidating the mechanism by which a sRNA, ChiX, down-regulates the distal portion of the bicistronic chiPQ operon of Salmonella by pairing at a unique site near the 5′ end of the transcript (Figueroa-Bossi et al. 2009). A search for mutations relieving the long-range repression yielded Rho factor mutants. Characterization of these isolates allowed us to infer that ChiX down-regulates chiQ “at a distance” by inducing premature Rho-dependent transcription termination within the early portion of the chiPQ operon. Mutational studies identified the chiP sequences responsible for Rho-dependent effects and revealed a requirement for NusG protein in the termination event. Overall, these findings add a novel twist that might have interesting implications for sRNA regulation.
Results
Rho-dependent polarity in the chiPQ operon
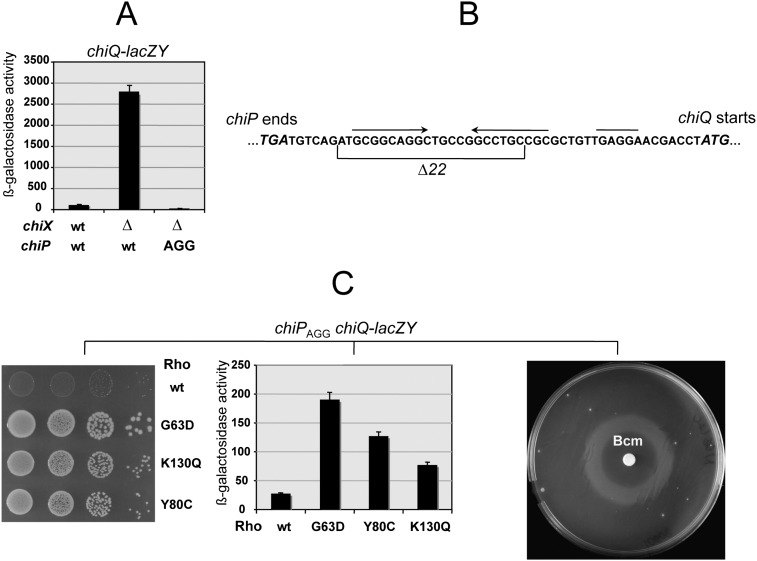
In Salmonella and Escherichia coli, the chiPQ operon encodes a porin (ChiP) involved in the uptake of chitin-derived di- and trisaccharides (chitobiose and chitotriose) and a putative lipoprotein (ChiQ) of unknown function (Figueroa-Bossi et al. 2009; Overgaard et al. 2009). The two cistrons are separated by a 49-nucleotide (nt) intercistronic region that includes a Shine-Dalgarno motif at the canonical distance from chiQ-initiating AUG. Under noninducing growth conditions (i.e., absence of chitooligosaccharides), constitutively made, Hfq-dependent ChiX sRNA prevents ChiP synthesis by pairing with a sequence in the 5′ UTR of the transcript (Figueroa-Bossi et al. 2009). Intriguingly, ChiX down-regulates chiQ at the same time even though no sequence where ChiX could pair is present at the chiP-chiQ boundary (Fig. 1A,B). As a result, ChiX activity renders a strain carrying a chiQ-lacZY fusion phenotypically Lac−. This offered the opportunity to gather insight into the mechanism responsible for the long-range chiQ repression through isolation of Lac+ derivatives. Previous experiments predicted that the Lac+ selection would yield a high background of chiX and hfq deletions/mutations. To avoid this, we made the assumption that the expression of chiQ would also be lost if chiP translation were blocked in a ChiX-independent manner. Mutating the chiP start codon from AUG to AGG was tested. The resulting strain was indeed Lac−, and this phenotype did not change upon deletion of the chiX gene (Fig. 1A). We then proceeded to select spontaneous mutants capable of using lactose as their sole carbon source. Mutants fell into two classes: one arising from mutations genetically linked to the chiQ-lacZ locus, and the other carrying changes elsewhere. Three linked mutations were found to be deletions removing portions of the chiP coding sequence. The mutant with the highest LacZ activity lacked the segment between nucleotides 242 and 932 of the chiP mRNA; the other two deletions removed segments 265–393 and 595–1203. The clue for explaining the phenotype of these mutants came from the analysis of unlinked Lac+ mutations. Isolates from this class were found to carry changes in the gene for transcription termination factor Rho (Fig. 1C). This suggested that when chiP translation is blocked, Rho prevents RNA polymerase from reaching chiQ, possibly acting at more than one site within chiP (see below). Consistent with this interpretation, the Lac+ phenotype could be reproduced by exposing rho+ cells to sublethal concentrations of Rho inhibitor Bicyclomycin (Bcm) (Fig. 1C).
Figure 1.
(A) Effect of chiX and rho mutations on chiQ-lacZY expression. ChiX sRNA down-regulates expression of a chiQ-lacZY fusion by pairing with a sequence upstream of the chiP gene (Figueroa-Bossi et al. 2009). Mutating the chiP initiation codon (from AUG to AGG) silences chiQ-lacZY expression regardless of the chiX allele (A) in spite of the fact that the intercistronic region between chiP and chiQ (B) contains a Shine-Dalgarno motif for independent chiQ translation (overlined in B). As a result, a strain carrying chiPAGG chiQ-lacZY is unable to grow on lactose as the sole carbon source (Lac− phenotype). (C) Mutations in the rho gene relieve chiQ-lacZY silencing to various degrees (middle) and restore growth on lactose (left). (Right) A similar effect is obtained by exposing cells to Rho inhibitor Bcm. In the latter test, wild-type rho bacteria carrying chiPAGG chiQ-lacZY were spread onto a minimal lactose plate, and a filter paper disc wet with 3 μL of a 50 mg/mL Bicyclomycin (Bcm) solution was applied in the center of the plate. Diffusion of the drug generated a concentration gradient. After a 2-d incubation, a ring of growth was observed in the region of the plate where the Bcm concentration was low enough not to inhibit growth but sufficiently high to interfere with Rho activity. Strains used were MA10608 (rhowt), MA10633 (rho G63D), MA10634 (rho K130Q), and MA10769 (rho Y80C). For the complete genotypes, see Supplemental Table S1.
Rho mutants included alleles G63D, R66C, K130Q, and S312I. We then tested the rho-111 mutation isolated in Salmonella in the early 1980s as a polarity suppressor (Housley and Whitfield 1982). This allele was also found to confer a Lac+ phenotype. Our sequence analysis showed that rho-111 corresponds to a Y80C change in the protein. Alleles G63D, Y80C, and K130Q were used for further study. The first two alleles affect the primary RNA-binding site (Gly 63 is part of “RNP1” domain [Gly 61–Phe 62–Gly 63–Phe 64]) (Martinez et al. 1996a,b; Chalissery et al. 2007); K130Q is in a region of Rho where no mutants have been described. The mutation affects a residue on the external surface of the hexamer, not far from residues proposed to contact NusG (Supplemental Fig. S1). The three Rho alleles differ in the extent to which they relieve polarity in chiPQ (Fig. 1C).
ChiX activity induces Rho-dependent polarity
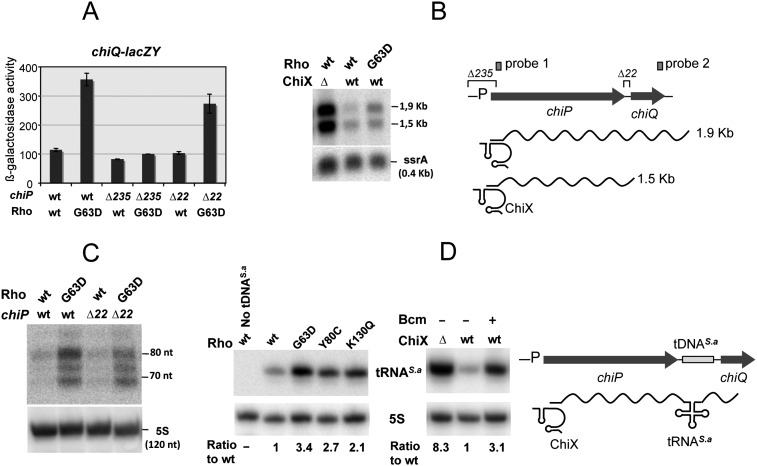
We next examined the effects of rho mutations in strains in which the chiP-initiating AUG is unaltered and ChiX is therefore solely responsible for the inhibition of chiP mRNA translation. As shown in Figure 2A, the introduction of Rho allele G63D into a strain carrying a chiQ-lacZ fusion leads to a significant increase in lacZ expression levels. No such effect is observed in a strain in which the chiP segment between −175 and +60 is deleted (chiP Δ235), indicating that the increase in lac expression in the rho mutant results from transcription originating at the chiP promoter. To obtain more direct evidence for the relief of transcriptional polarity, RNA from the distal portion of the chiPQ operon was subjected to Northern analysis. Our previous work showed that ChiX pairing causes rapid decay of the chiPQ transcript through a pathway initiated by RNase E (Figueroa-Bossi et al. 2009). Since the full-length chiPQ RNA is undetectable in rne wild-type cells, this analysis was performed by either detecting full length chiPQ RNA in strains carrying a temperature-sensitive rne allele (Fig. 2B) or measuring RNA processing intermediates from the 3′ end of the operon with a distal probe (Fig. 2C). The results from both approaches concurred in showing larger amounts of transcripts to reach the end of the operon in cells carrying Rho G63D as compared with the wild type (Fig. 2B,C). Quantification of the bands corresponding to the readthrough transcripts shows the increase to be on the order of threefold to fivefold. This parallels the results from the β-galactosidase determinations in Figure 2A. The smaller extent of this variation, compared with that measured in the chiP translation initiation mutant (sixfold) (Fig.1C), can be ascribed to the higher basal level of chiQ expression in the chiP+ chiX+ rho+ background (a likely indication that ChiX does not totally block chiP mRNA translation). Overall, it is apparent that even in the presence of rne and rho mutations, chiPQ RNA levels remain much lower than those measured in the ΔchiX strain (Fig. 2B). The eightfold difference in the intensities of the full-length bands in Figure 2B might reflect the activity of an RNase E-independent decay pathway in chiX wild-type cells. Alternatively, uncoupling transcription and translation in the chiP gene might lead to a generalized decline of transcriptional efficiency (e.g., stalling and/or backtracking of polymerase) that is only moderately suppressed by the rho mutation.
Figure 2.
ChiX sRNA-induced transcriptional polarity and its relief by rho mutations. (A) Effect of rho G63D on the expression of a chiQ-lacZ fusion. Stimulation of lacZ expression by rho G63D requires a functional chiP promoter and is independent of sequences in the intercistronic region between chiP and chiQ. Strains used were (from left to right) MA9655, MA9860, MA10235, MA10240, MA10277, and MA10295. (B) chiPQ transcription profile in strains carrying temperature-sensitive RNase E allele rne-3071 (Figueroa-Bossi et al. 2009). Exponentially growing cells were incubated for 15 min at 43°C prior to RNA extraction. RNA was separated on a 1.3% agarose-formaldehyde gel and probed with a 32P-labeled DNA oligonucleotide complementary to a sequence near the 5′ end of chiPQ mRNA (pp977; “probe 1” in the diagram). Strains used were (from left to right) MA9826, MA9816, and MA9886. (C) Transcription in the distal portion of the chiPQ operon. RNA turnover products from the 3′ end of the chiPQ mRNA are quantified by Northern blot hybridization. RNA was extracted from exponentially growing cultures of strains MA3409 (chiPwt, rhowt), MA9870 (chiPwt, rho G63D), MA10274 (chiPΔ22, rhowt), and MA10293 (chiPΔ22, rho G63D); fractionated in a polyacrylamide gel; and probed with a 32P-labeled oligonucleotide complementary to a sequence near the 3′ end of the chiPQ transcript (ppA83; “probe 2” in the diagram). (D) Effect of rho mutations or Bcm treatment on the synthesis of a S. aureus tRNA inserted in the intercistronic spacer of chiPQ mRNA. Bcm was used at a final concentration of 10 μg/mL. Total RNA extracted from stationary cultures was processed for Northern blotting as in C. The blot was hybridized to an oligonucleotide complementary to tRNAS.a (ppJ62). RNA bands were quantified by PhosphorImaging using the ImageQuant program; 5S was used for normalization. Strains used were MA3409 (wild-type: lacks tDNAS.a), MA11115 (rhowt), MA11150 (chiPwt, rho G63D), MA11168 (chiPwt, rho Y80C), and MA11169 (chiPwt, rho K130Q) (left panel); and MA11123 (chiPwt ΔchiX) and MA11115 (rhowt) without and with Bcm (right panel).
The intercistronic region between chiP and chiQ encodes a palindromic C/G-rich sequence (Fig. 1B) that is responsible for the presence of a shorter mRNA species in Northern blots (the 1.5-kb band in Fig. 2B). In the early stages of this study, we considered that this sequence might contribute to the polarity effects. A 22-base-pair (bp) deletion removing the palindrome (Δ22) (Fig. 1B) was included in the above analysis. Results showed that Δ22 does not increase chiQ-lacZY expression levels (Fig. 2A) or the amount of chiQ 3′ end RNA (Fig. 2C). On the contrary, the deletion appears to lower chiQ-lacZY expression in the rho G63D background slightly but reproducibly, suggesting that the putative secondary structure somehow exerts a positive role on chiQ translation. To date, the function of this structure and the origin of the 1.5-kb band—whether a stalled transcript or a processing intermediate—remain elusive and were not investigated further in this study.
To circumvent the problem inherent to the degradation of the untranslated mRNA, we resorted to the use of an exogenous tRNA as a transcriptional reporter. Previous work showed that tRNAs are rapidly and correctly excised from foreign RNA sequences and stably accumulate (Lopez et al. 1994; Espeli et al. 2001). A DNA sequence encoding a putative tRNA from Staphylococcus aureus was inserted in place of the chiPQ intercistronic region, and tRNAS.a production was measured by Northern blot hybridization. The results in Figure 2D show that ChiX activity represses tRNAS.a significantly and that rho mutations or inhibition by Bcm relieve this effect. Altogether, the above results suggest that ChiX action on the nascent mRNA induces Rho-dependent transcription termination within the chiP portion of the chiPQ operon.
Profiling the chiP polarity gradient by stop codon scanning
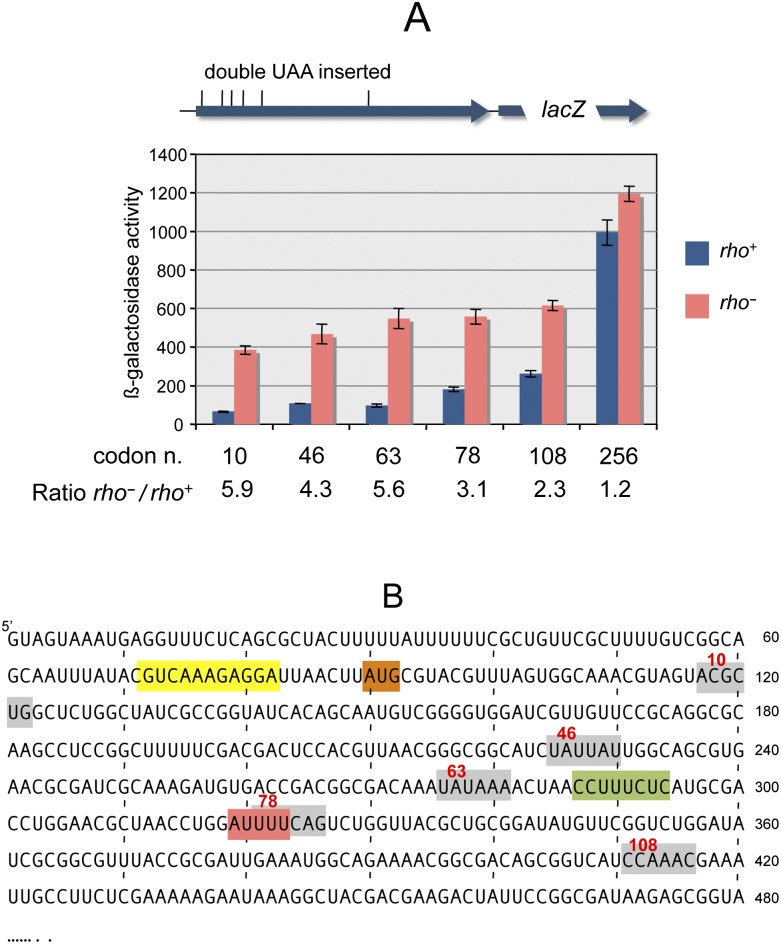
To map the presumptive sites of Rho-dependent transcription termination, tandem UAA stop codons were introduced at several positions in the chromosomal chiP gene, and chiQ-lacZ expression was measured in the resulting strains. The analysis was performed in a ΔchiX background to ensure that chiP translation would initiate normally. Results showed the polarity of the nonsense codons to inversely correlate with their distance from the initiating AUG (Fig. 3A). It is apparent that the Rho involvement is confined to the promoter-proximal half of the chiP gene: Stimulation of lacZ expression by Rho G63D declines from more than fivefold when translation stops before codon 63 to <20% for a stop at position 256. A significant part of this variation occurs within the interval between codons 63 and 108, corresponding to the +275 to +410 segment of chiP mRNA (Fig. 3B), suggesting the presence of a Rho-dependent polarity site within this segment.
Figure 3.
Polarity of tandem UAA codons in the chiP gene. The sequence UAAUAA was inserted in place of tandem sense codons at different positions of the chiP gene in the chromosome of a strain carrying chiQ-lacZ. (A) Effect of rho mutation on the polarity gradient. Loss of lacZ stimulation by rho G63D in the interval between UAA-63 and UAA-108 (from 5.6-fold to 2.3-fold) suggests the presence of a polarity site within this interval. Strains used were MA10802 (UAA-10), MA10960 (UAA-46), MA10961 (UAA-63), MA11046 (UAA-78), MA11047 (UAA-108), and MA10445 (UAA-256). (B) Nucleotide sequence of the 5′ portion of the chiPQ mRNA. The ChiX pairing sequence and chiP-initiating AUG are boxed in yellow and orange, respectively. Gray boxes denote the positions of the double UAA codons. An octameric rut site element and the sequences found at the 3′ ends of in vitro terminated transcripts (see the text) are boxed in green and red, respectively.
A functional rut site in the early portion of the chiP gene
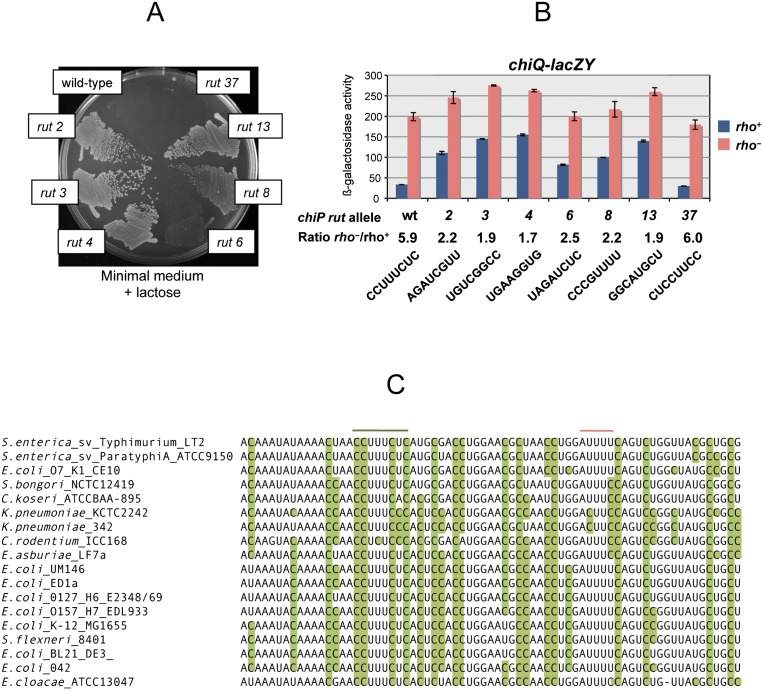
Examination of the sequence in the presumptive termination interval detected only a minor C>G skew centered around an 8-nt pyrimidine-only track (CCUUUCUC) (Fig. 3B, in green). To determine whether this element played a role in polarity, the octamer sequence was randomized by recombineering using PCR-amplified fragments carrying any of the 4 bp at each of the eight positions (see the Materials and Methods). This work was performed in a strain harboring the chiPQ-lacZ reporter so that mutant colonies could be directly screened on lactose indicator plates. The vast majority of recombinants were found to express lacZ at higher levels than the parental strain. In most instances, the increase conferred a Lac+ growth phenotype (Fig. 4).
Figure 4.
Mutational analysis of the chiP rut site. An octameric CCUUUCUC sequence, in the polarity interval identified by the UAA scanning analysis (see the text), was randomly mutagenized as described in the text in a strain carrying chiQ-lacZ. Most of the clones obtained expressed lacZ at higher levels than the parent strain, consistent with the role of the C-rich octamer in the polarity effects. (A) Growth phenotype of representative mutants on a minimal (NCE) lactose plate. (B) Effects of rut site changes on chiQ-lacZ expression in wild-type and rho G63D mutant backgrounds. The strains used in this experiment are listed in Supplemental Table S1. (C) Sequence alignment of the chiP rut region from members of the Enterobacteriaceae family. Cytosine residues are highlighted in green.
Thus, these data strongly suggested that the CCUUUCUC octamer is part of a rut site required for Rho binding to the nascent mRNA. Two out of 100 colonies that were analyzed, isolates #29 and #37, were indistinguishable from the parental strain. This could occur if the mutagenic procedure had regenerated a functional rut site. Sequence analysis showed that these two isolates harbor the sequence CACUCCUA (#29) and UCCUUCC (#37), thus corroborating the importance of the C>G skew in a rut sequence. Also consistent with this role of the C>G skew is the intermediate phenotype of mutants #6 and #8 in Figure 4. Similar sequence features are found at the corresponding position of the chiP gene from E. coli and of other members of the Enterobacteriaceae family. In these organisms, the C>G skew is conserved and, overall, considerably more pronounced than in Salmonella (Fig. 4C; Supplemental Fig. S3). Clearly, induction of Rho-dependent polarity appears to be a conserved feature of the mechanism by which ChiX sRNA down-regulates the chiPQ operon.
It is intriguing to notice that allowing chiP translation to proceed up to codon 63, which is only 8 nt upstream of the CU-rich octamer, does not interfere at all with the ability of Rho to function at the rut site (Fig. 3A). This could imply that the downstream edge of the ribosome stalling at the UAA codon does not extend enough to hamper Rho entry and action on the mRNA. Alternatively, Rho might bind to the rut site within the time interval between the release of the ribosome and the arrival of the next ribosome. Clearly, the latter scenario would seem more plausible if Rho were already associated with the EC at the time of ribosomal release (Mooney et al. 2009a; Epshtein et al. 2010).
The data in Figure 4B show that the presence of the Rho G63D mutation still causes chiQ-lacZY expression to increase in all of the rut mutants. These findings are consistent with various lines of evidence suggesting the presence of additional polarity sites elsewhere in the sequence. Among these data are the detection of residual Rho-dependent polarity in the interval between codons 108 and 256 (Fig. 3A), the increase in chiQ-lacZY expression in a chiP deletion mutant lacking a segment downstream from the rut site (between base pairs 595 and 1203) (see above), and the termination patterns from the in vitro transcription study described below.
NusG requirement for Rho-dependent termination at the chiP rut site in vitro
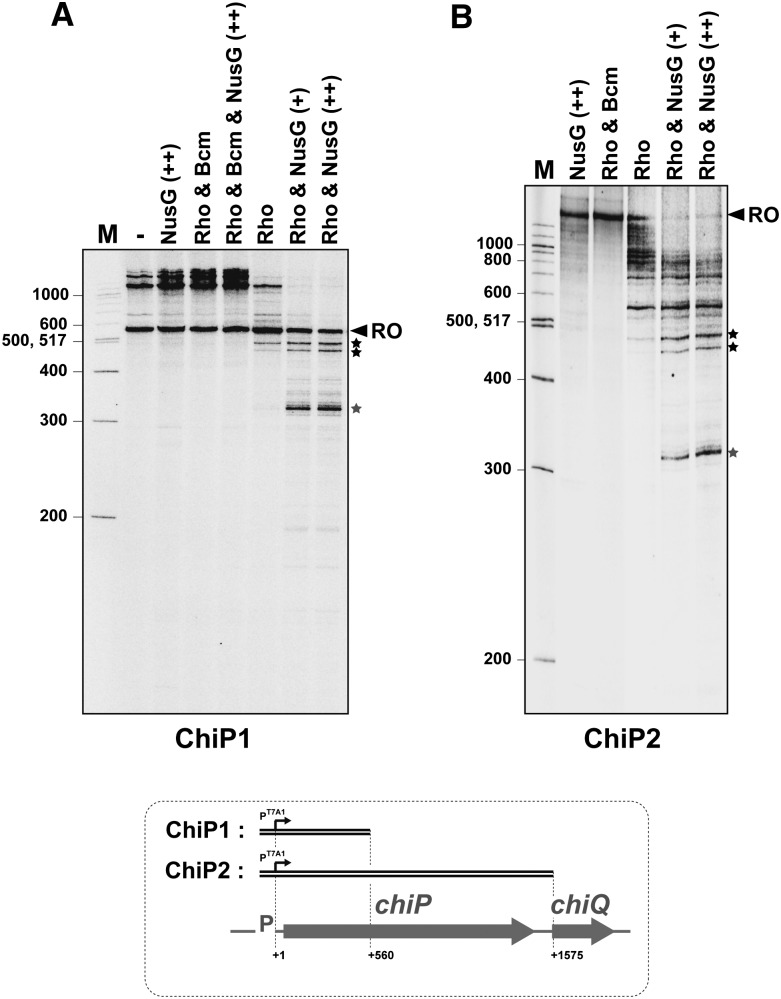
We next examined whether chiP would be similarly susceptible to Rho-dependent termination in vitro. We performed in vitro transcription experiments with E. coli RNA polymerase and a 600-bp DNA template containing the initial portion of the chiP gene fused to the pT7A1 promoter (Rabhi et al. 2011). These experiments showed the purified Rho factor to induce transcription termination in the rut region in the presence of the NusG protein (Fig. 5A, gray star). Neither Rho nor NusG caused any detectable termination at this site when present alone (Fig. 5A). Sequence analysis of the terminated transcript found its 3′ ends to fall within a 5-nt window located downstream from the rut octamer, between +319 and +323 (boxed in red in Fig. 3B; see also Supplemental Fig. S2). The experiments in Figure 5A detect two additional Rho-dependent termination sites in the interval between +450 and +500. These are not as strong or as dependent on NusG as the proximal site (Fig. 5A). Interestingly, the use of longer templates shows that, in the presence of NusG, collective termination activity in the promoter-proximal half of chiP is strong enough to completely prevent downstream transcription (Fig. 5B; data not shown). This strong termination activity is strictly Rho-dependent and can be totally inhibited by the Rho-specific inhibitor Bcm (Fig. 5, Bcm lanes).
Figure 5.
Effect of Rho and NusG on in vitro transcription of DNA templates containing the upstream section (ChiP1 template) or the entirety (ChiP2 template) of the chiP gene fused to the T7A1 promoter. Runoff (RO) transcripts are 560 and 1575 nt long for transcriptions with ChiP1 (A) and ChiP2 (B), respectively. Longer transcripts presumably result from RNA polymerase continuing transcription after reaching the end of a DNA template and “switching” to another template molecule (Nudler et al. 1996; Rabhi et al. 2011). The concentrations of Rho hexamers and Bcm were 70 nM and 150 μM, respectively. The concentration of NusG was 70 nM (+ lanes) or 140 nM (++ lanes).
NusG requirement for the ChiX-induced polarity in vivo
The findings from the in vitro study prompted us to examine whether NusG played any role in chiP polarity in vivo. Initial attempts to construct a nusG deletion replacing this gene with a cat cassette proved unsuccessful. Whenever chloramphenicol-resistant recombinants were obtained, they were found to contain a chromosomal duplication of the nusG region, suggesting that the deletion of nusG is a lethal event in Salmonella. Since the lethality of nusG mutations in E. coli was previously ascribed to the derepression of toxic prophage gene products (Cardinale et al. 2008), the above experiment was repeated in a strain cured for all four major prophages of Salmonella strain LT2 (Lemire et al. 2011). However, we were still unable to isolate the deletion mutant. We therefore changed our strategy and proceeded to isolate point mutations that might not completely inactivate the protein and still affect chiP polarity. A DNA fragment spanning the nusG gene and an adjacent cat cassette for selection was amplified by PCR under error-prone conditions and introduced by recombineering in the chromosome of a strain carrying the chiP initiator codon mutation and the chiQ-lacZ fusion. Recombinant clones were selected on LB plates supplemented with chloramphenicol and X-gal. The occurrence of rare blue-colored colonies suggested that these clones carried nusG mutations that stimulated transcriptional readthrough. Upon verifying that the mutations conferring the blue color were transductionally linked to the CmR marker, some representative mutants were subjected to DNA sequence analysis. This confirmed the presence of mutations in the nusG gene, all located within the portion of the gene encoding the C-terminal domain of the protein. One mutant carried a −1 frameshift mutation that resulted in multiple amino acid substitutions in the C-terminal sequence (DFSQVEKA to TSVRLRKRNRS, starting at position 174; this allele is referred to as nusG 174fs). Additional mutants contained changes F141S, F144I, and V162D. The effects of these three alleles on readthrough transcription in the chiP gene were assessed independently using the lacZ and S. aureus tRNA reporter systems. The results of both types of measurements showed that, like the rho mutations, the nusG mutations relieve transcriptional polarity in chiPQ (Fig. 6). Note, the experiment in Figure 6B was performed in the background of the chiPAGG mutant, where polarity is the strongest. This accounts for the stronger effects of the nusG mutations compared with the rho mutations in Figure 2D, where polarity results uniquely from the activity of ChiX. Overall, these data corroborate the in vitro study confirming the NusG requirement for transcription termination at the chiP rut site in vivo.
Figure 6.
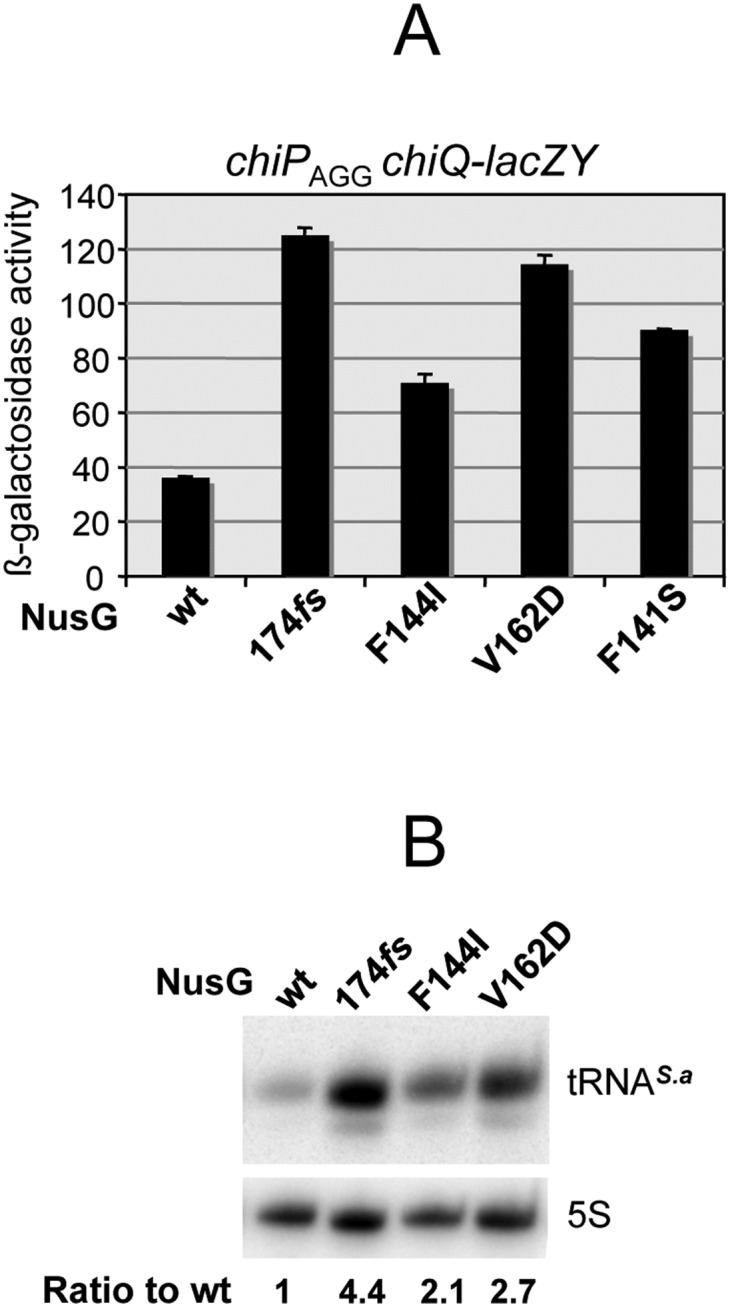
Mutations in nusG gene relieve chiP polarity. Mutants were obtained by combining random PCR mutagenesis and chromosomal recombineering as described in the text. (A) Effect of nusG mutations on the expression of chiQ-lacZ in a chiPAGG mutant background. Strains used were MA11167 (nusGwt), MA11158 (nusG 174fs), MA11159 (nusG F144I), MA11161 (nusG V162D), and MA11162 (nusG F141S). (B) Effect of nusG mutations on the synthesis of S. aureus tRNA (see the legend for Fig. 2D). Total RNA extracted from stationary cultures was processed for Northern blotting as in Figure 2D. The blot was hybridized to an oligonucleotide complementary to tRNAS.a (ppJ62). Strains used were MA11170 (nusGwt), MA11171 (nusG 174fs), MA11172 (nusG 144I), and MA11173 (nusG V162D).
Attempts to construct strains carrying nusG and rho mutations combined suggested that most of the combinations were incompatible. In particular, we were unable to combine rho allele Y80C with any of the nusG mutations, or nusG allele V162D with any of the rho mutations. In contrast, strains carrying rho G63D or K130K together with either nusG F141S or F144I proved viable and grew at rates comparable with the nusG single mutants (which typically grow more slowly than the rho mutants). Interestingly, these double mutants expressed the chiQ-lacZY fusion at higher levels than the corresponding single mutants. The rho G63D nusG F141S combination (the one with the highest activity) exhibited a 40% increase over the value measured in the rho G63D single mutant.
Discussion
In the present study, we showed that the activity of a regulatory sRNA induces Rho-dependent transcriptional polarity in the target gene. While cis-encoded sRNAs have long been known to promote factor-independent termination or anti-termination (Brantl 2007), to our knowledge, this is the first report of a trans-encoded sRNA triggering termination through the Rho/NusG pathway. In retrospect, it seems that these findings might have been predictable given the analogy between the mode of action of the sRNAs and conditions known to elicit transcriptional polarity. By inhibiting translation initiation, the sRNA allows the Rho protein to gain access to rut sites in the mRNA. A direct implication is that ChiX sRNA must act cotranscriptionally, presumably binding the mRNA target as soon as it emerges from the EC. The Rho recruitment makes good biological sense, as it would not be convenient for the cell economy to continuously make the full-length chiPQ mRNA under conditions where the mRNA will not be translated due to ChiX activity. Furthermore, the mechanism provides a solution for coordinating the expression of the two genes in the operon. This raises the possibility that a similar mechanism operates at other sRNA-controlled loci. A number of sRNA targets in E. coli and Salmonella are part of polycistronic transcripts. We notice that in many cases, cistrons lying downstream from the primary target are coregulated with the latter (Massé et al. 2005; Sharma et al. 2011). It seems possible that Rho-dependent polarity may contribute to some of these effects. We are currently testing this hypothesis on a few selected systems.
Rho-dependent transcription termination at the rut site identified in this study was found to be completely dependent on the NusG protein in an in vitro transcription system. Furthermore, nusG mutations promoted readthrough transcription like rho mutations, indicating that the NusG requirement for termination exists in vivo as well. All of the nusG mutations isolated in this study affect the portion of the protein thought to interact with Rho (Mooney et al. 2009b; Chalissery et al. 2011). Previous studies showed that the influence of NusG on Rho-dependent termination varies extensively among rut sites in vivo and/or in vitro (Sullivan and Gottesman 1992; Burns et al. 1999). The basis for such variability remains incompletely understood. The isolation of chiP rut variants that lose the nusG dependence and their characterization in vivo and in vitro should shed further light on this aspect.
The Rho involvement in regulation by trans-encoded Hfq-dependent sRNA evokes a link with the recent findings that Hfq protein from E. coli can bind Rho and interfere with its activity (Rabhi et al. 2011). Hfq was shown to compete with NusG in promoting anti-termination at several terminators, including the prototypical tR1 terminator of phage λ in vitro. Hfq also caused anti-termination in vivo, although this effect was only observed when Rho activity was partially inhibited with Bcm (Rabhi et al. 2011). In our system, Hfq clearly does not prevent termination when the Rho factor is fully functional (Fig. 2). However, in a way somewhat reminiscent of the in vivo data from the above study, we found that Hfq contributes to polarity suppression in the rho mutants. In particular, Rho allele K130Q loses the ability to relieve chiP polarity completely in an hfq deletion background (Supplemental Fig. S3). This effect does not depend on Hfq binding to the chiP mRNA leader region or to an A-rich stretch immediately 5′ to the rut site (data not shown). However, the effect appears to be specific to the chiPQ system, as it is not observed at a different Rho-dependent polarity site in the hisG gene of Salmonella (Supplemental Fig. S4; Ciampi and Roth 1988).
Currently, it is unclear whether the anti-terminator function of Hfq is exerted by the hexameric form of the protein or by the Hfq monomer. Intriguingly, a recent study (Argaman et al. 2012) proposed that the Hfq hexamer–monomer equilibrium can be affected by specific physiological conditions in the cell. In particular, the investigators showed the RelA protein to stimulate Hfq multimerization in vitro. Since RelA is specifically synthesized in response to amino acid starvation and these conditions also favor premature Rho-dependent termination, one may speculate that the Hfq monomer antagonizes Rho function and that RelA would counteract the anti-terminator by promoting Hfq hexamer formation. The study of Hfq mutants specifically affected in hexamer assembly might help clarify this issue.
Materials and methods
Strains and growth conditions
Strains used in this study were all derived from Salmonella enterica serovar Typhimurium strain MA3409, a strain LT2 derivative cured for the Gifsy-1 prophage (Figueroa-Bossi et al. 1997). The genotypes of strains used are listed in Supplemental Table S1. Bacteria were cultured at 37°C in liquid medium or in medium solidified by the addition of 1.5% Difco agar. LB broth (Bertani 2004) was used as complex medium. Carbon-free medium (NCE) (Maloy and Roth 1983) supplemented with 0.2% glycerol or 0.2% lactose was used as minimal medium. Antibiotics (Sigma-Aldrich) were included at the following final concentrations: 10 μg mL−1 chloramphenicol, 50 μg mL−1 kanamycin monosulphate, 100 μg mL−1 sodium ampicillin, 80 μg mL−1 spectinomycin dihydrochloride, and 25 μg mL−1 tetracycline hydrochloride. LB plates containing 40 μg mL−1 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal; from Sigma) were used to monitor lacZ expression in bacterial colonies. Liquid cultures were grown in New Brunswick gyratory shakers, and growth was monitored by measuring the optical density at 600 nm with a Shimazu UV-mini 1240 spectrophotometer.
Enzymes and chemicals
T4 polynucleotide kinase and Taq DNA polymerase were from New England Biolabs, Pfu-Turbo DNA polymerase was from Stratagene, T4 DNA ligase was from USB, and σ-saturated E. coli RNA polymerase was from Epicentre. DNA oligonucleotides were obtained from Sigma-Aldrich. Acrylamide-bisacrylamide and other electrophoresis reagents were from Bio-Rad or AMRESCO. Agarose was from Invitrogen. Hybond-N+ membranes and the hybridization buffer used for Northern blot analysis were from GE Healthcare and Applied Biosystems-Ambion, respectively. The rNTPs were from Promega, and the 32P-NTPs were from PerkinElmer or Hartmann Analytic. Rho and NusG proteins were overexpressed and purified as described previously (Rabhi et al. 2011). Bcm was a generous gift from Max Gottesman (Columbia University, New York).
Genetic techniques and chromosomal recombineering
Generalized transduction was performed using the high-frequency transducing mutant of phage P22, HT 105/1 int-201 (Schmieger 1972), as described (Lemire et al. 2011). Chromosomal engineering (recombineering) was carried out by the λ red recombination method (Datsenko and Wanner 2000; Murphy et al. 2000; Yu et al. 2000) implemented as in Datsenko and Wanner (2000). Donor DNA fragments were generated by PCR using plasmid DNA, chromosomal DNA, or DNA oligonucleotides as templates. DNA oligonucleotides used as primers for PCR amplification are listed in Supplemental Table S2. Amplified fragments were electroporated into appropriate strains harboring the conditionally replicating plasmid pKD46, which carries the λ red operon under the control of the PBAD promoter (Datsenko and Wanner 2000). Bacteria carrying pKD46 were grown at 30°C in the presence of ampicillin and exposed to arabinose (10 mM) for 3 h prior to preparation of electrocompetent cells. Electroporation was carried out using a Bio-Rad MicroPulser under the conditions specified by the manufacturer. Recombinant colonies were selected on LB plates containing the appropriate antibiotic. Constructs were verified by PCR and DNA sequence analysis (performed by GATC Co.).
‘Scarless' DNA recombineering
Scarless modification of chromosomal DNA sequences at the single-base-pair level was achieved with a two-step recombineering procedure adapted from Karlinsey (2007). The procedure involves (1) inserting a tetAR module (produced by PCR) at the chromosomal site to be modified and (2) replacement of the tetAR module by a DNA fragment carrying the desired changed through positive selection of tetracycline-sensitive recombinants (Bochner et al. 1980). Typically, the DNA fragment in the second step was also obtained by PCR using oligonucleotides with complementary sequences at their 3′ ends priming DNA synthesis on each other (“reciprocal priming”) (Supplemental Table S3).
Isolation and identification of rho mutants
Aliquots (0.1 mL) from overnight cultures of strain MA9679 (chiPAGG chiQ∷lacZY) were spread on NCE plates containing lactose as the sole carbon source. A number of Lac+ colonies were visible after a 2-d incubation period. Cells from the largest colonies were found to contain different-sized deletions in the chiP gene. Cells from smaller colonies carried mutations not linked to chiPQ. To characterize the latter class, a phage P22 lysate made on a pool of random Tn5-TPOP insertions in the Salmonella chromosome (Figueroa-Bossi et al. 2006) was used to transduce some of these isolates selecting tetracycline-resistance on lactose indicator plates. This allowed the identification of insertions genetically linked to the Lac+ mutations. Inverse PCR (Hartl and Ochman 1996) revealed the Tn5-TPOP insertion sites to be in proximity to the rho gene. DNA sequence analysis confirmed that the Lac+ phenotype from the small colony class results from rho gene mutations.
chiP rut site mutagenesis
An 87-bp DNA fragment with a randomized 8-bp sequence corresponding to the core portion of the chiP rut region was obtained by annealing oligonucleotides ppI83 and ppJ18 (Supplemental Table S3) and performing PCR amplification. The fragment was introduced into a strain harboring a tetAR insertion at the rut site (chiP135∷tetAR) and expressing λ red recombination functions (strain MA10974). Tetracycline-sensitive recombinants were selected as above. DNA sequence analysis of individual clones showed that each of them carries a mutagenized variant of the octameric rut core sequence. Presence of the chiQ∷lacZY fusion in the MA10974 background allowed it to visualize the phenotype of the recombinant clones directly on lactose indicator plates.
nusG mutagenesis
A cat gene cassette was introduced in the Salmonella chromosome at the 3′ side of the nusG gene. To avoid perturbing the expression of the adjacent rplK gene (coding for 50S ribosomal protein L11), the DNA insert contained only the cat ORF (including the Shine-Dalgarno motif) positioned 20 bp downstream from the end of the nusG coding sequence (thus expressed from the nusG promoter). A 1618-bp DNA fragment spanning the nusG-cat module was amplified by PCR under error-prone conditions (with primers ppK01 and ppK05) and introduced into strain MA11116 (chiPAGG chiQ∷lacZY/pKD46). CmR recombinants were selected on plates containing X-gal. Rare blue colonies were identified. Transductional crosses showed that the blue color results from mutations linked to the cat marker. DNA sequence analyses confirmed these mutations to lie within the nusG gene.
RNA extraction and Northern analysis
RNA was prepared by the acid–hot-phenol method from exponentially growing cells (OD600 of 0.35) as previously described (Bossi and Figueroa-Bossi 2007). RNA was fractionated on an 8% polyacrylamide–8 M urea gel and transferred to Hybond-N+ membranes. Blots were hybridized to DNA oligonucleotides (Supplemental Table S2) labeled at the 5′ ends with T4 polynucleotide kinase. Hybridization patterns were analyzed by PhosphorImaging, and, when appropriate, RNA bands were quantified using the ImageQuant or ImageJ programs.
In vitro transcription
DNA templates for in vitro transcription experiments were prepared by standard PCR procedures using genomic DNA from Salmonella and appropriate synthetic primers (Eurogentec). Standard transcription termination experiments were performed as described previously (Rabhi et al. 2011). Briefly, mixtures (18 μL) containing DNA template (5 nM final concentration), E. coli RNAP (22.5 nM), Rho (0 or 70 nM), SUPERase-In (0.5U/μL; Ambion), and NusG (0–210 nM) in transcription buffer (40 mM Tris-HCl at pH 8.0, 50 mM KCl, 5 mM MgCl2, 1.5 mM DTT) were preincubated for 10 min at 37°C. Then, 2 μL of transcription initiation mix (2 mM ATP, GTP, and CTP, 0.2 mM UTP, 2.5 μCi/μL of 32P-αUTP, 250 μg/mL rifampicin in transcription buffer) was added before incubation for 20 min at 37°C. Transcription reactions were stopped with 4 μL of EDTA (0.5M), 6 μL of tRNA (0.25 mg/mL), and 80 μL of sodium acetate (0.42 M) before ethanol precipitation at −20°C. Reaction pellets were dissolved in denaturing loading buffer (95% formamide, 5 mM EDTA) and analyzed by denaturing 6% (ChiP1 template) or 4.5% (ChiP2 template) polyacrylamide gel electrophoresis.
β-Galactosidase assays
β-Galactosidase activity was measured in toluene-permeabilized cells as described by Miller (1992) and is expressed in Miller units throughout this work. Unless specified otherwise, measurements were performed on cultures grown overnight in LB medium (OD600 ≈ 3).
Acknowledgments
We thank Fabienne Chevance and Kelly Hughes for advice on scarless DNA recombineering. Some of the major findings from this work were presented at the 2011 Meeting on Molecular Genetics of Bacteria and Phages, University of Wisconsin at Madison. This work was supported by a grant from the French National Research Agency (ANR-BLAN07-1_187785) to L.B.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.195412.112.
References
- Adhya S, Gottesman M 1978. Control of transcription termination. Annu Rev Biochem 47: 967–996 [DOI] [PubMed] [Google Scholar]
- Argaman L, Elgrably-Weiss M, Hershko T, Vogel J, Altuvia S 2012. RelA protein stimulates the activity of RyhB small RNA by acting on RNA-binding protein Hfq. Proc Natl Acad Sci 109: 4621–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Chalissery J, Bandey I, Sen R 2006. Rho-dependent transcription termination: More questions than answers. J Microbiol 44: 11–22 [PMC free article] [PubMed] [Google Scholar]
- Bertani G 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J Bacteriol 186: 595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner BR, Huang HC, Schieven GL, Ames BN 1980. Positive selection for loss of tetracycline resistance. J Bacteriol 143: 926–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L, Figueroa-Bossi N 2007. A small RNA downregulates LamB maltoporin in Salmonella. Mol Microbiol 65: 799–810 [DOI] [PubMed] [Google Scholar]
- Boudvillain M, Nollmann M, Margeat E 2010. Keeping up to speed with the transcription termination factor Rho motor. Transcription 1: 70–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantl S 2007. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr Opin Microbiol 10: 102–109 [DOI] [PubMed] [Google Scholar]
- Brennan RG, Link TM 2007. Hfq structure, function and ligand binding. Curr Opin Microbiol 10: 125–133 [DOI] [PubMed] [Google Scholar]
- Burmann BM, Schweimer K, Luo X, Wahl MC, Stitt BL, Gottesman ME, Rosch P 2010. A NusE:NusG complex links transcription and translation. Science 328: 501–504 [DOI] [PubMed] [Google Scholar]
- Burns CM, Nowatzke WL, Richardson JP 1999. Activation of Rho-dependent transcription termination by NusG. Dependence on terminator location and acceleration of RNA release. J Biol Chem 274: 5245–5251 [DOI] [PubMed] [Google Scholar]
- Cardinale CJ, Washburn RS, Tadigotla VR, Brown LM, Gottesman ME, Nudler E 2008. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science 320: 935–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis AJ 2007. The RNA degradosome of Escherichia coli: An mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol 61: 71–87 [DOI] [PubMed] [Google Scholar]
- Chalissery J, Banerjee S, Bandey I, Sen R 2007. Transcription termination defective mutants of Rho: Role of different functions of Rho in releasing RNA from the elongation complex. J Mol Biol 371: 855–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalissery J, Muteeb G, Kalarickal NC, Mohan S, Jisha V, Sen R 2011. Interaction surface of the transcription terminator Rho required to form a complex with the C-terminal domain of the antiterminator NusG. J Mol Biol 405: 49–64 [DOI] [PubMed] [Google Scholar]
- Ciampi MS 2006. Rho-dependent terminators and transcription termination. Microbiology 152: 2515–2528 [DOI] [PubMed] [Google Scholar]
- Ciampi MS, Roth JR 1988. Polarity effects in the hisG gene of Salmonella require a site within the coding sequence. Genetics 118: 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus M 2009. Killer and protective ribosomes. Prog Mol Biol Transl Sci 85: 423–466 [DOI] [PubMed] [Google Scholar]
- Epshtein V, Dutta D, Wade J, Nudler E 2010. An allosteric mechanism of Rho-dependent transcription termination. Nature 463: 245–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeli O, Moulin L, Boccard F 2001. Transcription attenuation associated with bacterial repetitive extragenic BIME elements. J Mol Biol 314: 375–386 [DOI] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Coissac E, Netter P, Bossi L 1997. Unsuspected prophage-like elements in Salmonella typhimurium. Mol Microbiol 25: 161–173 [DOI] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Lemire S, Maloriol D, Balbontín R, Casadesús J, Bossi L 2006. Loss of Hfq activates the σE-dependent envelope stress response in Salmonella enterica. Mol Microbiol 62: 838–852 [DOI] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Valentini M, Malleret L, Fiorini F, Bossi L 2009. Caught at its own game: Regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev 23: 2004–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Storz G 2011. Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol pii: a003798 doi: 10.1101/cshperspect.a003798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Ochman H 1996. Inverse polymerase chain reaction. Methods Mol Biol 58: 293–301 [DOI] [PubMed] [Google Scholar]
- Hollands K, Proshkin S, Sklyarova S, Epshtein V, Mironov A, Nudler E, Groisman EA 2012. Riboswitch control of Rho-dependent transcription termination. Proc Natl Acad Sci 109: 5376–5381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housley PR, Whitfield HJ 1982. Transcription termination factor rho from wild type and rho-111 strains of Salmonella typhimurium. J Biol Chem 257: 2569–2577 [PubMed] [Google Scholar]
- Karlinsey JE 2007. λ-Red genetic engineering in Salmonella enterica serovar Typhimurium. Methods Enzymol 421:199–209 [DOI] [PubMed] [Google Scholar]
- Lemire S, Figueroa-Bossi N, Bossi L 2011. Bacteriophage crosstalk: Coordination of prophage induction by trans-acting antirepressors. PLoS Genet 7: e1002149 doi: 10.1371/journal.pgen.1002149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez PJ, Iost I, Dreyfus M 1994. The use of a tRNA as a transcriptional reporter: The T7 late promoter is extremely efficient in Escherichia coli but its transcripts are poorly expressed. Nucleic Acids Res 22: 1186–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy SR, Roth JR 1983. Regulation of proline utilization in Salmonella typhimurium: Characterization of put:Mu d(Ap, lac) operon fusions. J Bacteriol 154: 561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Burns CM, Richardson JP 1996a. Residues in the RNP1-like sequence motif of Rho protein are involved in RNA-binding affinity and discrimination. J Mol Biol 257: 909–918 [DOI] [PubMed] [Google Scholar]
- Martinez A, Opperman T, Richardson JP 1996b. Mutational analysis and secondary structure model of the RNP1-like sequence motif of transcription termination factor Rho. J Mol Biol 257: 895–908 [DOI] [PubMed] [Google Scholar]
- Massé E, Vanderpool CK, Gottesman S 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol 187: 6962–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R 2009a. Regulator trafficking on bacterial transcription units in vivo. Mol Cell 33: 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RA, Schweimer K, Rosch P, Gottesman M, Landick R 2009b. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J Mol Biol 391: 341–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Mochizuki Y, Aiba H 2006. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc Natl Acad Sci 103: 4858–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KC, Campellone KG, Poteete AR 2000. PCR-mediated gene replacement in Escherichia coli. Gene 246: 321–330 [DOI] [PubMed] [Google Scholar]
- Nudler E, Gottesman ME 2002. Transcription termination and anti-termination in E. coli. Genes Cells 7: 755–768 [DOI] [PubMed] [Google Scholar]
- Nudler E, Avetissova E, Markovtsov V, Goldfarb A 1996. Transcription processivity: Protein–DNA interactions holding together the elongation complex. Science 273: 211–217 [DOI] [PubMed] [Google Scholar]
- Overgaard M, Johansen J, Moller-Jensen J, Valentin-Hansen P 2009. Switching off small RNA regulation with trap-mRNA. Mol Microbiol 73: 790–800 [DOI] [PubMed] [Google Scholar]
- Peters JM, Vangeloff AD, Landick R 2011. Bacterial transcription terminators: The RNA 3′-end chronicles. J Mol Biol 412: 793–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T 1986. Transcription termination and the regulation of gene expression. Annu Rev Biochem 55: 339–372 [DOI] [PubMed] [Google Scholar]
- Proshkin S, Rahmouni AR, Mironov A, Nudler E 2010. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328: 504–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabhi M, Espeli O, Schwartz A, Cayrol B, Rahmouni AR, Arluison V, Boudvillain M 2011. The Sm-like RNA chaperone Hfq mediates transcription antitermination at Rho-dependent terminators. EMBO J 30: 2805–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JP 1991. Preventing the synthesis of unused transcripts by Rho factor. Cell 64: 1047–1049 [DOI] [PubMed] [Google Scholar]
- Richardson JP 2002. Rho-dependent termination and ATPases in transcript termination. Biochim Biophys Acta 1577: 251–260 [DOI] [PubMed] [Google Scholar]
- Schmieger H 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet 119: 75–88 [DOI] [PubMed] [Google Scholar]
- Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf HJ, Hinton JC, Vogel J 2011. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol Microbiol 81: 1144–1165 [DOI] [PubMed] [Google Scholar]
- Storz G, Opdyke JA, Zhang A 2004. Controlling mRNA stability and translation with small, noncoding RNAs. Curr Opin Microbiol 7: 140–144 [DOI] [PubMed] [Google Scholar]
- Sullivan SL, Gottesman ME 1992. Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell 68: 989–994 [DOI] [PubMed] [Google Scholar]
- Vogel J 2009. A rough guide to the non-coding RNA world of Salmonella. Mol Microbiol 71: 1–11 [DOI] [PubMed] [Google Scholar]
- Vogel J, Luisi BF 2011. Hfq and its constellation of RNA. Nat Rev Microbiol 9: 578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner F 2012. A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J Mol Biol 417: 13–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci 97: 5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]