This study presents an antisense oligonucleotide methodology to phenocopy a disease—in this case, the motor neuron disease spinal muscular atrophy in mice. Sahashi et al. show that it is possible to fine-tune disease severity through dose-dependent effects on RNA splicing, making this a novel animal model for monitoring disease onset and progression as well as testing candidate therapeutics.
Keywords: spinal muscular atrophy, SMN2, antisense oligonucleotide, phenocopy, splicing modulation, mouse model
Abstract
Antisense oligonucleotides (ASOs) are versatile molecules that can be designed to specifically alter splicing patterns of target pre-mRNAs. Here we exploit this feature to phenocopy a genetic disease. Spinal muscular atrophy (SMA) is a motor neuron disease caused by loss-of-function mutations in the SMN1 gene. The related SMN2 gene expresses suboptimal levels of functional SMN protein due to alternative splicing that skips exon 7; correcting this defect—e.g., with ASOs—is a promising therapeutic approach. We describe the use of ASOs that exacerbate SMN2 missplicing and phenocopy SMA in a dose-dependent manner when administered to transgenic Smn−/− mice. Intracerebroventricular ASO injection in neonatal mice recapitulates SMA-like progressive motor dysfunction, growth impairment, and shortened life span, with α-motor neuron loss and abnormal neuromuscular junctions. These SMA-like phenotypes are prevented by a therapeutic ASO that restores correct SMN2 splicing. We uncovered starvation-induced splicing changes, particularly in SMN2, which likely accelerate disease progression. These results constitute proof of principle that ASOs designed to cause sustained splicing defects can be used to induce pathogenesis and rapidly and accurately model splicing-associated diseases in animals. This approach allows the dissection of pathogenesis mechanisms, including spatial and temporal features of disease onset and progression, as well as testing of candidate therapeutics.
RNA splicing requires recognition of pre-mRNA cis-acting elements by spliceosome components and auxiliary RNA-binding proteins (Cartegni et al. 2002). Synthetic antisense oligonucleotides (ASOs) can be designed to base-pair with high specificity to a cis-element on a given pre-mRNA so as to hinder binding of splicing activators or repressors, thereby altering splicing patterns. Based on these properties, ASOs are being developed as RNA-targeted therapeutics to correct disease-associated splicing defects or—in the case of Duchenne muscular dystrophy (DMD)—to skip exons and suppress frame-disrupting mutations (Bennett and Swayze 2010).
Spinal muscular atrophy (SMA) is characterized by skeletal muscle weakness due to progressive loss of α-motor neurons. A decrease in SMN protein, one of whose functions is to promote small nuclear ribonucleoprotein particle (snRNP) assembly, is responsible for SMA, although how this causes motor neuron degeneration is not understood (Burghes and Beattie 2009). SMA results from loss of SMN1; humans have a closely related SMN2 gene that predominantly expresses defective SMN due to alternative splicing of exon 7. SMN2 is a disease modifier, reducing disease severity with increased copy number (Burghes and Beattie 2009).
Mice have only one Smn gene, which is essential (Burghes and Beattie 2009). Introduction of a human SMN2 transgene rescues embryonic lethality, and the resulting mice (Smn−/− SMN2) have SMA-like phenotypes whose severity depends on the transgene copy number (Burghes and Beattie 2009; Park et al. 2010a). Severe “type I” SMA mice harboring two SMN2 copies, with or without an additional SMNΔ7 cDNA transgene, die shortly after birth (<1–2 wk) (Burghes and Beattie 2009; Park et al. 2010a), whereas mild “type III” SMA mice harboring four SMN2 copies survive normally without paralysis but develop tail and ear necrosis (Hsieh-Li et al. 2000; Park et al. 2010a). These strains, while extremely useful, fall short of providing phenotypically accurate models for intermediate “type II” SMA that would allow detailed analyses of phenotype, pathophysiology, and therapeutic efficacy. Other SMA models with point mutations (Smn2B/− mice) or exon deletions (SmnF7/Δ7, NSE-Cre mice) in murine Smn have intermediate phenotypes, including an ∼1-mo life span and progressive motor dysfunction with relevant pathological features (Park et al. 2010a); however, these genotypes are not present in SMA patients and are incompatible with therapeutic strategies involving splicing correction or up-regulation of the human SMN2 gene.
We previously showed that a 2′-O-(2-methoxyethyl) (MOE) therapeutic ASO (ASO-10-27) that promotes exon 7 inclusion rescues tail and ear necrosis in type III mice; this ASO is metabolically stable in CNS tissues and corrects SMN2 splicing in the spinal cord for >6 mo (Hua et al. 2010). Thus, ASOs can persistently control phenotypes through splicing modulation in the CNS. This property prompted us to attempt to generate a useful mouse model of intermediate SMA using ASOs complementary to a different site on SMN2 pre-mRNA so as to exacerbate exon 7 missplicing and promote pathogenesis.
Here we demonstrate that intracerebroventricular (ICV) injection of exon-skipping MOE ASOs in mild type III neonatal mice accurately phenocopies more severe SMA symptoms and histopathology as a result of SMN2 missplicing. Treatment with therapeutic ASO-10-27 corrects the splicing and ameliorates the SMA phenotypes. Furthermore, we report that the SMN2 splicing defect is exacerbated as a result of starvation, which correlates with prognosis. We propose that ASOs can serve as useful tools to develop various animal disease models by persistent modulation of splicing.
Results
ASO inhibition of SMN2 splicing
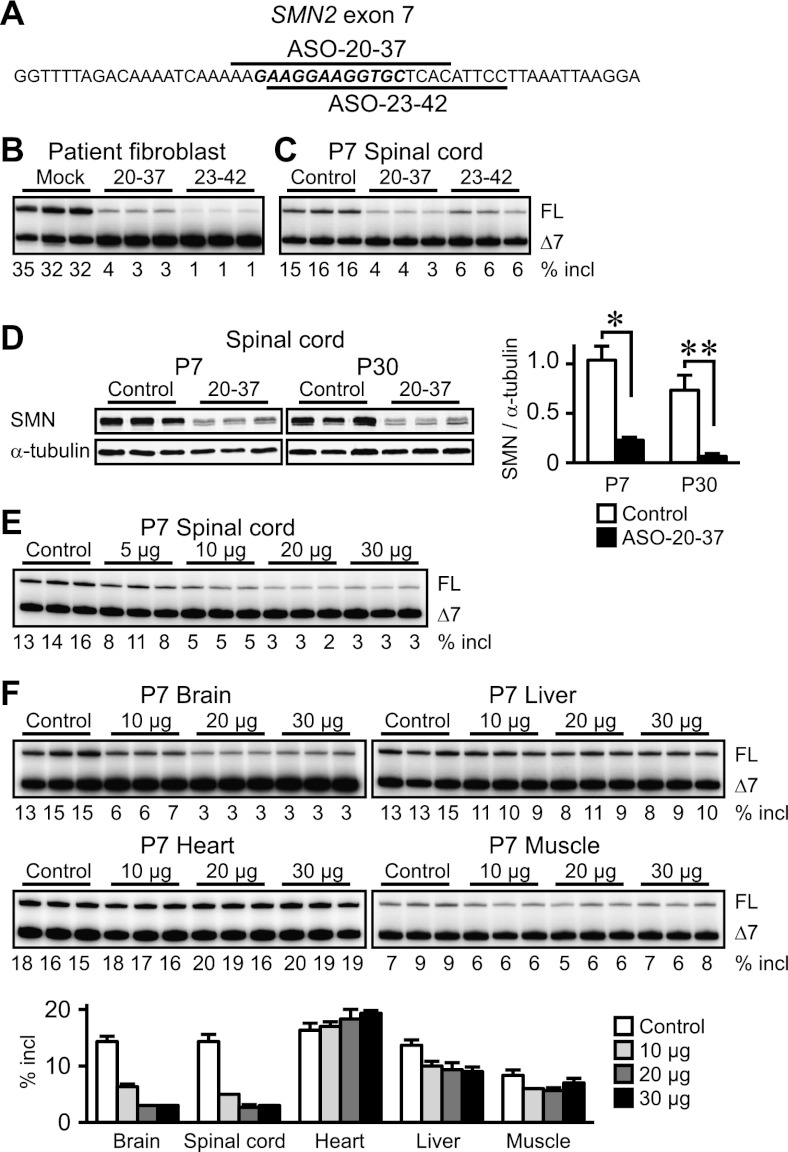
Based on our previous screen of ASOs tiled along SMN2 exon 7 (Hua et al. 2007), we designed two new ASOs: ASO-20-37 and ASO-23-42 (Fig. 1A; Supplemental Table S1). Their target sites in exon 7 comprise potent enhancer elements (Lunn and Wang 2008). We used MOE phosphorothioate ASOs, which do not trigger RNase H cleavage or RNAi (Crooke 2007). Their splicing inhibitory potencies were initially assessed by RT–PCR in type I SMA patient fibroblasts; both ASOs considerably decreased exon 7 inclusion (∼10-fold and ∼30-fold reduction by 100 nM ASO-20-37 and ASO-23-42, respectively) (Fig. 1B).
Figure 1.
ASO-induced inhibition of SMN2 splicing and SMN expression. (A) SMN2 exon 7 sequence and schematic representation of ASOs. The core splicing enhancer region is in bold italic, and bars depict ASOs complementary to the corresponding sequences. (B) RT–PCR shows strong inhibition of SMN2 exon 7 inclusion 48 h after transfection of 100 nM ASO into SMA patient fibroblasts. (C) Inhibition of exon 7 inclusion in the P7 mouse spinal cord. (D, left) Western analysis of three representative samples shows marked reduction in SMN expression at both P7 and P30. (Right) For full-length SMN, the top band in each doublet was quantitated (n = 4). (*) P = 0.0454; (**) P = 0.0049. (E) Dose response study of ASO-20-37 effects in the P7 spinal cord. (F) Dose response study of ASO-20-37 effects in P7 tissues. (FL) Full-length transcripts; (Δ7) exon 7-skipped transcripts; (% incl) the percentage of full-length transcripts in total transcripts. Control (20 μg of control ASO) or, for C and D, ASO-20-37 or ASO-23-42 (20 μg) was ICV-injected at P1. Error bars, SD.
These ASOs are specific for SMN1/2, so to determine whether we could phenocopy SMA pathology, we used them to target an SMN2 transgene in type III SMA mice (Smn−/− SMN2+/+) with normal motor function and life span (Hsieh-Li et al. 2000). We followed these phenotypes to determine whether they could be exacerbated by ASO-mediated splicing modulation. Severe SMA patients show initial symptoms during the newborn and early-infant periods (Lunn and Wang 2008). In addition, the blood–brain barrier (BBB) limits the access of systemically administered MOE ASO to the CNS (Crooke 2007; Hua et al. 2008, 2011). Therefore, we used direct administration into the cerebroventricular space in neonatal mice, which delivers ASOs via cerebrospinal fluid (CSF) to the brainstem and spinal cord, the loci of α-motor neurons (Hua et al. 2010). We used a single 20-μg (14.2 mg/kg) injection at postnatal day 1 (P1) and analyzed SMN2 splicing at P7 in the spinal cord. We focused our analysis on the thoracic cord region but observed similar effects throughout the spinal cord (Supplemental Fig. S1A). Both ASO-20-37 and ASO-23-42 potently decreased exon 7 inclusion (approximately fourfold and approximately threefold reduction, respectively), whereas a mismatch control ASO had no effect (Fig. 1C; Supplemental Fig. S1B; Supplemental Table S1). ASO-20-37 was slightly more effective than ASO-23-42, whereas the opposite was observed in patient fibroblasts (Fig. 1B), reflecting size-dependent differences in ASO uptake between cell transfection and spontaneous in vivo delivery, species-dependent pharmacokinetics, or different genetic backgrounds, cell types, and/or developmental stages, each of which can affect alternative splicing regulation (Cartegni et al. 2002). Immunoblotting revealed an approximately fivefold reduction in SMN expression at P7 by ASO-20-37 treatment (Fig. 1D).
Focusing on ASO-20-37, we found that it inhibited exon 7 inclusion in a dose-dependent manner (Fig. 1E). Because of CSF clearance, ICV-injected ASO partially distributes to the peripheral tissues (Hua et al. 2010, 2011); therefore, we analyzed its effects in other tissues. For muscle, we analyzed proximal quadriceps because motor tests pointed to a decline in hindlimb strength that limits essential behaviors (see below). Exon 7 splicing inhibition was comparable in the brain and spinal cord, whereas in peripheral tissues, ASO-20-37 had little or no effect (Fig. 1F).
MOE chemistry minimizes neuroinflammation and is well tolerated (Crooke 2007; Bennett and Swayze 2010; Hua et al. 2010), consistent with the very subtle increase in Aif1 mRNA expression—a macrophage activation marker—in the P14 spinal cord of mice injected with 20 μg of ASO-20-37 (Supplemental Fig. S2). Aif1 expression increased at P35 for the test ASO but not a control ASO, probably reflecting the disease process.
SMN2 splicing deterioration in terminal stage SMA
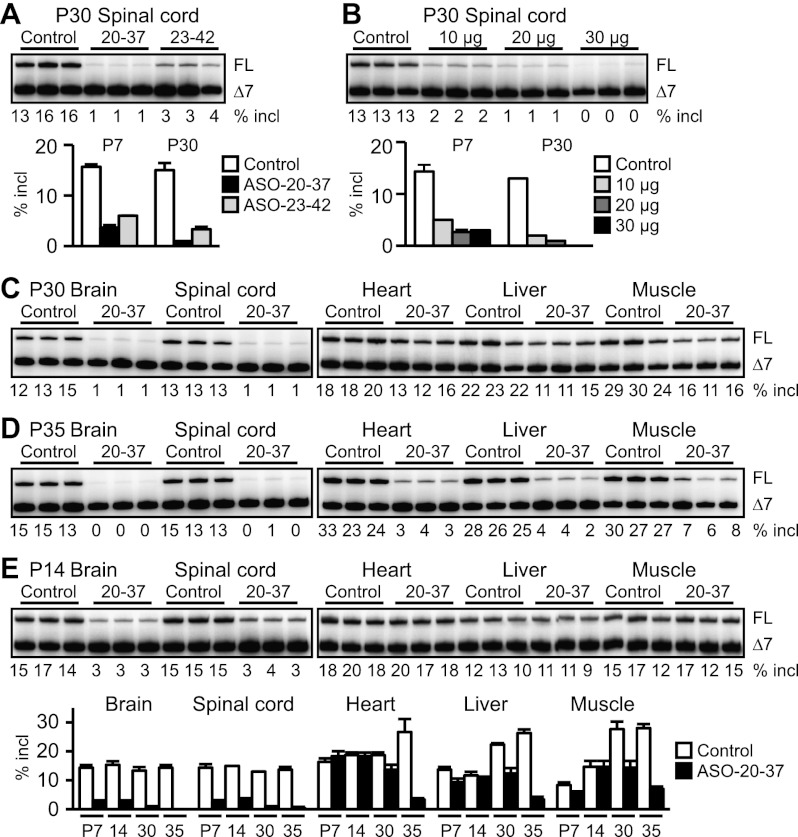
To determine the duration of the ASO effect, we measured SMN2 splicing at P30. Compared with the effects at P7, exon 7 inclusion was further suppressed in the spinal cord (∼15-fold and approximately fivefold reduction by treatment with 20 μg of ASO-20-37 and ASO-23-42, respectively) (Fig. 2A,B). Accordingly, there was a further reduction in SMN protein (∼12-fold) (Fig. 1D). In contrast to P7 mice, we observed increased missplicing in peripheral tissues at P30 (Fig. 2C) despite minimal ASO uptake (Supplemental Fig. S7B). Simultaneous skipping of exons 7 and 5 also increased in 20-μg or 30-μg ASO-20-37-treated mice (Supplemental Fig. S3A), suggesting indirect inhibition of SMN2 splicing in response to ASO treatment. Furthermore, extensive exon 7 skipping occurred systemically at P35 (Fig. 2D), the terminal stage for mice injected with 20 μg of ASO (see below). The finding that the level of SMN2 splicing at P14 (Fig. 2E) was similar to that at P7 (“20 μg” lanes in Fig. 1E,F) suggests that SMN2 splicing is subsequently suppressed at P30–P35 because of the end-stage disease state, rather than by a persistent ASO effect. Consistent with this interpretation, neonatal ICV injection of therapeutic ASO-10-27 with the same chemical modifications had reduced or no effect in the P30 spinal cord, liver, or heart compared with that at P7 (Supplemental Fig. S3B; Hua et al. 2010).
Figure 2.
Time-course RT–PCR analysis of SMN2 splicing inhibition. (A) Further inhibition of exon 7 inclusion in the P30 spinal cord. (B) Dose response study of ASO-20-37 effects at P30. For A and B, bar graphs also display P7 data from Figure 1, C and E, respectively. (C) Increased difference in SMN2 splicing level at P30 between control and ASO-20-37-treated mice. (D) Systemic exon 7 skipping in ASO-20-37-treated mice at P35. (E) SMN2 splicing level at P14 was similar to that at P7. Control (20 μg of control ASO) or, for A and C–E, ASO-20-37 or ASO-23-42 (20 μg) was ICV-injected at P1. Error bars, SD.
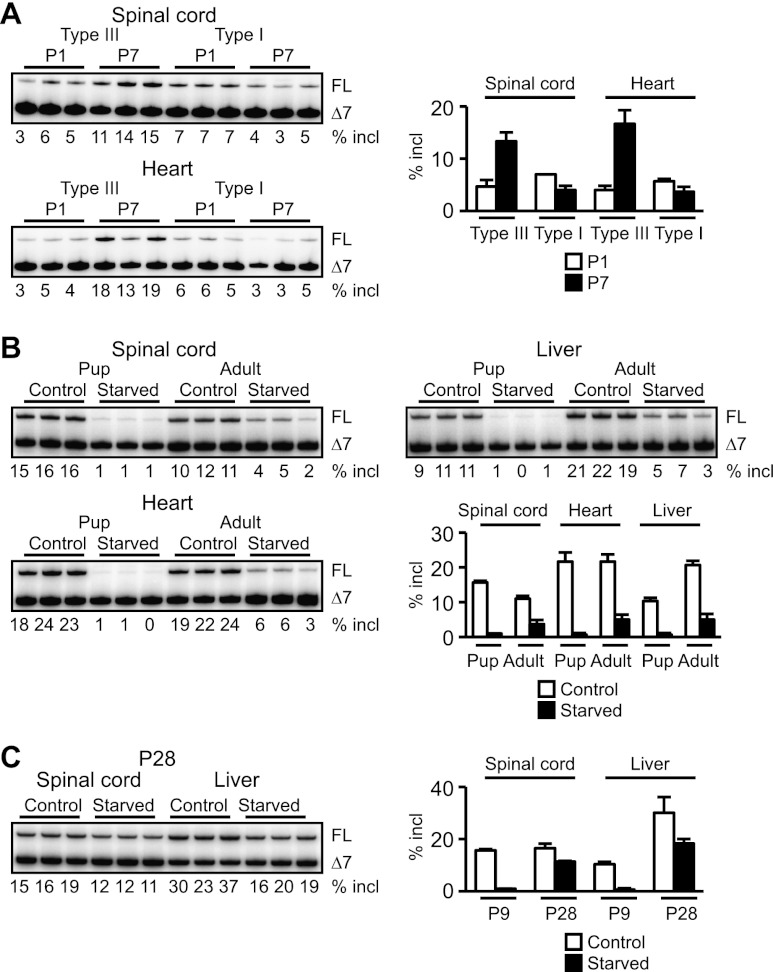
As a further test, we compared SMN2 splicing in untreated P1 or P7–P8 type I SMA mice (Smn−/− SMN2+/0) with that in type III mice (Smn−/− SMN2+/+) or healthy mice (Smn+/+ SMN2+/0 and Smn+/− SMN2+/0). At P7–P8, type I mice typically looked severely malnourished and moribund, and exon 7 inclusion in their spinal cord decreased at this time, whereas it increased in type III or healthy mice (Fig. 3A; Supplemental Fig. S4A). A similar trend was observed in the heart between type I and III mice (Fig. 3A).
Figure 3.
SMN2 splicing inhibition in end-stage SMA and as a result of starvation. (A) Changes in SMN2 splicing in the type I or type III SMA mouse spinal cord and heart between P1 and P7. (B) Forty-eight hours of fasting caused exon 7 skipping in both CNS and non-CNS tissues of type III SMA mouse pups or adults. (C) SMN2 splicing was restored at P28 once fasted pups were fed after food deprivation at P7–P9. Error bars, SD.
These results suggest that SMN2 splicing is suppressed as a result of nutritional deficiency, considering that dying SMA mice have poor food intake and extensive weight loss. To directly test this hypothesis, we subjected untreated type III mouse pups or adults to food deprivation; after 48 h, SMN2 splicing was markedly inhibited in both the CNS and peripheral tissues (Fig. 3B), especially in pups. Remarkably, when these pups were fed again, SMN2 splicing was restored, demonstrating a nutritional effect on splicing (Fig. 3C). Importantly, this inhibition of SMN2 splicing was also observed in the spinal cord of food-deprived wild-type transgenic mice (Smn+/+ SMN2+/+) (Supplemental Fig. S4B).
Widespread splicing abnormalities were reported in late stage type I mice, although whether these are the cause or consequence of SMA progression is controversial (Zhang et al. 2008; Baumer et al. 2009). We analyzed previously reported splicing alterations of Uspl1 and Chodl (Zhang et al. 2008; Baumer et al. 2009) in the spinal cord of P7 or P30 ASO-20-37-treated mice. RT–PCR demonstrated abnormal splicing of both pre-mRNAs, especially at P30 (Supplemental Fig. S5A). In contrast, splicing of these genes was not altered at P7 in normal mice (Smn+/+) treated with ASO-20-37 (Supplemental Fig. S5B). Splicing was also altered in food-deprived but otherwise untreated type III mice (Supplemental Fig. S5C), indicating that these splicing abnormalities are not a direct consequence of ASO-20-37 treatment.
ASO induction of SMA-like phenotypes
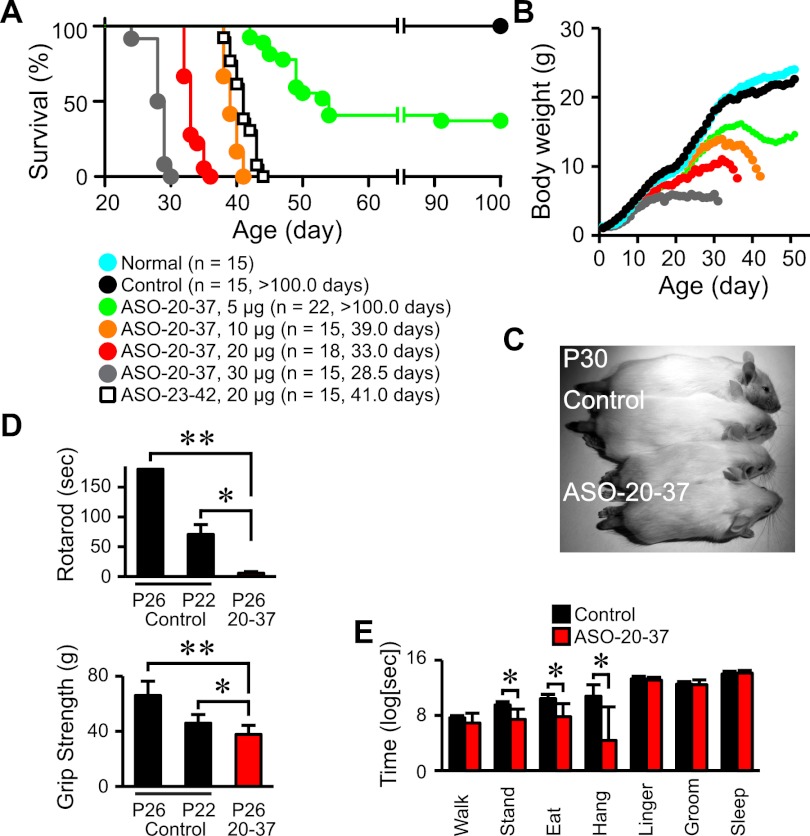
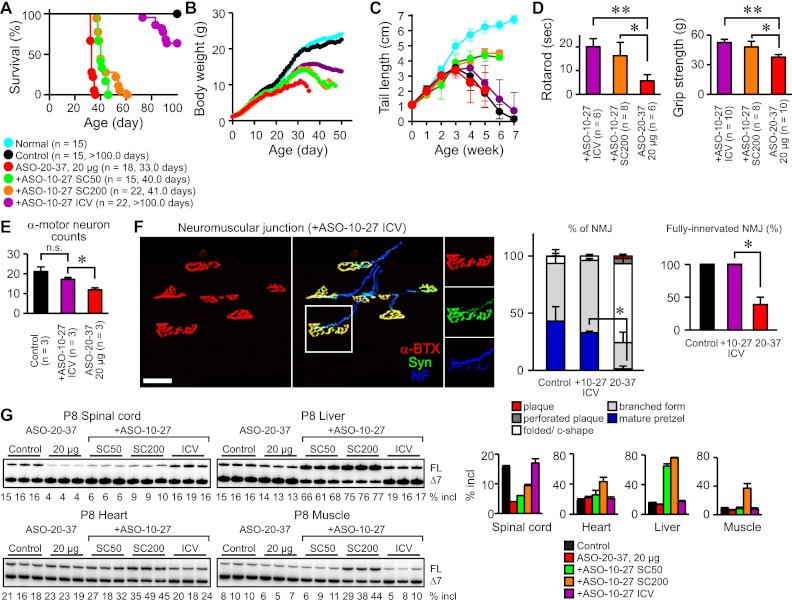
Both ASOs elicited “SMA-like” phenotypes. ICV injection of 20 μg of ASO at P1 strikingly shortened the life span of type III SMA mice, and ASO-20-37 was more potent than ASO-23-42 (median survival 33 vs. 41 d; P < 0.0001) (Fig. 4A). Focusing on ASO-20-37, we observed a dose-dependent reduction in survival and weight gain (Fig. 4A–C). There was no change in the onset or progression of tail necrosis (Fig. 6C, below). With respect to motor dysfunction—a cardinal SMA symptom—the mice exhibited a staggering gait with short strides (Supplemental Fig. S6A; Supplemental Movies) that became apparent around weaning. Dose-dependent motor impairment was evident in rotarod and grip strength tests (Fig. 4D; Supplemental Fig. S6B). Moreover, a video-based analysis of home cage behaviors showed declined locomotor activity, including decreases in standing, eating, and hanging (Fig. 4E). These physiological tests were mainly performed at P25–P26, when mice injected with 20 μg of ASO-20-37 grew large enough to perform the rotarod task. Although ASO-20-37-treated mice had a normal righting reflex until P12 (Supplemental Fig. S6C) and maintained grip strength at P15 and P22 (Supplemental Fig. S6D), once the abnormal gait occurred, their locomotor activity declined progressively, culminating in death. Both ASO-20-37- and ASO-23-42-treated mice also displayed systemic tremor (Supplemental Movies).
Figure 4.
ASO induction of SMA-like symptoms. (A) Shortened life span. The number of mice and median survival in days are given in parentheses for each group. (B) Decreased body weight gain. (C) Smaller ASO-20-37-treated mice. (D) Motor dysfunction. Rotarod task performance (n = 6; [*] P = 0.0002; [**] P < 0.0001) and grip strength (n = 10; [*] P = 0.0180; [**] P < 0.0001). P22 control mice of the same size as P26 ASO-20-37-treated mice were included. (E) Video-based analysis of home cage behaviors at P26 (n = 8). (*) P = 0.0143. (Normal) Normal mice. Control (20 μg of control ASO) or, for C–E, ASO-20-37 (20 μg) was ICV-injected at P1. Error bars, SD.
Figure 6.
Amelioration of ASO-20-37-treated mouse phenotype by therapeutic ASO-10-27. (A) Extended survival. The number of mice and median survival in days are given in parentheses for each group. (B) Increased body weight. (C) Delayed onset of tail necrosis. (D) Improved motor function. Rotarod task performance at P26 ([*] P = 0.0014; [**] P < 0.0001) and grip strength ([*] P = 0.0035; [**] P < 0.0001). (E) Retained α-motor neurons in the P30 lumbar spinal cord by ICV injection of ASO-10-27. (n.s.) P = 0.1252; (*) P = 0.0054. (F) Retained NMJs in the P30 LC. (Left) NMJs were labeled as in Figure 5D. Bar, 40 μm. (Middle) Recovered topological complexity of AChR clusters (elaborate pretzels; n = 3). (*) P = 0.0335. (Right) Intact NMJ innervation (n = 3). (*) P = 0.0003. (G) Restoration of SMN2 exon 7 splicing in P8 tissues. (+ASO-10-27 ICV) ICV injection of 20 μg of ASO-10-27 at P2; (+ASO-10-27 SC50 or SC200) SC injection of 50 or 200 μg/g per day ASO-10-27 at P2 and P3, respectively. Error bars, SD.
Supportive SMA histological features
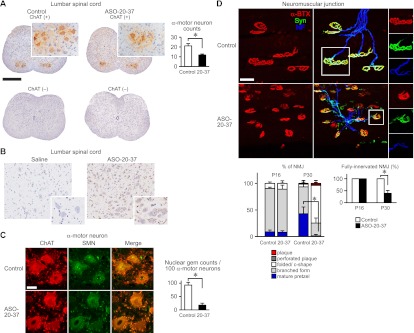
α-Motor neuron degeneration is the pathological hallmark of SMA (Lunn and Wang 2008). In ASO-20-37-treated mice, immunohistochemistry (IHC) of the lumbar spinal cord showed significant loss of choline acetyltransferase (ChAT)-positive α-motor neurons at P30 but not at P16 (Fig. 5A; Supplemental Fig. S7A). In contrast, there was a small but not significant loss in the cervical spinal cord at P30 (Supplemental Fig. S7A). IHC with antibody against the phosphorothioate backbone showed that motor neurons in the lumbar spinal cord still have internalized ASO at P30 (Fig. 5B). SMN-positive nuclear gem counts, which correlate directly with functional SMN protein level and inversely with SMA severity (Lunn and Wang 2008), decreased markedly (Fig. 5C). This SMN decrease probably triggers neuronal dysfunction and subsequent degeneration.
Figure 5.
ASO effects on mouse histopathology. (A) α-Motor neuron loss. (Left) DAB staining with or without ChAT antibody of α-motor neurons in P30 lumbar spinal cord. Bar, 500 μm. (Right) α-Motor neuron counts (n = 3). (*) P = 0.0171. (B) Detection of ASO uptake in P30 spinal cord cells, including α-motor neurons, by IHC. For A and B, α-motor neurons are displayed at higher magnification in the insets. (C) Decreased nuclear gems. (Left) Dual ChAT and SMN staining of α-motor neurons in the P30 lumbar spinal cord. Bar, 50 μm. (Right) Nuclear gem counts in α-motor neurons (n = 4). (*) P = 0.0001. (D) NMJ defects in the P30 LC. (Top) NMJs were labeled with anti-neurofilament (NF; blue) for nerves, anti-synaptophysin (Syn; green) for nerve terminals, and α-bungarotoxin (α-BTX; red) for AChRs on the postsynaptic muscle. Bar, 40 μm. (Bottom left) Reduced topological complexity of AChR clusters (elaborate pretzels; n = 3). (*) P = 0.0300. (Bottom right) Denervated NMJs at P30 but not at P16 (n = 3). (*) P = 0.0335. Control (20 μg of control ASO), saline (2 μL of saline), or ASO-20-37 (20 μg of ASO-20-37) was ICV-injected at P1. Error bars, SD.
Groups of atrophic fibers are seen in quadriceps of type I mice (Hsieh-Li et al. 2000). However, we did not observe this feature (Supplemental Fig. S7D), which would have been indicative of a prepathological stage preceding rapid disease progression (Dubowitz and Sewry 2007). Recent reports described structural and electrophysiological defects in the neuromuscular junction (NMJ) in severe SMA (Burghes and Beattie 2009). We analyzed NMJ pathology in several vulnerable muscles that are affected in SMA Δ7 mice (Ling et al. 2012). Acetylcholine receptor (AChR) clusters at the NMJ are initially plaque-shaped; later, perforated plaque, fold/C-shaped, branched, and pretzel-shaped structures become successively more prevalent as development proceeds (Kummer et al. 2004). In ASO-20-37-treated mice at P30, immunofluorescence staining for NMJs in the longissimus capitus (LC) and serratus posterior inferior (SPI) showed smaller AChR clusters with reduced topological complexity; there were fewer mature pretzel and branched forms and more of the less mature forms (Fig. 5D; Supplemental Fig. S7C). Moreover, fully innervated NMJs (see the Materials and Methods) were markedly reduced in the LC (Fig. 5D). There was a small reduction in the extent of innervation in the splenius capitus, but little or no reduction in the SPI and extensor digitorum longus (Supplemental Fig. S7C). The preservation of α-motor neuron counts, AChR topology, and NMJ innervation in the LC at P16 (Fig. 5D; Supplemental Fig. S7A) is consistent with the progressive nature of the disease. Taken together, these results show that ASO-20-37-treated mice recapitulated the characteristic severe SMA pathology.
Finally, there is cardiac involvement in severe SMA (Rudnik-Schoneborn et al. 2008; Bevan et al. 2010), but we detected no evidence of inflammation in heart sections at P30, such as cell infiltration and increased interstitial fibrosis (Supplemental Fig. S7E). At the terminal stage, however, some ASO-20-37-treated mice manifested paw and face edema and tachypnea, indicative of cardiac and/or respiratory failure, despite only faint ASO uptake in the heart, lung, and diaphragm at P30 (Supplemental Fig. S7B). To characterize the cardiac phenotype, hearts were harvested and weighed at P30. Consistent with a study of type I mice (Bevan et al. 2010), the heart mass was reduced, even when normalized to femoral length (Supplemental Fig. S7E). In addition, there was an increase in interventricular septal thickness, normalized to the heart mass (Supplemental Fig. S7E), suggesting cardiac involvement.
Amelioration of SMA-like symptoms by a therapeutic ASO
We previously reported an ASO complementary to positions +10 to +27 in SMN2 intron 7 (ASO-10-27) (Supplemental Table S1) that blocks a splicing silencer element, strongly increasing exon 7 inclusion (Hua et al. 2008). This therapeutic ASO-10-27, when administered by neonatal ICV and/or systemic subcutaneous (SC) injection, effectively rescues both type I and type III mouse phenotypes (Hua et al. 2010, 2011; Passini et al. 2011). Here we tested whether it could also rescue SMN2 splicing in ASO-20-37-treated mice and thus influence the phenotype. The binding site of therapeutic ASO-10-27 in intron 7 does not overlap with that of ASO-20-37 in exon 7, and thus these ASOs do not directly affect each other's binding to SMN2 pre-mRNA. After ICV injection of 20 μg of ASO-20-37 at P1, we administered ASO-10-27 by ICV or systemic SC injection. ICV injection of 20 μg of ASO-10-27 at P2 markedly extended the life span (median survival >100 d, P < 0.0001) (Fig. 6A) with improved weight gain (Fig. 6B) and slightly delayed onset of tail necrosis (Fig. 6C). The treatment also improved motor function (Fig. 6D) and correspondingly prevented α-motor neuron loss (Fig. 6E) and preserved normal AChR topology and NMJ innervation (Fig. 6F). In contrast, SC injection of ASO-10-27 (50 or 200 μg/g per day at both P2 and P3) only had a slight effect on survival (median survival, 50 μg/g/ per day: 40 d, P < 0.0001; 200 μg/g per day: 41 d, P < 0.0001) (Fig. 6A). However, it moderately improved weight gain and motor function (Fig. 6B,D) and markedly rescued tail necrosis (Fig. 6C). RT–PCR showed that ICV-injected ASO-10-27 restored SMN2 splicing in the spinal cord to the control level but had little or no effect in the liver, heart, and muscle, whereas SC injection only had a small effect in the spinal cord but significant effects in peripheral tissues, especially in the liver, as we previously reported (Fig. 6G; Hua et al. 2008, 2010, 2011). These results indicate that the extent of exon 7 skipping in the spinal cord is the main determinant of the SMA-like symptoms induced by ICV administration of ASO-20-37 and also support ASO-10-27 as a drug candidate.
Discussion
We applied exon-skipping ASO technology to phenocopy a motor neuron disease by postnatally inhibiting SMN2 splicing in transgenic mice. We successfully elicited phenotypes resembling core SMA symptoms, thus exacerbating the phenotype of a mild SMA mouse model. To promote exon skipping, we selected ASOs targeting splicing enhancer sequences within exon 7 of SMN2, in part to avoid interference with the target site of ASO-10-27 in intron 7. This made it possible to perform splicing rescue experiments using ASO-10-27.
Because SMA primarily affects α-motor neurons, we assumed that SMN2 splicing in the brainstem and spinal cord would be critical. We used ICV administration for direct ASO delivery to the CNS, and a single injection had significant, dose-dependent effects on both SMN2 splicing and the phenotypes (Figs. 1, 4). As with type I mice (Park et al. 2010a) or severe SMA patients (Lunn and Wang 2008; Rudnik-Schoneborn et al. 2008), growth impairment and cardiac involvement likely influenced the prognosis of ASO-20-37-treated mice (Supplemental Fig. S7). With an ∼1-mo life span and overt SMA-like phenotypes that are partly shared by Smn2B/− or SmnF7/Δ7, NSE-Cre mice (Park et al. 2010a), these mice are especially useful for analyses of disease progression, physiological tests, and therapeutic efficacy.
Although ICV-administered MOE ASOs distribute throughout the CNS, they are partly cleared out of the CNS and accumulate in peripheral tissues (Supplemental Fig. S7). However, because of a combination of low dose, ASO dilution in plasma and tissues, much shorter half-life in peripheral tissues, and renal excretion (Crooke 2007), any pharmacological effects outside the CNS are likely minimal. Indeed, ASO-20-37 predominantly inhibited SMN2 splicing in the CNS and resulted in α-motor neuron loss (Fig. 5). The nuclear gem number, an indicator of SMN abundance (Lunn and Wang 2008), decreased markedly in α-motor neurons (Fig. 5), which could cause neuronal dysfunction. On the other hand, similarly to SMA Δ7 mice (Ling et al. 2012), ASO-20-37-treated mice exhibited NMJ defects (Fig. 5). Until P16, the LC NMJs attained full innervation, and the AChR clusters continued to mature normally. The reduced complexity of AChR topology at P30 might be attributed to rapid AChR turnover and/or impairment of further maturation due to NMJ denervation (Fambrough 1979) under conditions of α-motor neuron dysfunction after P16. Alternatively or additionally, ASO-20-37 might have a direct effect in muscle that alters endplate maturity. The ASO-mediated postnatal induction of SMA pathology reveals that SMN is required for α-motor neurons after completion of their genesis around embryonic day 11 (Nornes and Carry 1978). Conversely, restoring SMN levels shortly after birth is therapeutically effective for severe SMA mice (Hua et al. 2011; Le et al. 2011; Lutz et al. 2011), consistent with a role of SMN in the postnatal development of motor neurons.
Compared with SC administration of therapeutic ASO-10-27, ICV administration considerably extended the life span and alleviated motor dysfunction of ASO-20-37-treated mice, implying that SMN2 splicing correction in the CNS is necessary and sufficient for phenotypic and histological amelioration (Fig. 6). In contrast, we previously found that neonatal SC administration of ASO-10-27 markedly extends the life span of type I mice (Hua et al. 2011). In that case, SMN restoration in peripheral tissues was necessary for efficient rescue, although an additional direct effect in the CNS was not excluded.
The apparent inconsistency between the two studies may reflect the fact that here SMN2 splicing was predominantly inhibited in the CNS, in contrast to the ubiquitous SMN2 splicing defect in type I mice (or human SMA). Another important difference is that here SMN depletion was induced postnatally, whereas it occurs embryonically in type I mice. The BBB in our mice might be more intact compared with that in type I mice, which in the present study might reduce the CNS effects of systemically administered ASO-10-27. However, the present results suggest that the SMN level in the CNS is critical for the SMA phenotype as well as its severity. On the other hand, SC administration of ASO-10-27 markedly rescued tail necrosis, whereas ICV administration only had a subtle effect, implying that peripheral SMN levels are more relevant for distal necrosis. Incomplete closure of the BBB in neonates (Stewart and Hayakawa 1987) and/or retrograde axonal transport of ASO (Crooke 2007) to spinal cord neurons might also contribute to the rescue of tail necrosis by peripheral ASO administration, which would be consistent with our previous finding that ICV administration of therapeutic ASO-10-27 rescues tail necrosis in type III SMA mice (Hua et al. 2010).
SMN depletion in motor neuron progenitors results in relatively mild SMA-like phenotypes, suggesting that other cells also contribute to SMA pathogenesis (Park et al. 2010b). As our ASOs do not exclusively target α-motor neurons (Fig. 5; Supplemental Fig. S7; Hua et al. 2010; Passini et al. 2011), their effects in other cells could also have contributing roles in this SMA model. Considering the potential developmental component of SMA, the ASO effects at different developmental stages also need to be addressed. Further analysis of the relationship between the spatial and/or temporal distribution of ASOs that promote correct or incorrect SMN2 splicing, and their pharmacological effects, should yield new insights into the roles of SMN in SMA pathogenesis as well as its normal physiological functions.
SMN2 splicing deteriorated further in dying SMA mice (Figs. 2, 3), and we demonstrated that nutritional status affected splicing even in mild type III or wild-type transgenic mice in the absence of any ASO (Fig. 3; Supplemental Fig. S4). Widespread splicing changes were reported in late stage type I mice (Zhang et al. 2008; Baumer et al. 2009), and indeed we confirmed altered splicing of two representative genes, Uspl1 and Chodl, both in our ASO-induced SMA mice—especially at the terminal stage—and in food-deprived but untreated type III mice (Supplemental Fig. S5). The splicing changes that we uncovered in SMN2 transgene transcripts during late stage SMA were not described in these studies, although in other studies, SMN2 splicing was shown to decrease in type I mice at P1 (Jodelka et al. 2010; Ruggiu et al. 2012). Dying mice likely suffer from hypoxia; hypoxic stress induces reactive oxygen species generation, which inactivates the SMN complex (Wan et al. 2008). This complex plays fundamental roles in assembling snRNPs, which are required for splicing (Burghes and Beattie 2009), and low SMN levels result in decreased SMN2 exon 7 splicing through a feedback loop (Jodelka et al. 2010; Ruggiu et al. 2012). Thus, under critical dying conditions, including malnutrition and hypoxia, SMA may progress with gradual, widespread splicing alterations in which the resulting SMN dysfunction and deficiency could be partly involved.
RNA-targeted methods for generating animal disease models have largely focused on RNAi-based or antisense knockdown approaches (Sandy et al. 2005; Crooke 2007). Compared with these approaches, our method, which we dubbed TSUNAMI (for targeting splicing using negative ASOs to model illness) retains the primary transcript, allowing the testing of therapeutics that correct missplicing. Splicing-modulating ASOs do not cause RNA cleavage and thus have fewer potential off-target hybridization effects than knockdown ASOs or RNAi. Furthermore, ICV-injected MOE ASO at the effective doses induces minimal or no chemistry-related neuroinflammation, as demonstrated by the limited extent of Aif1 mRNA expression (Supplemental Fig. S2). Indeed, our ASOs did not affect normal or heterozygote Smn+/− SMN2+/+ mice, reflecting their species-specific effects on splicing and ruling out off-target effects (Supplemental Movies). In addition, two different ASOs targeting SMN2 exon 7 elicited the same SMA-like phenotype, again ruling out off-target effects. Moreover, the phenotypic amelioration of ASO-20-37-treated mice with therapeutic ASO-10-27 definitively proved that the SMA-like phenotypes were induced through SMN2 splicing inhibition.
Although genetic strategies are also very useful for temporal and spatial regulation of SMN expression in mice (Park et al. 2010b; Le et al. 2011; Lutz et al. 2011), TSUNAMI has several advantages: (1) It circumvents the need to engineer mouse strains, which requires expertise and time-consuming genetic crosses and characterization of individual lines. (2) It provides great flexibility in fine-tuning disease severity simply by changing the ASO dose or choosing ASOs with different potencies. (3) It can help to define the relevant tissues and cell types in a given disease by taking advantage of ASO pharmacokinetic properties, which depend on the administration route, chemical modifications, and formulation. (4) It facilitates the temporal regulation of target gene expression simply by delivering the ASO at various prenatal or postnatal stages, varying the dosing frequency, or using different chemical modifications to adjust the ASO half-life. (5) It enables the testing of splicing-correcting therapeutics to prevent, delay, or rescue the disease phenotype.
Exon-skipping ASOs are being used therapeutically, notably to restore the translational reading frame in DMD (Aartsma-Rus 2010). In contrast, we demonstrate here how to use them to generate a disease model. Previously, systemically administered exon-skipping ASOs that alter Dmd splicing were shown to decrease dystrophin expression in the mouse diaphragm and were used to map functional protein domains of dystrophin (Fletcher et al. 2010). An important distinction with the present study is that DMD is mostly caused by intragenic deletions (Muntoni and Wood 2011) and not typically by missplicing. Thus, although ASO-induced missplicing might to some extent phenocopy DMD because of the reduction in full-length protein, it does not model the typical genetic defect and is therefore not suitable for testing some of the current candidate therapeutics for DMD; namely, exon-skipping ASOs and translational readthrough compounds (Muntoni and Wood 2011).
In short, we show here that a single ICV injection of ASO in neonatal mice can phenocopy a neurodegenerative disease, and disease severity can be fine-tuned through dose-dependent effects on splicing. MOE ASOs induced SMA-like symptoms and histopathology that progressed irreversibly in the absence of therapy. TSUNAMI represents a useful alternative to genetic models based on stable or conditional knockout/rescue of disease-associated genes. When transgenic animal models are available, targeting the human transgene—as we did here—offers the advantage that TSUNAMI can be used to test the same candidate therapeutics that would be used in the clinic. On the other hand, TSUNAMI should be applicable in principle to any mouse strain or any animal species, although this requires the careful development of species-specific ASOs that appropriately modulate splicing of the target gene. Although here we used exon-skipping ASOs to elicit a splicing defect, and an exon-including ASO to correct it, our method can be readily adapted to promote other types of changes in splicing, as appropriate for each disease and target gene (Kole et al. 2012). Our method should be compatible with other oligonucleotide chemistries as long as the ASOs are internalized by the appropriate cell types, are able to reach the cell nucleus, have favorable hybridization properties, do not promote RNase H cleavage or RNAi, and do not elicit substantial immune or inflammatory responses or other adverse effects. Last but not least, TSUNAMI is not at all limited to modeling CNS diseases: It should also be possible to use it to alter splicing of target genes expressed in various tissues using the appropriate administration route and, if necessary, multiple dosing.
Materials and methods
Oligonucleotide synthesis
2′-MOE oligonucleotides with phosphorothioate backbone and all 5-methylcytosines were synthesized and purified as described (Hua et al. 2010). ASOs were dissolved in saline and adjusted to the desired concentration. The ASO sequences are listed in Supplemental Table S1.
Animals
Mouse protocols were approved by Cold Spring Harbor Laboratory's Institutional Animal Care and Use Committee. Type III SMA mice (Smn−/− SMN2+/+) with four SMN2 copies were purchased from The Jackson Laboratory [strain FVB.Cg-Tg(SMN2)2Hung Smn1tm1Hung/J]. Type I SMA mice (Smn−/− SMN2+/0) harboring two SMN2 copies were generated as described (Gogliotti et al. 2010; Riessland et al. 2010). After weaning, mice were fed a normal chow diet (PicoLab Rodent Diet 20, LabDiet) and a nutrient-fortified water gel diet (DietGel Recovery, ClearH2O).
Neonatal injection
For ICV injection, P1 or P2 neonatal mice were cryo-anesthetized on ice, and 2 μL of ASO in saline was injected into the right lateral ventricle using a 5-μL microsyringe (Hamilton) and a 33-gauge needle. The coordinates for injection were 1 mm lateral from the sagittal suture, 2 mm anterior from the lambdoid suture, and 2 mm deep. ICV injections were well tolerated. Fast Green FCF (0.01% [w/v]; Sigma-Aldrich) was included in the ASO solution so that the shape of both the lateral and fourth ventricles could be visualized when the ICV injection was successful. No leakage was observed around the skull surface. For SC injection, 1.5–8.0 μL of 50 μg/g ASO in saline—with the volume adjusted according to the body weight—was injected into the dorsal skin using a 10-μL microsyringe (Hamilton) and a 33-gauge needle.
Radioactive RT–PCR and Western blotting
For each group, RT–PCR with 32P-dCTP was performed with tissues from three mice. Total RNA extraction and RT–PCR were performed to analyze SMN2 transcripts as described (Hua et al. 2007, 2008). We previously used both radioactive RT–PCR and real-time PCR to quantitate SMN2 exon 7-included and -skipped isoforms with entirely consistent results (Hua et al. 2010). To analyze Uspl1 or Chodl splicing patterns or Aif1 expression level, the following PCR primers were used: Uspl1, forward (5′-AGTTCGGGTCCACTGTATGC-3′) and reverse (5′-GGCTTACTTGGAGTGGGACA-3′), Chodl, forward (5′-AAGCCGTATCTTACAAACCAACCT-3′) and reverse (5′-CCACTTTCCTTCCTCGTGCT-3′), and Aif1, forward (5′-CAGCAATGATGAGGATCTGC-3′) and reverse (5′-GTTTCTCCAGCATTCGCTTC-3′). Gapdh mRNA levels were analyzed using the following primers: forward (5′-CGTCCCGTAGACAAAATGGT-3′) and reverse (5′-GAATTTGCCGTGAGTGGAGT-3′). PCR products were analyzed by native PAGE and PhosphorImage detection (FLA-5100, Fujifilm). Band intensities were quantitated with Multi Gauge software (Fujifilm). The signal intensity of each band was normalized according to its GC content.
Total protein was extracted from the thoracic spinal cord and analyzed by Western blotting. The blots were probed with a monoclonal antibody (mAb) specific for human SMN (SMN-KH) (Hua et al. 2010) or anti-α-tubulin mAb (Sigma-Aldrich), followed by IRDye-conjugated secondary antibody (LI-COR Biosciences). Protein bands were detected and quantitated with an Odyssey infrared imaging system (LI-COR Biosciences) as described (Hua et al. 2010, 2011).
Cell culture and transfection
Type I SMA patient fibroblasts (GM03813, Coriell Institute) were cultured as described (Hua et al. 2008). ASOs were transfected using Lipofectamine 2000 (Invitrogen).
Neurological and behavioral tests
The righting reflex was assessed at P8, P10, and P12. Pups were placed on their side, and the time elapsed until they placed all four paws stably on the ground was recorded.
A rotarod task was carried out using a RotaRodIV instrument (AccuScan). The time to fall from a 7-rpm rotating rod was measured until an arbitrary limit of 180 sec. Ten sequential trials were performed, and the longest time was recorded.
Grip strength was measured using a Grip Strength Meter (Columbus Instruments). Mouse palms were placed on a triangle bar, and once they grasped it, the mice were pulled back horizontally until they released it. Six sequential trials were performed, and the highest value was recorded.
Phenotypic characterization of mouse behavior was performed using a video-based behavior recognition platform (Steele et al. 2007). Mice were recorded throughout a 12-h dark phase, and the data were analyzed using HomeCageScan software (Clever Sys.).
Histology
The formalin-fixed, paraffin-embedded spinal cord was cut into 6-μm sections. After antigen retrieval with citrate buffer (pH 6.0), each section was incubated with goat anti-ChAT antibody (1:500; Millipore). Immunoreactivity was detected by ImmPress anti-goat IgG antibody (Vector Laboratories) and DAB substrate chromogen (Dako), followed by hematoxylin counterstaining. ChAT-positive α-motor neurons in the anterior horn with a single nucleolus within the nucleus (>350 μm2) (Friese et al. 2009) were counted in 6-μm, 24 serial sections of L1–L2 spinal cord. To ensure consistent and nonredundant counting, the total neuron counts were determined in every eighth section for three sections, and the counts of the eight groups were averaged. All of the steps from harvesting through staining of spinal cords were processed in parallel so as to reduce artifactual variation between experimental samples. ASO cellular uptake was assessed with antibody against the phosphorothioate backbone. For dual staining for SMN and ChAT, the section was incubated with anti-ChAT antibody (1:500), followed by Alexa Fluor 594 IgG antibody (Invitrogen) and anti-SMN antibody (1:500; BD Biosciences), followed by Alexa Fluor 488 IgG antibody (Invitrogen). Nuclear gems located within 100 α-motor neurons were counted in the L1–L2 spinal cord.
For NMJ staining, after perfusing and post-fixing with 4% paraformaldehyde, whole muscles were dissected and teased into layers five to 10 fibers thick. NMJs were immunolabeled with anti-neurofilament (1:2000; Chemicon) for nerves, anti-synaptophysin (1:200; Invitrogen) for presynaptic terminals, and Alexa Fluor 594-conjugated α-bungarotoxin (Invitrogen) for AChRs. The proportion of fully innervated NMJs was quantified. Fully innervated NMJs were defined by the complete overlap of presynaptic (i.e., synaptophysin) and post-synaptic (α-bungarotoxin) labeling. Endplate maturity was assessed by AChR topology, as described (Kummer et al. 2004), and the percentage of endplates with mature AChR topology was quantified. For each sample, at least 100 NMJs were evaluated from random visual fields of the whole mount. Flash-frozen quadriceps muscle and formalin-fixed heart were cut into 14- and 7-μm sections, respectively, at the level of each maximum transection for H&E staining. Maximum heart interventricular septal thickness was measured in these sections. Images were acquired with an Axio Observer.Z1 microscope and an LSM 710 confocal microscope (Carl Zeiss) for bright-field and immunofluorescence imaging, respectively.
Statistical analysis
We analyzed data using two-tailed t-tests and considered P-values ≤0.05 to be statistically significant. For analysis of home cage behaviors, we used the Friedman test, a nonparametric version of ANOVA implemented in Matlab. Kaplan-Meier survival curves were prepared with Prism 5 (GraphPad), and statistical significance was calculated with the log-rank (Mantel-Cox) test. The various histograms and plots show mean values ± standard deviation (SD).
Acknowledgments
We thank Jie Bu and Marco Passini for advice on NMJ staining, Lisa Bianco for assistance with mouse studies, Raisa Puzis and Aigoul Nourjanova for help with histological analysis, and Stephen Hearn for assistance with microscope imaging. We acknowledge support from the National Institutes of Health, the Muscular Dystrophy Association, the SMA Foundation, and the St. Giles Foundation.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.197418.112.
References
- Aartsma-Rus A 2010. Antisense-mediated modulation of splicing: Therapeutic implications for Duchenne muscular dystrophy. RNA Biol 7: 453–461 [DOI] [PubMed] [Google Scholar]
- Baumer D, Lee S, Nicholson G, Davies JL, Parkinson NJ, Murray LM, Gillingwater TH, Ansorge O, Davies KE, Talbot K 2009. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet 5: e1000773 doi: 10.1371/journal.pgen.1000773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CF, Swayze EE 2010. RNA targeting therapeutics: Molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol 50: 259–293 [DOI] [PubMed] [Google Scholar]
- Bevan AK, Hutchinson KR, Foust KD, Braun L, McGovern VL, Schmelzer L, Ward JG, Petruska JC, Lucchesi PA, Burghes AH, et al. 2010. Early heart failure in the SMNΔ7 model of spinal muscular atrophy and correction by postnatal scAAV9-SMN delivery. Hum Mol Genet 19: 3895–3905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghes AH, Beattie CE 2009. Spinal muscular atrophy: Why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci 10: 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR 2002. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat Rev Genet 3: 285–298 [DOI] [PubMed] [Google Scholar]
- Crooke ST. 2007 Antisense drug technology: Principles, strategies, and applications. CRC Press, Boca Raton, FL. [Google Scholar]
- Dubowitz V, Sewry CA 2007. Muscle biopsy: A practical approach. Elsevier, Amsterdam, Netherlands [Google Scholar]
- Fambrough DM 1979. Control of acetylcholine receptors in skeletal muscle. Physiol Rev 59: 165–227 [DOI] [PubMed] [Google Scholar]
- Fletcher S, Adams AM, Johnsen RD, Greer K, Moulton HM, Wilton SD 2010. Dystrophin isoform induction in vivo by antisense-mediated alternative splicing. Mol Ther 18: 1218–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friese A, Kaltschmidt JA, Ladle DR, Sigrist M, Jessell TM, Arber S 2009. γ and α motor neurons distinguished by expression of transcription factor Err3. Proc Natl Acad Sci 106: 13588–13593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogliotti RG, Hammond SM, Lutz C, Didonato CJ 2010. Molecular and phenotypic reassessment of an infrequently used mouse model for spinal muscular atrophy. Biochem Biophys Res Commun 391: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh-Li HM, Chang JG, Jong YJ, Wu MH, Wang NM, Tsai CH, Li H 2000. A mouse model for spinal muscular atrophy. Nat Genet 24: 66–70 [DOI] [PubMed] [Google Scholar]
- Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR 2007. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol 5: e73 doi: 10.1371/journal.pbio.0050073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR 2008. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet 82: 834–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Sahashi K, Hung G, Rigo F, Passini MA, Bennett CF, Krainer AR 2010. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev 24: 1634–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Sahashi K, Rigo F, Hung G, Horev G, Bennett CF, Krainer AR 2011. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature 478: 123–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodelka FM, Ebert AD, Duelli DM, Hastings ML 2010. A feedback loop regulates splicing of the spinal muscular atrophy-modifying gene, SMN2. Hum Mol Genet 19: 4906–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole R, Krainer AR, Altman S 2012. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov 11: 125–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummer TT, Misgeld T, Lichtman JW, Sanes JR 2004. Nerve-independent formation of a topologically complex postsynaptic apparatus. J Cell Biol 164: 1077–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, McGovern VL, Alwine IE, Wang X, Massoni-Laporte A, Rich MM, Burghes AH 2011. Temporal requirement for high SMN expression in SMA mice. Hum Mol Genet 20: 3578–3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling KK, Gibbs RM, Feng Z, Ko CP 2012. Severe neuromuscular denervation of clinically relevant muscles in a mouse model of spinal muscular atrophy. Hum Mol Genet 21: 185–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn MR, Wang CH 2008. Spinal muscular atrophy. Lancet 371: 2120–2133 [DOI] [PubMed] [Google Scholar]
- Lutz CM, Kariya S, Patruni S, Osborne MA, Liu D, Henderson CE, Li DK, Pellizzoni L, Rojas J, Valenzuela DM, et al. 2011. Postsymptomatic restoration of SMN rescues the disease phenotype in a mouse model of severe spinal muscular atrophy. J Clin Invest 121: 3029–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntoni F, Wood MJ 2011. Targeting RNA to treat neuromuscular disease. Nat Rev Drug Discov 10: 621–637 [DOI] [PubMed] [Google Scholar]
- Nornes HO, Carry M 1978. Neurogenesis in spinal cord of mouse: An autoradiographic analysis. Brain Res 159: 1–6 [DOI] [PubMed] [Google Scholar]
- Park GH, Kariya S, Monani UR 2010a. Spinal muscular atrophy: New and emerging insights from model mice. Curr Neurol Neurosci Rep 10: 108–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park GH, Maeno-Hikichi Y, Awano T, Landmesser LT, Monani UR 2010b. Reduced survival of motor neuron (SMN) protein in motor neuronal progenitors functions cell autonomously to cause spinal muscular atrophy in model mice expressing the human centromeric (SMN2) gene. J Neurosci 30: 12005–12019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini MA, Bu J, Richards AM, Kinnecom C, Sardi SP, Stanek LM, Hua Y, Rigo F, Matson J, Hung G, et al. 2011. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci Transl Med 3: 72ra18 doi: 10.1126/scitranslmed.3001777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riessland M, Ackermann B, Forster A, Jakubik M, Hauke J, Garbes L, Fritzsche I, Mende Y, Blumcke I, Hahnen E, et al. 2010. SAHA ameliorates the SMA phenotype in two mouse models for spinal muscular atrophy. Hum Mol Genet 19: 1492–1506 [DOI] [PubMed] [Google Scholar]
- Rudnik-Schoneborn S, Heller R, Berg C, Betzler C, Grimm T, Eggermann T, Eggermann K, Wirth R, Wirth B, Zerres K 2008. Congenital heart disease is a feature of severe infantile spinal muscular atrophy. J Med Genet 45: 635–638 [DOI] [PubMed] [Google Scholar]
- Ruggiu M, McGovern VL, Lotti F, Saieva L, Li DK, Kariya S, Monani UR, Burghes AH, Pellizzoni L 2012. A role for SMN exon 7 splicing in the selective vulnerability of motor neurons in spinal muscular atrophy. Mol Cell Biol 32: 126–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandy P, Ventura A, Jacks T 2005. Mammalian RNAi: A practical guide. Biotechniques 39: 215–224 [DOI] [PubMed] [Google Scholar]
- Steele AD, Jackson WS, King OD, Lindquist S 2007. The power of automated high-resolution behavior analysis revealed by its application to mouse models of Huntington's and prion diseases. Proc Natl Acad Sci 104: 1983–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PA, Hayakawa EM 1987. Interendothelial junctional changes underlie the developmental ‘tightening’ of the blood–brain barrier. Brain Res 429: 271–281 [DOI] [PubMed] [Google Scholar]
- Wan L, Ottinger E, Cho S, Dreyfuss G 2008. Inactivation of the SMN complex by oxidative stress. Mol Cell 31: 244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G 2008. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133: 585–600 [DOI] [PMC free article] [PubMed] [Google Scholar]