Abstract
Cilia are organelles found on most eukaryotic cells, where they serve important functions in motility, sensory reception, and signaling. Recent advances in electron tomography have facilitated a number of ultrastructural studies of ciliary components that have significantly improved our knowledge of cilium architecture. These studies have produced nanometer‐resolution structures of axonemal dynein complexes, microtubule doublets and triplets, basal bodies, radial spokes, and nexin complexes. In addition to these electron tomography studies, several recently published crystal structures provide insights into the architecture and mechanism of dynein as well as the centriolar protein SAS-6, important for establishing the 9-fold symmetry of centrioles. Ciliary assembly requires intraflagellar transport (IFT), a process that moves macromolecules between the tip of the cilium and the cell body. IFT relies on a large 20-subunit protein complex that is thought to mediate the contacts between ciliary motor and cargo proteins. Structural investigations of IFT complexes are starting to emerge, including the first three‐dimensional models of IFT material in situ, revealing how IFT particles organize into larger train-like arrays, and the high-resolution structure of the IFT25/27 subcomplex. In this review, we cover recent advances in the structural and mechanistic understanding of ciliary components and IFT complexes.
Abbreviations: 2D, two‐dimensional; 3D, three‐dimensional; DIC, differential interference contrast; DRC, dynein regulatory complex; EM, electron microscopy; ET, electron tomography; IDA, inner dynein arm; IFT, intraflagellar transport; MT, microtubule; MTBD, microtubule binding domain; ODA, outer dynein arm; RS, radial spoke; RSP, radial spoke protein
Keywords: cilium, intraflagellar transport, electron tomography, IFT complex, flagellum
Graphical Abstract
Highlights
► Electron tomographic structures of axonemal components. ► High‐resolution structures of dynein and SAS-6. ► Electron tomographic reconstruction of IFT particles. ► High‐resolution crystal structure of IFT complex 25/27.
Introduction
The presence of specialized organelles for specific cellular functions is one of the hallmarks of eukaryotic cells. One of these organelles is the cilium/flagellum (interchangeable terms), a membrane-surrounded cellular extension found on many types of eukaryotic cells, with the notable exceptions of fungi and higher plants. Cilia function in a wide variety of biological processes, including motility, sensory reception, and signaling.1 The basic design of the cilium is conserved throughout evolution and consists of a microtubule (MT)-based internal structure (the axoneme), which grows from a modified centriole (the basal body), and an overlying ciliary membrane (Fig. 1). The MTs of the axoneme are highly organized, with the two most common axoneme architectures being ‘9 + 2’ (9 peripheral MT doublets and 2 single central MTs) and ‘9 + 0’ (only 9 peripheral MT doublets).
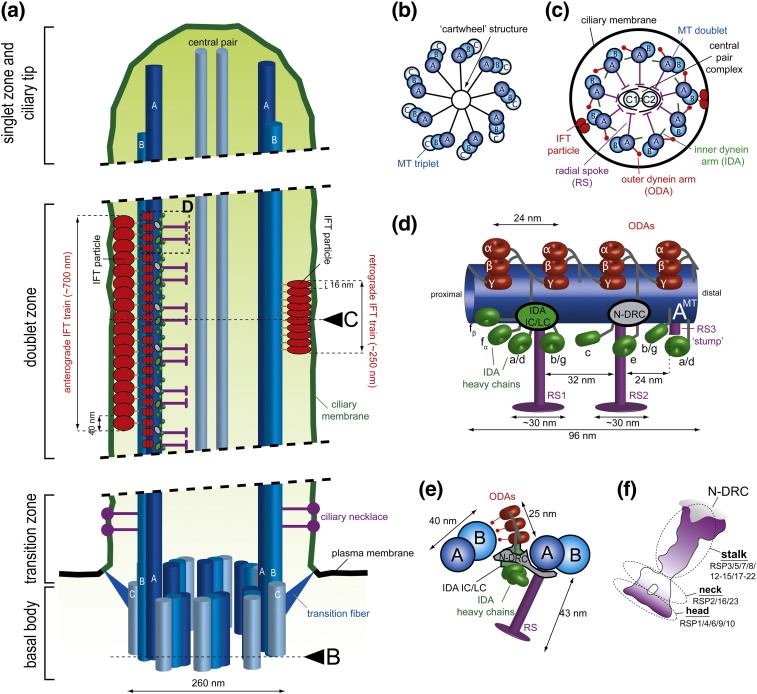
Fig. 1.
Simplified representations of a cilium and various axonemal structures. (a) Longitudinal section through a motile cilium. The various regions of the cilium are indicated on the left (for detailed information on these regions, the reader is referred to a recent review by Fisch and Dupuis-Williams2). The A and B MTs of the basal body triplets are extended to form the outer doublets of the axoneme. Motility-related axonemal structures are attached to the A-tubule. IFT particles move between the B-tubule and the overlying ciliary membrane. A long anterograde IFT train (moving towards the ciliary tip) is shown on the left; a short retrograde IFT train (moving back to the base) is shown on the right. These two types of trains differ not only in their overall length but also in the length of their repeating units. Triangles marked with B and C indicate the planes of cross sections shown in (b) and (c), respectively. The dashed black rectangle indicates the region shown in detail in (d). (b) Cross section through the proximal part of the basal body. The MTs in the triplets are designated as A, B, and C, with the A-tubule being closest to the center. The ‘cartwheel’ structure can be found within the ring of triplets and connects to the A-tubules. (c) Cross section through the doublet zone of the ciliary axoneme. Nine MT doublets form a ring that is connected to the central pair complex by RSs. Outer dynein arms (ODAs) and inner dynein arms (IDAs) extend from the A-tubule towards the B-tubule of the adjacent doublet. Two IFT trains are shown, moving between the B-tubule and the overlying ciliary membrane. (d) Schematic illustration of data from ET on the structure of the 96‐nm axonemal repeat. ODAs, represented by the three dynein heavy chains (α, β, γ), are shown as red circles. IDA heavy chains are shown as green circles and the IDA intermediate and light chains (IDA IC/LC) are shown as a green oval. The gray oval indicates the location of the nexin/dynein regulatory complex (N-DRC). Two RSs (RS1 and RS2) are represented in purple and extend towards the central pair complex. An additional ‘stump’ (RS3 ‘stump’), as observed in C. reinhardtii flagella, is also indicated as a short purple structure. (e) View of the structure shown in (d) from distal to proximal. Colors and shapes of the various structures are the same as in (d). (f) The RS consists of three distinct regions (head, stalk, and neck). The RS proteins (RSPs) found in each region are indicated.
The 9 + 2 and 9 + 0 MT patterns are usually found in motile and immotile cilia, respectively, although there are exceptions to this rule, such as the motile 9 + 0 cilia in the mouse embryonic node3 and the non-motile 9 + 2 kinocilia of fish.4 A better indicator for motile cilia is the presence of additional axonemal structures required for motility, such as axonemal dyneins. Motile cilia carry out a number of functions, including the movement of protists towards environmental cues (e.g., light or nutrients), the locomotion of sperm cells towards the egg, the directed movement of extracellular material (e.g., the removal of mucus from the respiratory tract by multi-ciliated cells), and sensory reception.5 Immotile cilia function primarily as cellular sensors of the extracellular environment and can detect mechanical forces (e.g., fluid flow in kidney tubules), signaling molecules (e.g., hedgehog ligands and growth factors), chemical compounds (e.g., olfactory molecules), or physical cues (e.g., light on the retina).6
Almost all types of cilia are assembled by a highly conserved transport pathway called intraflagellar transport (IFT), the bidirectional movement of proteinaceous particles from the base to the tip of the cilium (anterograde transport) and from the tip back to the base (retrograde transport).7,8 This transport was first discovered by Kozminski et al.9 and is carried out by the kinesin 210–12 and cytoplasmic dynein 2/1b (also known as IFT dynein) motors,13–16 which provide the force for movement along the MTs of the axoneme. This transport is mediated by IFT complexes, which consist of at least 20 different proteins and are believed to act as adaptors between the motors and cargoes.10 Interest in ciliary research increased dramatically when it was shown that the perturbation of ciliary assembly by mutating IFT components can cause polycystic kidney disease.17 Since this landmark publication, several groups have identified ciliary defects (both in ciliary assembly and in ciliary motility) as the underlying cause for a growing number of human genetic diseases, which are now commonly referred to as ‘ciliopathies’.18–23
Early structural studies of cilia were primarily carried out using conventional transmission electron microscopy, yielding two‐dimensional (2D) projections of cilia architecture. However, to understand how the components of this sophisticated machinery work together to produce the coordinated ciliary beat, it became necessary to visualize how axonemal structures are positioned relative to each other in three dimensions. Recent advances in electron tomography (ET)24 have enabled researchers to characterize the three‐dimensional (3D) architecture of axonemal structures in situ from cryo-preserved flagella of Chlamydomonas reinhardtii and sea urchin sperm (see Table 1). In addition, the first 3D view of IFT particles in situ was recently acquired by ET of plastic-embedded flagella.38 Other structural biology methods, such as X-ray crystallography and nuclear magnetic resonance (NMR) have also begun to contribute important high-resolution information for reconstituted complexes. In this review, we summarize recent structural studies on cilia and IFT that provide novel insights into the overall organization of MT-associated structures of the axoneme, the structure of IFT particles, and the molecular basis for the 9-fold symmetry of basal bodies.
Table 1.
Summary of the recent structural studies of flagellar components using cryo‐ET
| Organism | Strain(s) | Missing component(s) | Focus | Resolution (Å) | Reference |
|---|---|---|---|---|---|
| Sea urchin | wt | — | Axonemal arrangement | 58 | Nicastro et al.25 |
| C. reinhardtii Sea urchin | wt | — | ODAs/IDAs, MT inner proteins (MIPs) | ~ 40 | Nicastro et al.26 |
| pf9-3 | I1 inner arm complex | ||||
| wt | — | ||||
| C. reinhardtii | wt | — | ODAs | 45 | Ishikawa et al.27 |
| oda11 | ODAα | ||||
| oda4-s7 | ODAβ (head) | ||||
| C. reinhardtii | wt | — | IDAs | 41 | Bui et al.28 |
| oda1 | ODA | 38 | |||
| ida1 | Dynein f | ||||
| ida2–6 | Dynein f (β) | ||||
| ida4 | Dynein a, c, d | ||||
| ida5 | Dynein a, c, d, e | ||||
| ida9 | Dynein c | ||||
| pf2 | Dynein b(−), e(−), DRC (partial) | ||||
| pf3 | Dynein e, DRC3/4/5/6/7 | ||||
| sup-pf3 | DRC3(−)/4(−)/5/6/7(−), dynein e(−) | ||||
| C. reinhardtii | wt | — | IDAs, outer arm docking complex, asymmetrical arrangement of axonemal components | 41 | Bui et al.29 |
| oda4, 4-s7, 1, 11 | ODAβ, ODA, ODAα | ||||
| pf3 | Dynein e, DRC3/4/5/6/7 | ||||
| oda1 | ODA, DC | ||||
| ida4 | Dynein a, c, d | ||||
| C. reinhardtii | wt | — | DRC, nexin | 40 | Heuser et al.30 |
| p-wt (PF2-GFP) | — | 33 | |||
| sup-pf4 | DRC5/6 | 38 | |||
| sup-pf3 | DRC3(−)/4(−)/5/6/7(−), dynein e(−) | 37 | |||
| pf2–4 | DRC1/2/3(−)/5/6/7(−), RSP13d, dynein e | 39 | |||
| pf3 | DRC3/4/5/6/7 | 38 | |||
| C. reinhardtii | wt | — | ODA/IDA conformational change | 39 | Movassagh et al.31 |
| C. reinhardtii | p-wt (PF2-GFP) | — | MIPs | 34 | Nicastro et al.32 |
| p-wt (PF2-GFP) | — | 34 | |||
| p-wt (PF2-GFP) | — | 39 | |||
| pf3 | Dynein e, DRC3/4/5/6/7 | 33 | |||
| ida6 | Dynein e | ||||
| 4d6 | CaM-IP2(−) | ||||
| Sea urchin | wt | — | 36 | ||
| Isolated doublet | — | 38 | |||
| C. reinhardtii | wt | — | RSs | 39 | Pigino et al.33 |
| pf1 | RSP1/4/6/9/10 | ||||
| pf24 | RSP1-23 | ||||
| pf14 | RSP1/2/4/6/9/10/16/23(−) | ||||
| ida4 | Dynein a, c, d | ||||
| T. thermophila | wt | — | |||
| C. reinhardtii | wt | — | Basal body triplet | 33 | Li et al.34 |
| C. reinhardtii | wt | — | RSs | 39 | Barber et al.35 |
| p-wt (PF2-GFP) | — | 33 | |||
| pf17 | RSP1/4/6/9/10 | 40 | |||
| Sea urchin | wt | — | RSs | 36 | Lin et al.36 |
| T. thermophila | wt | — | |||
| C. reinhardtii | wt | — | MIPs | 34 | Pigino et al.37 |
| T. thermophila | wt | — | 42 | ||
| Sea urchin | wt | — | 43 | ||
| C. reinhardtii | wt/pWT | — | dynein f | 39/31 | Heuser et al, 2012; in press |
| bop5-1 | Intermediate chain/light chain complex (ICLC) (−) | 38 | |||
| bop5-2 | ICLC (−) | 38 | |||
| pf9-3 | dynein f, ICLC tether head | 35 |
(−), reduced amounts of the protein are expressed.
Visualizing Cilia by Electron Microscopy
Although cilia were discovered on protists soon after the development of the light microscope,39 their internal structure remained invisible to researchers until important advances in electron microscopy (EM) in the middle of the 20th century. Since then, a large number of studies have focused on defining the exact axoneme structure as well as the distinct regions of cilia such as the transition zone, the basal body, and the ciliary tip (extensively discussed in a recent review, see Ref. 2). EM studies in the early 1950s revealed ‘fibrils’ with a 9 + 2 arrangement in axonemes of different ciliated cell types.40–42 Fawcett and Porter also drew attention to the fact that the diameter of the central fibrils was different from that of the peripheral fibrils (MT singlets versus doublets). A later study identified these fibrils as MTs,43 with similar properties to the ‘fibrils’ of the mitotic apparatus. The two adjacent MTs in the peripheral doublets (the A- and B-tubules) and the associated dynein arms and radial spokes (RSs) (see Fig. 1) were detailed in separate studies.44,45 It was shown that glycerol-extracted cilia from Polytoma uvella become motile upon ATP treatment,46 and elegant work by Gibbons subsequently revealed that the ATPase activity of axonemes resides in the ‘dynein arms’.47 Removal of these structures resulted in axoneme immotility, and in a convincing demonstration, both ATPase activity and axonemal beating were restored after adding back the extracted dynein arms to the axoneme.47 Furthermore, the fractionation of outer dynein arms (ODAs) revealed 14S and 30S complexes with similar ATPase activity but distinct structures corresponding to monomeric and multimeric ODAs, respectively.48 It was later shown that axoneme motility is achieved by dynein-mediated sliding of adjacent MT doublets relative to each other.49,50 The activities of individual dyneins along the length of the cilium, as well as around its circumference, need to be regulated in order to move in a controlled fashion. Work from several groups has shown that additional axonemal structural elements, such as nexin links, the dynein regulatory complex (DRC), and RSs, play critical roles in this regulation (reviewed in Ref. 51).
These early EM studies, as important as they were in the initial structural characterization of cilia, were technically limited primarily by the need for chemical fixation methods that limit resolution and can distort the structures of the macromolecules under investigation. Current EM studies have benefited from advances in sample preparation, such as the freezing techniques that preserve the sample in a near-native state (see chapter by Nicastro in Ref. 52). Additionally, modern high-voltage electron microscopes equipped with field-emission guns allow electron beams to penetrate thicker specimens encountered during ET (e.g., axonemes and basal bodies), where sample thickness increases at the high tilt angles required for 3D reconstructions. Electron microscopes used for the acquisition of such tilt series are equipped with an energy filter that removes noise caused by inelastic electron scattering. Furthermore, high‐sensitivity recording devices such as CCD and direct detection cameras have helped in the retention of the high-resolution information. Together with significant improvements in automatic data collection and processing,24 these advances have resulted in numerous high‐quality 3D reconstructions of ciliary structures by ET (summarized in Table 1), including MT doublets and triplets, dynein and RS complexes, and IFT particles.
General Structural Organization of Cilia: The MT Backbone
MT filaments are an integral component of the cellular cytoskeleton and play key roles in a number of processes, including intracellular transport, cell motility, and cell division. Therefore, much effort has gone into understanding MT structure (for a comprehensive review on cryo-EM of tubulin and MTs, see Ref. 53). MTs are dynamically built in cycles of polymerization and depolymerization from αβ-tubulin heterodimers, forming a repetitive helical tube-like structure most commonly containing 13 protofilaments.54 The first structures of αβ-tubulin that allowed the polypeptide chains to be traced (3.5 Å resolution) were obtained by electron crystallography on zinc-induced 2D crystals.55,56 These structures revealed the overall architecture of αβ-tubulin heterodimers as well as the binding sites for guanine nucleotides and the MT stabilizing chemical taxol.57 Subsequent X-ray crystallographic analysis of αβ-tubulin bound to the dimer-sequestering protein RB3-SLD (also at 3.5 Å resolution) showed a curved conformation of the tubulin heterodimer incompatible with incorporation into MT filaments.58,59 For studying the structure of assembled MTs, helical reconstruction techniques as well as methods based on a combination of single‐particle reconstruction and crystallography have been very successful in producing high‐quality maps.53 Around the same time that the molecular structure of tubulin was determined, cryo-EM maps of MTs at ~ 20 Å resolution became available, which was sufficient to assemble a pseudo-atomic MT model.60 Subsequent studies have improved the resolution of the MT reconstructions to about 8 Å, which allows for accurate fitting of atomic coordinates, providing molecular models that reveal the intermolecular interactions between tubulins (and various bound factors) in polymerized MTs.61–63
However, MTs in cilia exist not only as single filaments but also as doublets (A‐ and B-tubules) and triplets (A‐, B‐, and C-tubules, see Fig. 1). Early EM analysis of isolated C. reinhardtii flagella revealed that the A-tubule is a complete MT containing 13 protofilaments, whereas the B-tubule is incomplete and contains only 10 protofilaments.64,65 More recently, cryo-ET has provided 3D reconstructions of MT doublets32,66 and triplets,34 confirming that both B- and C-tubules contain 10 protofilaments (discussed in the following sections).
General Structural Organization of Cilia: Basal Bodies and their 9-Fold Symmetry
Centrioles are large macromolecular structures that serve a number of important cellular functions involving the organization of MTs.67 During cell division, centrioles recruit pericentriolar material to form centrosomes, which facilitate assembly of the bipolar spindle, required for accurate segregation of chromosomes to the daughter cells.67 During interphase, centrioles migrate to the cell periphery and anchor underneath the plasma membrane, where they are referred to as basal bodies and serve as a template for cilia formation. Here, the basal body is a nucleation point for MT doublets that grow out of the cell body underneath an extension of the membrane to form a hairlike projection known as the cilium (Fig. 1). In a recent cryo-ET study, the structure of purified C. reinhardtii basal bodies was determined by subtomogram averaging, resulting in a high‐quality map of MT triplets at 3.3 nm resolution.34 This cryo-ET structure enabled visualization of individual protofilaments and docking of αβ-tubulin into the map, thus providing an atomic model for MT triplets.34 Once the MT portion of the electron tomogram was accounted for, significant densities remained, primarily towards the center of the basal body ring. These densities included a long tail projecting from the C-tubule and, most prominently, a large (estimated molecular mass of 1.1 MDa) Y-shaped structure linking the A- and B-tubules.34 Although this Y-shaped density could have a role in stabilizing the A- and B-tubules, its exact function in the basal body is currently unknown.
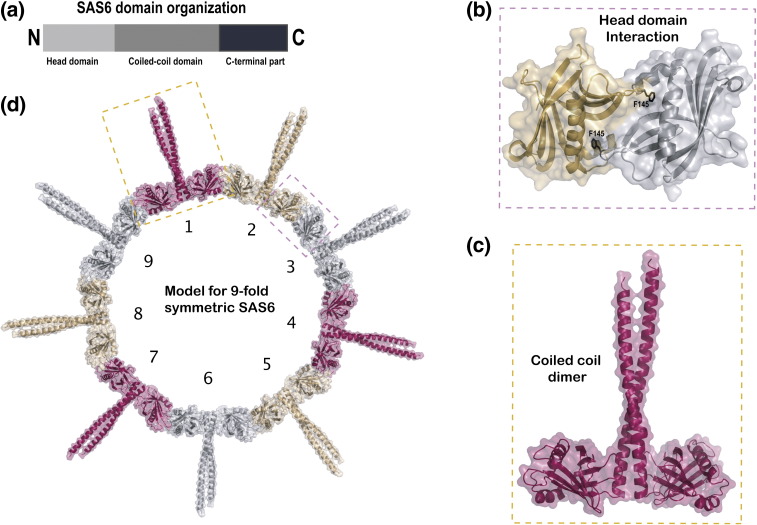
Basal body biogenesis is an intricate process and C. reinhardtii basal bodies contain about 50 different proteins in addition to tubulins.68 Early biogenesis is known to rely on the formation of a cartwheel structure (see Fig. 1b), which is an important determinant of the basal body's 9-fold symmetry.69 The 3D structure by Li et al. did not reveal anything about the cartwheel as this complex was presumably lost during the purification.34 However, recent structural analyses of the centriolar protein SAS-6 have provided novel insights into the molecular basis of the basal body's 9-fold symmetry.70,71 The depletion of SAS-6 has previously been shown to result in centrioles with an aberrant number of MTs in C. reinhardtii,72 Drosophila,73 and Paramecium.73 In C. reinhardtii, the null mutant for SAS-6 is known as bld12 (bald cells), as 90% of the cells are not flagellated.72 Two independent studies have now revealed the atomic structure of different truncated SAS-6 constructs.70,71 SAS-6 contains an N-terminal head domain, a central coiled-coil region, and a non-conserved C-terminal region (Fig. 2a). Crystal structures were determined for the N-terminal head domain of SAS-6 from Danio rerio,70 Caenorhabditis elegans, and C. reinhardtii71 (Fig. 2b). The structures show that two head domains interact via the insertion of a hydrophobic residue (Ile or Phe depending on the organism) from one monomer into a hydrophobic pocket of the neighboring monomer (Fig. 2b). Although this interaction is rather weak (Kd ~ 60–110 μΜ), it is biologically significant as mutation of the hydrophobic residue prevents rescue following RNAi knockdown in C. elegans71 and causes mislocalization of SAS-6 in human cells.70 In addition to the structure of the SAS-6 head domain, both research groups also determined structures of longer constructs encompassing portions of the central coiled-coil domain. These structures reveal an additional interaction via dimerization of the coiled-coil helices from neighboring monomers (Fig. 2c). This coiled-coil dimer appears to be rather stable, with a Kd of 0.9 μM.71 The two different structures (Fig. 2b and c) provide important insights into how the 9-fold symmetry of basal bodies could be established, as models taking both structures into account result in ring‐shaped assemblies most consistent with 9-fold symmetry (Fig. 3d).70,71 Furthermore, both research groups showed by EM that SAS-6 constructs containing both head and coiled-coil domains indeed display the propensity for ring formation in vitro, thus corroborating the model. Although these structural studies go a long way in explaining how SAS-6 self-assembles to dictate the 9-fold symmetry of centrioles and basal bodies, other factors appear to be required for faithful and stable formation of 9-fold symmetric rings.70 Future studies will likely focus on how additional proteins associate with SAS-6 during basal body biogenesis.
Fig. 2.
SAS-6 crystal structures: molecular basis for the 9-fold symmetry of centrioles. (a) Schematic representation of the domain organization of SAS-6. The protein contains an N-terminal head domain, a central coiled-coil domain, and a C-terminal region that is not well conserved in sequence. (b) Crystal structure of the N-terminal head domain of SAS-6 from C. reinhardtii. Each monomer is shown in a different color and the phenylalanine (F145) crucial for dimer formation is labeled and shown in black stick representation. (c) Crystal structure of a longer N-terminal construct of SAS-6 from C. reinhardtii [compared to (b)] containing part of the central coiled-coil domain. This structure shows a second dimerization interface mediated by the coiled coil. (d) Model for 9-fold symmetrical multimerization of SAS-6. Modeling of the two dimer structures shown within the dashed rectangles of (b) and (c) results in a ring-like structure with exactly nine copies of SAS-6 dimers. Such an arrangement likely provides a molecular rationale for the 9-fold symmetry of centrioles. All structures shown in this figure were determined from C. reinhardtii proteins.
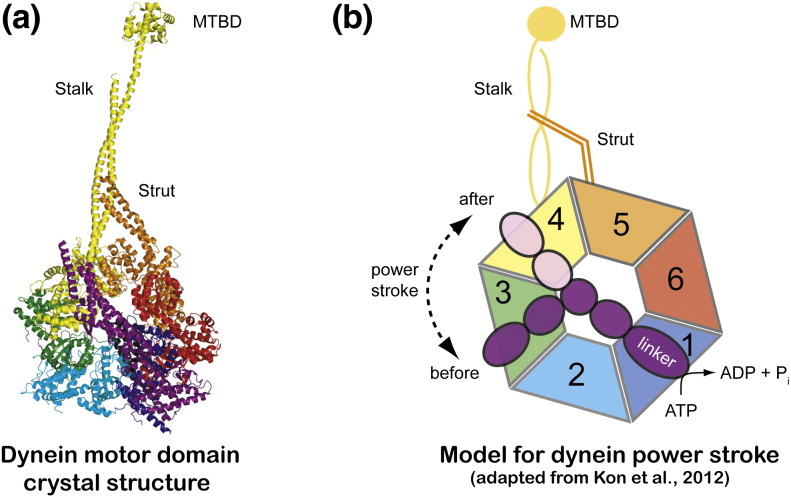
Fig. 3.
Dynein motor domain crystal structure. (a) Crystal structure of the dynein motor domain from Dictyostelium discoideum shown in cartoon representation. The various structural elements such as stalk, strut, linker region, and AAA domains are shown in different colors. (b) Model for the dynein power stroke (drawn based on Fig. 5 from Kon et al.74). The large conformation change in the linker region during power stroke is indicated by dark purple (pre-power stroke) and light purple (post-power stroke) colors. The AAA1–6 domains are labeled 1–6. MTBD, microtubule binding domain.
Periodical Arrangement (96 nm) of Axonemal Complexes
Because of the repetitive nature of the MT doublets, associated axonemal proteins often display repetition along the axoneme. A periodicity of 96 nm is found for many of the axonemal complexes involved in ciliary motility75,76 with an exception of asymmetric components as discussed below. This feature is advantageous for structural studies of the cilium as it facilitates averaging of cryo-ET reconstructions by extracting subtomograms every 96 nm along a doublet MT. However, one caveat to this method is that heterogeneous components not obeying the 96‐nm repeat structure will be missed in the analyses. Nevertheless, this averaging technique results in a substantial increase in signal-to-noise ratio and, thus, resolution and have proven to be very powerful in resolving the molecular organization of axonemal dyneins, RSs, nexins, and other cilia-related components, as discussed in the following sections.
Dynein
Dyneins are motor proteins responsible for MT-based intracellular transport and the force generation required for motile cilia beating and wave formation.77,78 Dynein complexes consist of heavy chains containing six AAA + (ATPases associated with diverse cellular activities) domains, together with several intermediate, light-intermediate, and light chains. The dynein heavy chain contains the actual motor domain responsible for force generation and MT binding, whereas the intermediate and light chains regulate dynein activity and mediate cargo binding. Currently, 54 polypeptides have been identified as flagellar dynein complex components or as factors required for dynein complex assembly and/or transport into the flagella of C. reinhardtii.79 Analyses of dynein mutants have revealed that each dynein species appears to play a specific role for the orchestrated wave formation of motile cilia.77 Axonemal dyneins occupy the space between neighboring MT doublets and are subcategorized into outer- and inner-arm dyneins depending on their specific locations (Fig. 1c–e).
ODAs are located at the outer periphery of the cylindrical axoneme, and a total of four ODAs are found within one 96‐nm repeat unit (Fig. 1d). ODAs consist of two or three heavy chains (α, β, and γ dyneins), each with a molecular mass of ~ 500 kDa, along with several intermediate and light chains, bringing the total molecular mass of ODA complexes to ~ 2 MDa. Based on mutant studies and in vitro motility assays using extracted ODAs, these complexes have been suggested to control the speed at which the axoneme bends to create motility waves.77,80 Cryo-ET studies were performed on axonemes from different organisms to map ODAs along the 96‐nm axonemal unit and to understand the geometry of their association (see Table 1). The reconstructed tomograms showed stacked arrangements of the dynein rings whose surfaces are facing each other, with the axis of this stack positioned perpendicular to the axis of the attached A-tubule.25–27,81 Reconstructions of axonemes from ODA mutants revealed the locations of individual dynein heavy chains (Fig. 1d and e). Early cryo-ET reconstructions at lower resolutions raised a question about the exact orientation of the γ-dynein ring, providing a possibility that the stalk of γ-dynein may not extend to the adjacent B-tubule.27 However, recent higher‐resolution data appear to verify that all dynein rings are oriented in a way that enables them to reach the B-tubule.31 In addition, the improved resolution of the latter studies revealed an ordered arrangement of ODAs as well as conformational changes of heavy chains associated with their various nucleotide states (see below).
Inner dynein arms (IDAs) are, as the name suggests, located at the inner periphery of the doublet MT (Fig. 1c). In C. reinhardtii, there are seven inner arm dynein species, of which six (a, b, c, d, e, and g) are monomeric and one (f) is dimeric. ET reconstructions using mutants lacking specific dynein chains revealed the positions of IDAs along the A-tubule, with f-dyneins closer to the surface of the A-tubule.28 Based on observations of the beating motion of mutant flagella lacking certain IDAs, these complexes are thought to regulate wave formation and curve propagation along the axoneme to produce a flagellar ‘swimming’ motion.82–84
Structural Studies of Dynein Components
Given the large size of dynein heavy chains, they have been extensively studied by EM.85–88 These observations established that the dynein heavy chains organize into rings of AAA + domains, extended by a linker and a tail at the N-terminus, as well as a 15‐nm‐long stalk with a small globular MT binding domain (MTBD) at its tip. Crystallographic analysis of dynein heavy chains has been hampered by difficulties in recombinant protein production, but recently, several crystal structures of cytoplasmic dynein motor domains were reported (see Fig. 3a).74,89–91 The first structures of dynein motors were determined at relatively modest resolutions (4.5–6 Å), revealing the overall architecture of secondary‐structure elements but not the exact register of amino acids.90,91 After improving the resolution to about 3 Å, amino acid side chains could be assigned and nucleotides bound to AAA1–4 were localized. Both structures captured the post-power stroke and revealed an asymmetric ring of the AAA1–6 domains, of which only four bind nucleotides, in agreement with previous biochemical studies.92 Furthermore, the linker traverses the AAA ring and an α-helical ‘strut’ (or ‘buttress’) emerges from the AAA5 domain to interact with the stalk region (Fig. 3a). Structure-based mutagenesis at the interface between the linker and the AAA ring demonstrated that this interaction is key to motor function (see Fig. 3b for model).74,89 Additionally, mutations of the strut revealed that the strut–stalk interaction observed in crystal structures is also important for allosteric communication within the dynein motor. The current data support a model in which ATP hydrolysis results in a large conformational change communicated via the linker, strut, and stalk to the MTBD at the tip of the stalk, thus changing its affinity for MTs (Fig. 3b). Further insights could be gleaned from crystal structures of pre-power stroke dynein and from fitting the dynein crystal structure into cryo-ET maps of dynein in different nucleotide states.
Crystallographic studies have also been carried out for several dynein light chains. In ODA complexes, light chain 1 (LC1) associates with the nucleotide-binding region of the γ heavy chain93 and adopts the fold of a leucine-rich repeat protein with a pronounced hydrophobic patch that likely mediates the interaction with the γ heavy chain.94,95 3D structures are also available for the dynein light chain Tctex196,97 and LC8,98,99 showing that the two proteins are homologues as they adopt very similar homodimeric structures. Both LC8 and Tctex1 are known to bind to dynein intermediate chain (IC), and several structural studies have mapped the binding site to a prominent groove that is conserved between LC8 and Tctex1.96,100,101 Surprisingly, the cargo-binding groove is identical to the IC-binding groove, indicating that the binding to IC and cargo is mutually exclusive100,101 (for a comprehensive review, see the chapter by Williams et al. in Ref. 77).
Relating Dynein Conformational Changes to Motility
Information about dynein conformations in different nucleotide states is required to understand how ATP hydrolysis in dynein motor domains relates to motility. In the case of axonemal dynein, the previously mentioned 96‐nm subaveraging technique was used to obtain cryo-ET reconstructions of axonemes both in the presence and in the absence of nucleotide.31 This study employed statistical image analyses to examine the conformational changes of dynein during ATP hydrolysis and the propagation of these conformational changes along the axoneme. By comparing tomograms of axonemes in the absence of nucleotide (apo) with axonemes in the presence of ADP-vanadate (mimicking the ADP*Pi transition state) or ADP, two distinct conformations of ODAs were observed.31 While dyneins in apo and with ADP displayed a similar conformation (reflecting the post-power stroke state), dyneins in the presence of ADP-vanadate showed mixed populations of the apo conformation and one additional conformation. This likely represents dynein's nucleotide-dependent conformational change and indicates, in agreement with kinetic data,102 that force generation occurs during the phosphate release step after ATP hydrolysis. Notably, both of these ADP-vanadate dynein conformations clustered along a single doublet, suggesting a cooperative movement of dynein that could explain the propagation of axonemal wave patterns. The positional displacement between the ODA AAA rings and the IDA c/e AAA rings during force generation was measured to be 8 nm towards the distal end of the flagellum, which is consistent with single‐molecule observations for cytoplasmic dynein.103 This is the first direct in situ observation of dynein's conformational change relative to the MT and supports a model in which dynein moves along the B-tubule by winching, using the stalk as a contact point.86,104,105
Cytoplasmic and IFT dyneins contain two copies of the same heavy chain that form a homodimer. Unlike the axonemal dyneins, which slide along the neighboring MTs using the A-tubule as an anchor point, cytoplasmic/IFT dyneins move processively along MTs. The dynein heads must coordinate their actions to achieve this processive movement. Furthermore, it is critical that the energy from ATP hydrolysis in the AAA + head is transmitted along the ~ 15‐nm distance to the stalk tip where dynein directly interacts with MTs. Electron microscopic studies have been performed on dynein in isolation87 as well as on dyneins bound to MTs,87,106 but it is still unclear how the conformational changes in the motor domains translate into a processive motility of dynein along MTs.
RSs and the DRC
RSs are T-shaped protein complexes that protrude from doublet MTs towards the central MT pair (see Fig. 1).107,108 At least 23 polypeptides have been identified as components of the RSs and named RS proteins (RSPs).109–114 Mutant studies of RSPs have shown that they function together with the central MT pair to regulate dynein activity and that the failure to assemble RSs causes flagellar paralysis.107,115 The stalk of the RS is anchored at the A-tubule and its orthogonal head is located near the central pair tubules. The interaction between the central pair tubules and the orthogonal head is thus believed to be key to the propagation of axonemal movement, although the details of this mechanism are not well understood.
Within the 96‐nm longitudinal repeat unit of axonemes, either two RSs (e.g., C. reinhardtii; RS1 and RS2) or three RSs (e.g., Tetrahymena and sea urchin; RS1, RS2, and RS3) are found with alternating periodicities of 32/64 nm or 32/24/40 nm, respectively.75,116 However, recent cryo-ET studies also identified an ‘RS3 stump’ in the 96‐nm axonemal repeat of C. reinhardtii.33,35–37 Comparison of proteins from wild‐type C. reinhardtii with those from pf-24 (lacking the spoke) and pf-1 (lacking the spoke head) mutants by 2D electrophoresis showed that RSP1, RSP4, RSP6, RSP9, and RSP10 are parts of the orthogonal head.109–111 Using these mutants, cryo-ET reconstructions further enabled mapping of the positions of RSPs within the RSs (Fig. 1f).33,35 Combining these data, the current view of the organization of RSPs is illustrated in Fig. 1f. The detailed cryo-ET structure revealed that the orthogonal head likely possesses 2-fold symmetry (i.e., contains a dimer), which led to the hypothesis that the identified 12S units pre-assemble in the cytoplasm117 and then undergo dimerization within the flagellum to become a mature RS (20S).33,118 Furthermore, mapping the RSPs into the RS structure revealed the locations of important components for CP-IDA signal transduction, including calmodulin (RSP20), HSP40 (RSP16), LC8 (RSP22), and NDK (nucleoside diphosphate kinase) (RSP23). These proteins are localized to the RS-neck, which suggests that the neck/stalk may be responsible for mechanochemical signal propagation, while the orthogonal head may function as a simpler mechanical anchor point. The functions of individual components need to be further characterized within the context of the fully assembled RS to comprehensively understand how IDA movement is controlled through the central pair.
The nexin linker was first identified by conventional EM as a structure connecting two adjacent MT doublets.47 Nexin has been suggested to be a source of elasticity required for axonemal movement,119,120 as the corresponding density was seen to stretch substantially upon MT doublet sliding.50,119 Nexin's molecular identity remained enigmatic, as no proteins corresponding to the nexin density could be identified. The DRC was originally identified as a crescent-shaped structure that spans a large area between MT doublets.121–123 Both nexin and the DRC are located at the RS2 of the 96‐nm repeat, connecting to IDAs. The shared position of nexin and the DRC suggested that these two structures may be subcomponents of a bigger complex. Recent cryo-ET reconstructions comparing C. reinhardtii wild‐type and DRC mutants (pf-2/3 and sup-pf-3/4) elegantly demonstrated that the mysterious nexin linker density actually is a part of the DRC (referred to as N-DRC).30 In an effort to characterize the components of the N-DRC complex, proteomic analysis of N-DRC mutants was used to identify 12 N-DRC proteins,124 of which only 7 had been previously shown to be components of the DRC.121,125 Future research will likely seek to understand how these N-DRC proteins assemble into a large macromolecular complex and how N-DRC stabilizes the structure of the axoneme.
Asymmetric Components on Axonemes
Although the axoneme is highly symmetric and many axonemal complexes are found in 96‐nm repeat units, several components are asymmetrically localized. While such components are missed when global averaging is applied, they can be locally classified into subgroups and compared by using statistic-based averaging techniques or by manually selecting target units via knowledge from previous studies. Examples of asymmetry include ODAs with multiple conformations (discussed above), missing ODAs126 and IDA dynein c at MT doublet 1,29 and missing IDA b/g at MT doublets 1 and 9.29 Additional asymmetric densities are seen bridging adjacent MT doublets as well as within the interior lumen of MT doublets.32 Furthermore, asymmetric structures project from the C1 and C2 tubules of the central MT pair.127 However, with the exception of the central pair apparatus proteins KLP1128 and Hydin,129 the molecular identities of these asymmetric axonemal densities are currently unknown.
3D Structure of IFT Trains In Situ
IFT, the bidirectional movement of macromolecules along the cilium, was discovered in 1993 by Kozminski et al. by differential interference contrast (DIC) imaging of flagella from live immobilized C. reinhardtii cells. Granule particles, large enough to be seen by light microscopy, were observed to move underneath the flagellar membrane at velocities of about 2–3 μm/s.9,130 Although the first observation of IFT particles by EM is usually attributed to Ringo,131 it was not until the seminal work by Kozminski was published that the significance of this observation was realized.9,11 2D EM studies revealed IFT particles as electron‐dense material spanning the ~ 30‐nm space between the outer axoneme MTs and the flagellar membrane.9 The IFT particles are organized into so-called IFT trains (previously called IFT rafts) with variable lengths in the range of hundreds of nanometers.9 An appealing idea at the time was that molecular motors power IFT, moving material such as flagellar precursors along the axonemal MT tracks. The involvement of motors in IFT was proven by showing that IFT does not occur in the temperature-sensitive fla10 mutant (FLA10 encodes a kinesin 2 motor domain) at the restrictive temperature.11 Furthermore, this study correlated IFT observed by light microscopy with the electron‐dense material seen in electron micrographs by imaging the same C. reinhardtii cells by DIC and EM.11 As kinesin 2 walks towards the plus ends of MTs, it serves as the anterograde IFT motor.132 It was later proven that cytoplasmic dynein 2/1b is the motor that powers retrograde transport from the flagellar tip back to the cell body.13,14,133 The fact that IFT particles disappear in the fla10 mutant at the restrictive temperature facilitated the biochemical isolation of a large 16–17S protein complex that corresponds to the IFT particles.10,134,135 Further evidence that the electron‐dense material observed by EM indeed contains IFT proteins was provided by an EM experiment using immunogold-labeled polyclonal antibodies raised against IFT46.136 This study demonstrated that IFT46 is present in IFT particles all along the length of the flagellum, as expected for a protein traveling between the flagellar base and tip. In addition, direct observations of green fluorescent protein fusion proteins have shown that KAP (a subunit of kinesin 2), IFT27, and IFT20 undergo flagellar transport with similar kinetics to DIC-measured IFT.137–140
Until recently, 2D EM and biochemical studies provided the only information on IFT particle organization.10,134,141–145 Although 2D EM revealed the overall dimensions of the IFT trains and suggested an ordered repeat structure within the trains, ultimately high-resolution 3D information is needed for a detailed understanding of how the ~ 20 IFT proteins assemble into large complexes that facilitate IFT. 3D information is also required to elucidate the molecular basis for the interactions of IFT particle components with ciliary motors and cargoes. The first and so far only 3D structure of IFT particles in situ was published by Pigino et al. in 2009.38 To obtain longitudinal sections of C. reinhardtii flagella, live WT and mutant cells were placed on glass coverslips, where the cells adhere to the solid surface and start a gliding movement with both flagella stretched out along their entire length. Cells treated this way were then chemically fixed, flat-embedded, thin and thick sectioned by ultramicrotomy, and imaged using ET to reconstruct volumes of the flagella containing IFT particles.38,146 The authors first carried out a careful 2D study to classify several hundred IFT trains by size and ultrastructure. IFT trains fall into two distinct categories, one of longer (700 nm ± 244 nm) but less electron‐dense material and one of shorter (251 nm ± 45 nm) but denser material. The longer trains show an intrinsic repeat of about 40 nm, whereas the shorter trains appear to have a 16‐nm repeat unit. To investigate what these different classes of IFT trains correspond to, the authors examined the fla14 mutant (LC8 dynein light‐chain deletion) that disrupts retrograde but not anterograde IFT.133 In fla14 flagella, the longer IFT trains were still observed but the shorter trains were absent, leading to the conclusion that the longer trains represent anterograde IFT material and the shorter trains represent retrograde IFT material.38 Although this interpretation is certainly plausible, the authors also offer the alternative explanation that the different train sizes and periodicities could represent different degrees of cargo loading.38 In a separate study published alongside the Pigino et al. paper, total internal reflection fluorescence microscopy was used to observe fluorescently labeled anterograde IFT components in regenerating C. reinhardtii flagella.139 This experiment revealed that IFT train size is inversely proportional to the length of the flagellum, with short rapidly growing flagella containing the longest trains. This finding is consistent with the observation by Pigino et al. that the short swollen flagella of fla14 cells contain mostly longer IFT trains, but it suggests that IFT train length may be influenced by the state or length of the flagellum. Additional experiments are currently being performed to provide conclusive evidence of the functions of short versus long IFT trains (P. Lupetti, personal communication). Expanding the study to different flagellar mutants may cast more light on these issues.
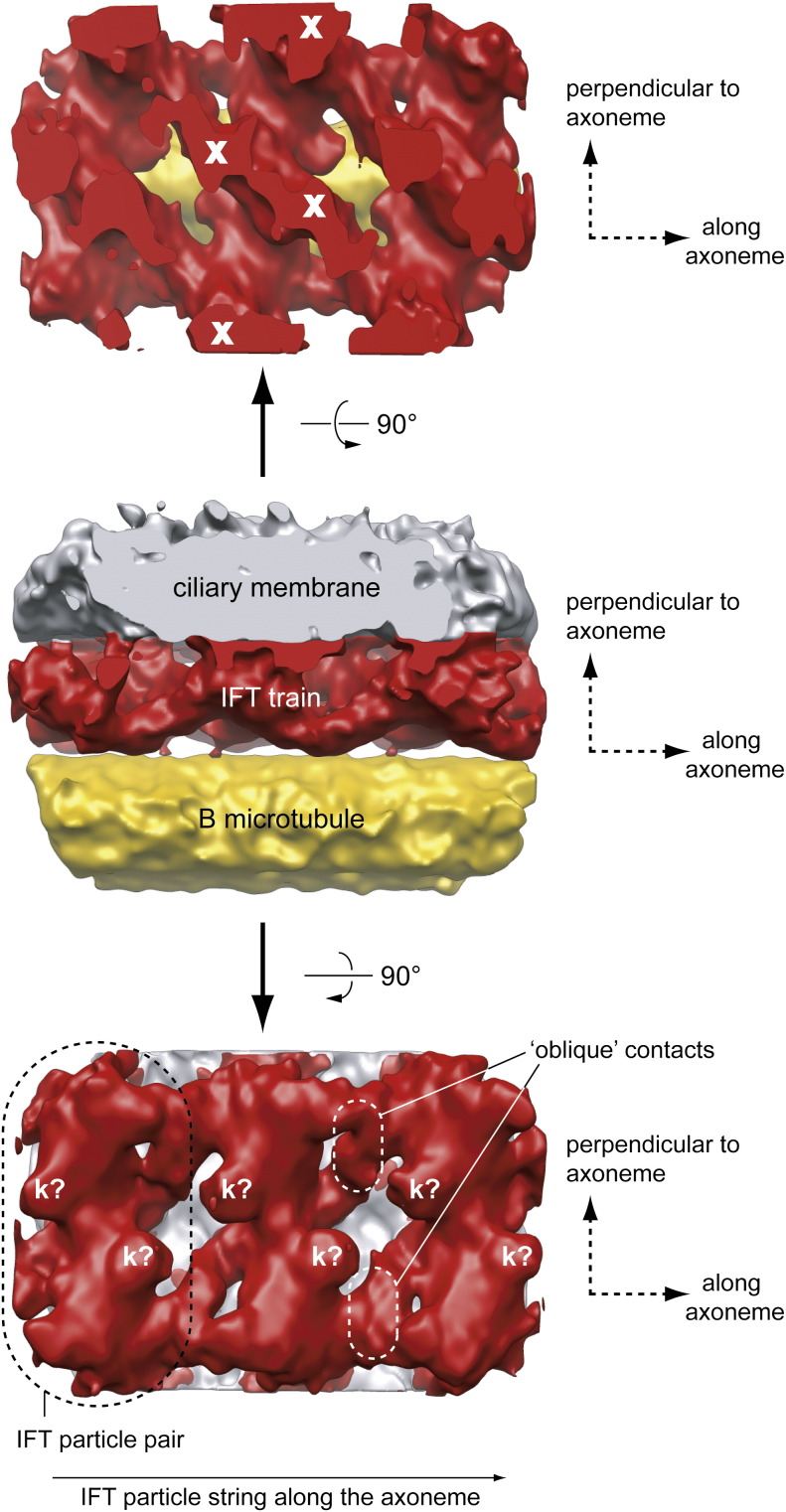
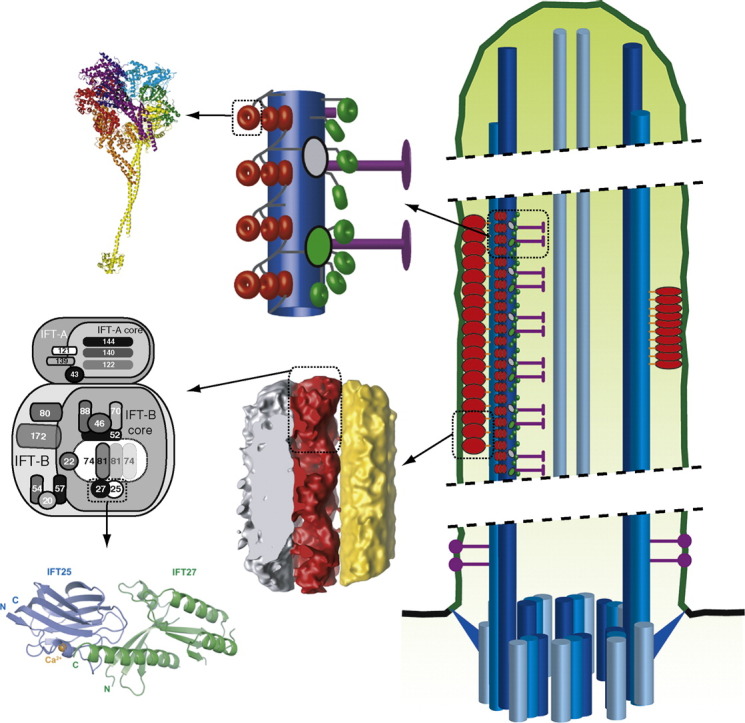
In addition to the extensive 2D EM analysis of IFT material, Pigino et al. also reconstructed 3D volumes using ET. In these reconstructions, the repetitive arrangement of the IFT particles was leveraged for averaging procedures to improve the resolution of the resulting map. The 3D reconstruction of IFT particles revealed direct contacts to the axoneme MT doublets and the flagellar membrane along each IFT particle (Fig. 4). As previously observed by 2D EM,9,26,27 contact with the MT doublet appears to occur via the B-tubule, with no discernible contacts to the A-tubule. The densities connecting IFT trains to the axonemal doublet have features that suggest that they could be anterograde kinesin 2 motors (marked with a ‘k?’ in Fig. 4), although this remains to be proven either by immuno-EM labeling or by comparative 3D reconstructions of IFT trains using mutants of IFT motor domains. The contacts to the ciliary membrane are extensive, consistent with the evidence that ciliary membrane proteins are transported by IFT.28,147–151 As expected from the 2D EM images of long IFT trains, individual IFT particles (with diameters of 30–40 nm) contact each other along the length of the flagellum to form strings of particles (see Figs. 4 and 1a for schematics). Interestingly, the reconstructions reveal that the repeating units of IFT trains are actually pairs of particles that contact each other not only along the long axis of the axoneme but also along an axis perpendicular to the axoneme, as well as by oblique links (see Fig. 4). In addition to the EM reconstructions of longer, likely anterograde, IFT trains from fla14 mutant flagella, ET reconstructions were also carried out for shorter trains from wild‐type flagella, potentially representing retrograde IFT material.38,82,83 However, the individual particles from this reconstruction were not similar enough to allow for subtomogram averaging, and thus more work is required to reveal the detailed structure of the shorter IFT trains.
Fig. 4.
EM structure of IFT trains in situ. Electron tomographic reconstruction of IFT trains using subtomographic averaging. The center image shows an IFT train (red) sandwiched between the flagellar membrane (gray) and an MT doublet (yellow). The top image is a 90° rotation (as indicated by the arrow) with the membrane removed. ‘X’ denotes points of contact between the IFT particles and the flagellar membrane. The bottom image is a 90° rotation (as indicated by the arrow) with the MT removed. ‘k?’ denotes points of contact between the MT and the IFT particles and may represent kinesin motors. From this image, it is clear that IFT trains are composed of strings of IFT particle dimers rather than monomers.
Although the work by Pigino et al. represents a major step forward in our structural understanding of IFT particles, it is clear that further progress is needed both on in situ IFT trains and on purified IFT components to elucidate the molecular interactions and mechanisms of the IFT machinery. For example, it is currently not known which parts of the reconstructed volumes represent IFT polypeptides, ciliary cargoes, and molecular motors.38,93 Ultimately, higher‐resolution studies of purified IFT complexes using X-ray crystallography, NMR, and single‐particle cryo-EM methods will be required to obtain atomic models.
Towards Atomic Models of IFT Complexes
The first biochemical characterizations of IFT complexes from C. reinhardtii were reported ~ 15 years ago and showed that IFT material purifies as two loosely associated subcomplexes, referred to as IFT-A (6 subunits) and IFT-B (14 subunits).10,94,95,134,135 Subsequent biochemical work demonstrated that the IFT-B complex has a salt stable core96,97,141 that consists of nine members (IFT22, IFT25, IFT27, IFT46, IFT52, IFT70, IFT74, IFT81, and IFT88).152 Of the 20 identified IFT subunits, high-resolution structures are only available for IFT25 (also known as C1orf41, Hsp16.2, or FAP232) and IFT27.153,154 IFT25 and IFT27 interact with each other and, curiously, are not conserved in Drosophila melanogaster and C. elegans, suggesting that their functions are not essential for IFT.155–157 Consistent with this idea, the recent analysis of an IFT25 null mouse revealed that IFT25 is not needed for ciliogenesis but is required for proper hedgehog signaling, an important developmental pathway that is mediated by the cilium in mammals.158 Interestingly, Western blots of cells from the IFT25 null mouse did not detect IFT27 (other IFT proteins were unaffected), indicating that IFT27 is degraded in the absence of IFT25.158 These observations support the notion that IFT25/27 functions as a complex and performs a specialized function in the hedgehog signaling of mammalian cilia. However, the fact that IFT25/27 is conserved in unicellular organisms such as C. reinhardtii, where hedgehog signaling does not occur, indicates that additional functions for IFT25/27 remain to be identified.
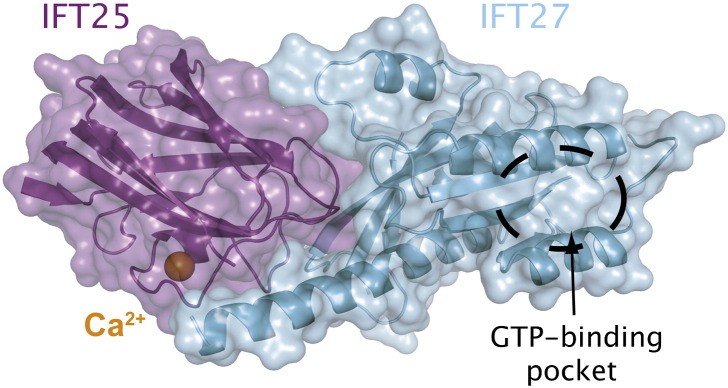
The structure of IFT25 was determined by both NMR and X-ray crystallography (1.6 Å resolution) as part of a high-throughput project by the Northeast Structural Genomics Consortium.153 It revealed a jelly-roll fold and an overall structural similarity to the galactose-binding domain of sialidases. However, it is clear from the structure that the specific site responsible for galactose binding in sialidases is not conserved in IFT25. The IFT25 structure also revealed a bound calcium ion coordinated by conserved residues located in a loop region, although the functional implications of calcium binding, if any, are currently unknown. A molecular structure is also available for the C. reinhardtii IFT25/27 complex (Fig. 5; 2.6 Å resolution), which shows how IFT25 and IFT27 interact to form a stable complex.154 In contrast to IFT25, which is highly soluble and can easily be purified in large quantities, IFT27 is insoluble (both human and C. reinhardtii proteins tested).154 However, C. reinhardtii IFT27 is solubilized when co-expressed with its binding partner IFT25, allowing for the structure determination of the complex. The structure of IFT25/27 shows that IFT27 adopts the fold of small GTPases and is most similar to structures of the Rab family of GTPases involved in membrane trafficking.85–87,137,159 Biochemical analysis of the IFT25/27 complex revealed low but detectable GTPase activity, indicating that an unidentified activator could be required for robust GTP turnover.88,154 Furthermore, the IFT25/27 complex was shown to bind GDP and GTP nucleotides with relatively low affinity (20–40 μM), significantly weaker than what is typically observed for small GTPases (picomolar to nanomolar affinity), suggesting that IFT27 may not require an exchange factor to replace GDP with GTP.154 Currently, no effectors for IFT27 have been identified and it is not known if a particular nucleotide state is needed to assemble higher‐order IFT complexes containing IFT27.
Fig. 5.
Crystal structure of the IFT25/27 complex. Crystal structure at 2.6 Å resolution of the IFT25/27 complex from C. reinhardtii. IFT25 (purple) binds one divalent calcium ion (shown as an orange ball), and IFT27 (cyan) is a GTPase with the putative GTP-binding pocket indicated with the black circle. The GTP-binding site is located far from the IFT25/27 interface, indicating that IFT25 likely does not influence the GTP binding or GTPase activity of IFT27 directly.
A complete structural understanding of IFT will require an integrated approach utilizing different structural techniques. Cellular ET will be essential in imaging the 3D structures of IFT particles in situ, as already demonstrated by Pigino et al.38 However, in the near future, this approach is unlikely to deliver the resolution needed to model individual IFT polypeptides. Fortunately, recent advances in recombinant production and purification of larger IFT complexes have made it possible to determine these structures using X-ray crystallography and cryo-EM.145 Single‐particle cryo-EM has proven to be a valuable technique for determining structures of large macromolecular complexes. Given the high molecular weight of IFT complexes, which sediment at ~ 16–17S,10 it is feasible to obtain structural insights into the architecture of IFT complexes via cryo-EM. In combination with high-resolution structures such as the crystal structure of IFT25/27, this approach could produce atomic models for larger IFT assemblies. Such atomic models of larger IFT complexes might then in turn be modeled into densities from cellular ET reconstructions, yielding atomic models for IFT trains in situ.
It is still early days for structural studies of IFT, and no structural information is currently available for most of the involved factors. Structural biology methods will likely be important not only in outlining the architecture of IFT complexes but also in revealing the molecular basis for their interaction with IFT motors and ciliary cargoes. Such studies will significantly broaden our understanding of IFT at the molecular level, which in turn will undoubtedly inspire new cell biological experiments.
Acknowledgements
We are grateful to Natacha Olieric and to Gaia Pigino for providing us with the models/pictures used in Figs. 2 and 4, respectively, and for many useful comments on the manuscript. Additionally, we would also like to thank Pietro Lupetti, Takashi Ishikawa, Stephen M. King, Stefan Geimer, and Daniela Nicastro for many improvements to the manuscript. This work was funded by an Emmy Noether grant (Deutsche Forschungsgemeinschaft; LO1627/1-1) and the EMBO Young Investigator program. M.T. is the recipient of an Erwin-Schroedinger-stipend granted Austrian Science Fund J 3148-B12. B.E. is the recipient of a postdoctoral research fellowship granted by the Alexander von Humboldt Foundation.
Edited by P. Wright
Contributor Information
Naoko Mizuno, Email: mizuno@biochem.mpg.de.
Esben Lorentzen, Email: lorentze@biochem.mpg.de.
References
- 1.Goodenough U.W. Cilia, flagella and the basal apparatus. Curr. Opin. Cell Biol. 1989;1:58–62. doi: 10.1016/s0955-0674(89)80037-7. [DOI] [PubMed] [Google Scholar]
- 2.Fisch C., Dupuis-Williams P. Ultrastructure of cilia and flagella—back to the future! Biol. Cell. 2012;103:249–270. doi: 10.1042/BC20100139. [DOI] [PubMed] [Google Scholar]
- 3.Nonaka S., Tanaka Y., Okada Y., Takeda S., Harada A., Kanai Y. Randomization of left–right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- 4.Flock A., Duvall A.J. The ultrastructure of the kinocilium of the sensory cells in the inner ear and lateral line organs. J. Cell. Biol. 1965;25:1–8. doi: 10.1083/jcb.25.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloodgood R.A. Sensory reception is an attribute of both primary cilia and motile cilia. J. Cell. Sci. 2010;123:505–509. doi: 10.1242/jcs.066308. [DOI] [PubMed] [Google Scholar]
- 6.Satir P. Cilia biology: stop overeating now! Curr. Biol. 2007;17:R963–R965. doi: 10.1016/j.cub.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Rosenbaum J.L., Witman G.B. Intraflagellar transport. Nat. Rev., Mol. Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 8.Ishikawa H., Marshall W.F. Ciliogenesis: building the cell's antenna. Nat. Rev., Mol. Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- 9.Kozminski K.G., Johnson K.A., Forscher P., Rosenbaum J.L. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl Acad. Sci. USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole D.G., Diener D.R., Himelblau A.L., Beech P.L., Fuster J.C., Rosenbaum J.L. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell. Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozminski K.G., Beech P.L., Rosenbaum J.L. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J. Cell. Biol. 1995;131:1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walther Z., Vashishtha M., Hall J.L. The Chlamydomonas FLA10 gene encodes a novel kinesin-homologous protein. J. Cell. Biol. 1994;126:175–188. doi: 10.1083/jcb.126.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pazour G.J., Dickert B.L., Witman G.B. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell. Biol. 1999;144:473–481. doi: 10.1083/jcb.144.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porter M.E., Bower R., Knott J.A., Byrd P., Dentler W. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell. 1999;10:693–712. doi: 10.1091/mbc.10.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perrone C.A., Tritschler D., Taulman P., Bower R., Yoder B.K., Porter M.E. A novel dynein light intermediate chain colocalizes with the retrograde motor for intraflagellar transport at sites of axoneme assembly in Chlamydomonas and mammalian cells. Mol. Biol. Cell. 2003;14:2041–2056. doi: 10.1091/mbc.E02-10-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou Y., Pazour G.J., Witman G.B. A dynein light intermediate chain, D1bLIC, is required for retrograde intraflagellar transport. Mol. Biol. Cell. 2004;15:4382–4394. doi: 10.1091/mbc.E04-05-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pazour G.J., Dickert B.L., Vucica Y., Seeley E.S., Rosenbaum J.L., Witman G.B., Cole D.G. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell. Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoetzel C., Laurier V., Davis E.E., Muller J., Rix S., Badano J.L. BBS10 encodes a vertebrate-specific chaperonin-like protein and is a major BBS locus. Nat. Genet. 2006;38:521–524. doi: 10.1038/ng1771. [DOI] [PubMed] [Google Scholar]
- 19.Badano J.L., Mitsuma N., Beales P.L., Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 20.Fliegauf M., Benzing T., Omran H. When cilia go bad: cilia defects and ciliopathies. Nat. Rev., Mol. Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 21.Zariwala M.A., Knowles M.R., Omran H. Genetic defects in ciliary structure and function. Annu. Rev. Physiol. 2007;69:423–450. doi: 10.1146/annurev.physiol.69.040705.141301. [DOI] [PubMed] [Google Scholar]
- 22.Sharma N., Berbari N.F., Yoder B.K. Ciliary dysfunction in developmental abnormalities and diseases. Curr. Top. Dev. Biol. 2008;85:371–427. doi: 10.1016/S0070-2153(08)00813-2. [DOI] [PubMed] [Google Scholar]
- 23.Pazour G.J., Rosenbaum J.L. Intraflagellar transport and cilia-dependent diseases. Trends Cell. Biol. 2002;12:551–555. doi: 10.1016/s0962-8924(02)02410-8. [DOI] [PubMed] [Google Scholar]
- 24.Lučić V., Förster F., Baumeister W. Structural studies by electron tomography: from cells to molecules. Annu. Rev. Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- 25.Nicastro D., McIntosh J.R., Baumeister W. 3D structure of eukaryotic flagella in a quiescent state revealed by cryo-electron tomography. Proc. Natl Acad. Sci. USA. 2005;102:15889–15894. doi: 10.1073/pnas.0508274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicastro D., Schwartz C., Pierson J., Gaudette R., Porter M.E., McIntosh J.R. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 27.Ishikawa T., Sakakibara H., Oiwa K. The architecture of outer dynein arms in situ. J. Mol. Biol. 2007;368:1249–1258. doi: 10.1016/j.jmb.2007.02.072. [DOI] [PubMed] [Google Scholar]
- 28.Bui K.H., Sakakibara H., Movassagh T., Oiwa K., Ishikawa T. Molecular architecture of inner dynein arms in situ in Chlamydomonas reinhardtii flagella. J. Cell. Biol. 2008;183:923–932. doi: 10.1083/jcb.200808050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bui K.H., Sakakibara H., Movassagh T., Oiwa K., Ishikawa T. Asymmetry of inner dynein arms and inter-doublet links in Chlamydomonas flagella. J. Cell. Biol. 2009;186:437–446. doi: 10.1083/jcb.200903082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heuser T., Raytchev M., Krell J., Porter M.E., Nicastro D. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J. Cell. Biol. 2009;187:921–933. doi: 10.1083/jcb.200908067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Movassagh T., Bui K.H., Sakakibara H., Oiwa K., Ishikawa T. Nucleotide-induced global conformational changes of flagellar dynein arms revealed by in situ analysis. Nat. Struct. Mol. Biol. 2010;17:761–767. doi: 10.1038/nsmb.1832. [DOI] [PubMed] [Google Scholar]
- 32.Nicastro D., Fu X., Heuser T., Tso A., Porter M.E., Linck R.W. Cryo-electron tomography reveals conserved features of doublet microtubules in flagella. Proc. Natl Acad. Sci. USA. 2011;108:E845–E853. doi: 10.1073/pnas.1106178108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pigino G., Bui K.H., Maheshwari A., Lupetti P., Diener D., Ishikawa T. Cryoelectron tomography of radial spokes in cilia and flagella. J. Cell. Biol. 2011 doi: 10.1083/jcb.201106125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S., Fernandez J.-J., Marshall W.F., Agard D.A. Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. EMBO J. 2011 doi: 10.1038/emboj.2011.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barber C.F., Heuser T., Carbajal-González B.I., Botchkarev V.V., Nicastro D. Three-dimensional structure of the radial spokes reveals heterogeneity and interactions with dyneins in Chlamydomonas flagella. Mol. Biol. Cell. 2012;23:111–120. doi: 10.1091/mbc.E11-08-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin J., Heuser T., Carbajal-González B.I., Song K., Nicastro D. The structural heterogeneity of radial spokes in cilia and flagella is conserved. Cytoskeleton (Hoboken) 2012;69:88–100. doi: 10.1002/cm.21000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pigino G., Maheshwari A., Bui K.H., Shingyoji C., Kamimura S., Ishikawa T. Comparative structural analysis of eukaryotic flagella and cilia from Chlamydomonas, Tetrahymena, and sea urchins. J. Struct. Biol. 2012 doi: 10.1016/j.jsb.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 38.Pigino G., Geimer S., Lanzavecchia S., Paccagnini E., Cantele F., Diener D.R. Electron-tomographic analysis of intraflagellar transport particle trains in situ. J. Cell. Biol. 2009;187:135–148. doi: 10.1083/jcb.200905103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Leewenhoeck A. Concerning little animals by Him observed in rain–well–sea and snow water; as also in water wherein pepper had lain infused. Philos. Trans. 1677;12:821–831. [Google Scholar]
- 40.Fawcett D.W., Porter K.R. A study of the fine structure of ciliated epithelia. J. Morphol. 1954;94:221–281. [Google Scholar]
- 41.Sedar A.W., Beams H.W., Janney C.D. Electron microscope studies on the ciliary apparatus of the gill cells of Mya arenaria. Proc. Soc. Exp. Biol. Med. 1952;79:303–305. doi: 10.3181/00379727-79-19359. [DOI] [PubMed] [Google Scholar]
- 42.Manton I., Clarke B. An electron microscope study of the spermatozoid of sphagnum. J. Exp. Bot. 1952;3:265. [Google Scholar]
- 43.Pease D.C. The ultrastructure of flagellar fibrils. J. Cell. Biol. 1963;18:313–326. doi: 10.1083/jcb.18.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Afzelius B. Electron microscopy of the sperm tail; results obtained with a new fixative. J. Biophys. Biochem. Cytol. 1959;5:269–278. doi: 10.1083/jcb.5.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibbons I.R., Grimstone A.V. On flagellar structure in certain flagellates. J. Biophys. Biochem. Cytol. 1960;7:697–716. doi: 10.1083/jcb.7.4.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brokaw C. Movement and nucleoside polyphosphatase activity of isolated flagella from Polytoma uvella. Exp. Cell. Res. 1961 [Google Scholar]
- 47.Gibbons I.R. Studies on the protein components of cilia from Tetrahymena pyriformis. Proc. Natl Acad. Sci. USA. 1963;50:1002–1010. doi: 10.1073/pnas.50.5.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gibbons I.R., Rowe A.J. Dynein: a protein with adenosine triphosphatase activity from cilia. Science. 1965;149:424–426. doi: 10.1126/science.149.3682.424. [DOI] [PubMed] [Google Scholar]
- 49.Satir P. Studies on cilia. 3. Further studies on the cilium tip and a “sliding filament” model of ciliary motility. J. Cell. Biol. 1968;39:77–94. doi: 10.1083/jcb.39.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Summers K.E., Gibbons I.R. Adenosine triphosphate-induced sliding of tubules in trypsin-treated flagella of sea-urchin sperm. Proc. Natl Acad. Sci. USA. 1971;68:3092–3096. doi: 10.1073/pnas.68.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Porter M.E. Axonemal dyneins: assembly, organization, and regulation. Curr. Opin. Cell Biol. 1996;8:10–17. doi: 10.1016/s0955-0674(96)80042-1. [DOI] [PubMed] [Google Scholar]
- 52.King S.M., Pazour G.J. Methods in cell biology. Cilia. 2009;91:1–39. [Google Scholar]
- 53.Downing K.H., Nogales E. Cryoelectron microscopy applications in the study of tubulin structure, microtubule architecture, dynamics and assemblies, and interaction of microtubules with motors. Methods Enzymol. 2010;483:121–142. doi: 10.1016/S0076-6879(10)83006-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ledbetter M.C., Porter K.R. Morphology of microtubules of plant cell. Science. 1964;144:872–874. doi: 10.1126/science.144.3620.872. [DOI] [PubMed] [Google Scholar]
- 55.Nogales E., Wolf S.G., Downing K.H. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 56.Löwe J., Li H., Downing K.H., Nogales E. Refined structure of alpha beta-tubulin at 3.5 Å resolution. J. Mol. Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- 57.Nogales E., Wolf S.G., Khan I.A., Ludueña R.F., Downing K.H. Structure of tubulin at 6.5 Å and location of the taxol-binding site. Nature. 1995;375:424–427. doi: 10.1038/375424a0. [DOI] [PubMed] [Google Scholar]
- 58.Gigant B., Curmi P.A., Martin-Barbey C., Charbaut E., Lachkar S., Lebeau L. The 4 Å X-ray structure of a tubulin:stathmin-like domain complex. Cell. 2000;102:809–816. doi: 10.1016/s0092-8674(00)00069-6. [DOI] [PubMed] [Google Scholar]
- 59.Ravelli R.B.G., Gigant B., Curmi P.A., Jourdain I., Lachkar S., Sobel A., Knossow M. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- 60.Nogales E., Whittaker M., Milligan R.A., Downing K.H. High-resolution model of the microtubule. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- 61.Li H., DeRosier D.J., Nicholson W.V., Nogales E., Downing K.H. Microtubule structure at 8 Å resolution. Structure. 2002;10:1317–1328. doi: 10.1016/s0969-2126(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 62.Alushin G.M., Ramey V.H., Pasqualato S., Ball D.A., Grigorieff N., Musacchio A., Nogales E. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–810. doi: 10.1038/nature09423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fourniol F.J., Sindelar C.V., Amigues B., Clare D.K., Thomas G., Perderiset M. Template-free 13-protofilament microtubule-MAP assembly visualized at 8 Å resolution. J. Cell. Biol. 2010;191:463–470. doi: 10.1083/jcb.201007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Witman G.B., Carlson K., Berliner J., Rosenbaum J.L. Chlamydomonas flagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J. Cell. Biol. 1972;54:507–539. doi: 10.1083/jcb.54.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Warner F.D., Satir P. The substructure of ciliary microtubules. J. Cell. Sci. 1973;12:313–326. doi: 10.1242/jcs.12.1.313. [DOI] [PubMed] [Google Scholar]
- 66.Sui H., Downing K.H. Molecular architecture of axonemal microtubule doublets revealed by cryo-electron tomography. Nature. 2006;442:475–478. doi: 10.1038/nature04816. [DOI] [PubMed] [Google Scholar]
- 67.Nigg E.A., Raff J.W. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 68.Keller L.C., Romijn E.P., Zamora I., Yates J.R., Marshall W.F. Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 2005;15:1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 69.Bettencourt-Dias M., Glover D.M. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev., Mol. Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- 70.van Breugel M., Hirono M., Andreeva A., Yanagisawa H.A., Yamaguchi S., Nakazawa Y. Structures of SAS-6 suggest its organization in centrioles. Science. 2011;331:1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- 71.Kitagawa D., Vakonakis I., Olieric N., Hilbert M., Keller D., Olieric V. Structural basis of the 9-fold symmetry of centrioles. Cell. 2011;144:364–375. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakazawa Y., Hiraki M., Kamiya R., Hirono M. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr. Biol. 2007;17:2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 73.Jerka-Dziadosz M., Gogendeau D., Klotz C., Cohen J., Beisson J., Koll F. Basal body duplication in Paramecium: the key role of Bld10 in assembly and stability of the cartwheel. Cytoskeleton (Hoboken) 2010;67:161–171. doi: 10.1002/cm.20433. [DOI] [PubMed] [Google Scholar]
- 74.Kon T., Oyama T., Shimo-Kon R., Imamula K., Shima T., Sutoh K., Kurisu G. The 2.8 Å crystal structure of the dynein motor domain. Nature. 2012 doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- 75.Linck R.W. Advances in the ultrastructural analysis of the sperm flagellar axoneme. In: Fawcett D.W., Bedford J.M., editors. The Spermatozoon. Urban & Schwarzenberg; Baltimore-Munich: 1979. pp. 99–115. [Google Scholar]
- 76.Fawcett D.W., Bedford J.M. Urban & Schwarzenberg; Baltimore-Munich: 1979. The Spermatozoan. [Google Scholar]
- 77.King S.M. Elsevier; 2011. Dyneins. [Google Scholar]
- 78.Wickstead B., Gull K. Dyneins across eukaryotes: a comparative genomic analysis. Traffic. 2007;8:1708–1721. doi: 10.1111/j.1600-0854.2007.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hom E.F.Y., Witman G.B., Harris E.H., Dutcher S.K., Kamiya R., Mitchell D.R. A unified taxonomy for ciliary dyneins. Cytoskeleton (Hoboken) 2011;68:555–565. doi: 10.1002/cm.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Elam C.A., Sale W.S., Wirschell M. The regulation of dynein-driven microtubule sliding in Chlamydomonas flagella by axonemal kinases and phosphatases. Methods Cell Biol. 2009;92:133–151. doi: 10.1016/S0091-679X(08)92009-4. [DOI] [PubMed] [Google Scholar]
- 81.Lupetti P., Lanzavecchia S., Mercati D., Cantele F., Dallai R., Mencarelli C. Three-dimensional reconstruction of axonemal outer dynein arms in situ by electron tomography. Cell Motil. Cytoskeleton. 2005;62:69–83. doi: 10.1002/cm.20084. [DOI] [PubMed] [Google Scholar]
- 82.Brokaw C.J., Kamiya R. Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil. Cytoskeleton. 1987;8:68–75. doi: 10.1002/cm.970080110. [DOI] [PubMed] [Google Scholar]
- 83.Kamiya R., Kurimoto E., Muto E. Two types of Chlamydomonas flagellar mutants missing different components of inner-arm dynein. J. Cell. Biol. 1991;112:441–447. doi: 10.1083/jcb.112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Porter M.E., Sale W.S. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J. Cell. Biol. 2000;151:F37–F42. doi: 10.1083/jcb.151.5.f37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goodenough U., Heuser J. Structural comparison of purified dynein proteins with in situ dynein arms. J. Mol. Biol. 1984;180:1083–1118. doi: 10.1016/0022-2836(84)90272-9. [DOI] [PubMed] [Google Scholar]
- 86.Burgess S.A., Walker M.L., Sakakibara H., Knight P.J., Oiwa K. Dynein structure and power stroke. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- 87.Roberts A.J., Numata N., Walker M.L., Kato Y.S., Malkova B., Kon T. AAA + ring and linker swing mechanism in the dynein motor. Cell. 2009;136:485–495. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Samsó M., Koonce M.P. 25 Angstrom resolution structure of a cytoplasmic dynein motor reveals a seven-member planar ring. J. Mol. Biol. 2004;340:1059–1072. doi: 10.1016/j.jmb.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 89.Schmidt H., Gleave E.S., Carter A.P. Insights into dynein motor domain function from a 3.3-Å crystal structure. Nat. Struct. Mol. Biol. 2012 doi: 10.1038/nsmb.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Carter A.P., Cho C., Jin L., Vale R.D. Crystal structure of the dynein motor domain. Science. 2011;331:1159–1165. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kon T., Sutoh K., Kurisu G. X-ray structure of a functional full-length dynein motor domain. Nat. Struct. Mol. Biol. 2011;18:638–642. doi: 10.1038/nsmb.2074. [DOI] [PubMed] [Google Scholar]
- 92.Mocz G., Gibbons I.R. Phase partition analysis of nucleotide binding to axonemal dynein. Biochemistry. 1996;35:9204–9211. doi: 10.1021/bi960662u. [DOI] [PubMed] [Google Scholar]
- 93.Benashski S.E., Patel-King R.S., King S.M. Light chain 1 from the Chlamydomonas outer dynein arm is a leucine-rich repeat protein associated with the motor domain of the gamma heavy chain. Biochemistry. 1999;38:7253–7264. doi: 10.1021/bi990466y. [DOI] [PubMed] [Google Scholar]
- 94.Wu H., Maciejewski M.W., Marintchev A., Benashski S.E., Mullen G.P., King S.M. Solution structure of a dynein motor domain associated light chain. Nat. Struct. Biol. 2000;7:575–579. doi: 10.1038/76804. [DOI] [PubMed] [Google Scholar]
- 95.Wu H., Blackledge M., Maciejewski M.W., Mullen G.P., King S.M. Relaxation-based structure refinement and backbone molecular dynamics of the dynein motor domain-associated light chain. Biochemistry. 2003;42:57–71. doi: 10.1021/bi026762j. [DOI] [PubMed] [Google Scholar]
- 96.Wu H., Maciejewski M.W., Takebe S., King S.M. Solution structure of the Tctex1 dimer reveals a mechanism for dynein–cargo interactions. Structure. 2005;13:213–223. doi: 10.1016/j.str.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 97.Williams J.C., Xie H., Hendrickson W.A. Crystal structure of dynein light chain TcTex-1. J. Biol. Chem. 2005;280:21981–21986. doi: 10.1074/jbc.M414643200. [DOI] [PubMed] [Google Scholar]
- 98.Fan J., Zhang Q., Tochio H., Li M., Zhang M. Structural basis of diverse sequence-dependent target recognition by the 8 kDa dynein light chain. J. Mol. Biol. 2001;306:97–108. doi: 10.1006/jmbi.2000.4374. [DOI] [PubMed] [Google Scholar]
- 99.Liang J., Jaffrey S.R., Guo W., Snyder S.H., Clardy J. Structure of the PIN/LC8 dimer with a bound peptide. Nat. Struct. Biol. 1999;6:735–740. doi: 10.1038/11501. [DOI] [PubMed] [Google Scholar]
- 100.Williams J.C., Roulhac P.L., Roy A.G., Vallee R.B., Fitzgerald M.C., Hendrickson W.A. Structural and thermodynamic characterization of a cytoplasmic dynein light chain–intermediate chain complex. Proc. Natl Acad. Sci. USA. 2007;104:10028–10033. doi: 10.1073/pnas.0703614104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Benison G., Karplus P.A., Barbar E. Structure and dynamics of LC8 complexes with KXTQT-motif peptides: swallow and dynein intermediate chain compete for a common site. J. Mol. Biol. 2007;371:457–468. doi: 10.1016/j.jmb.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 102.Johnson K.A. Pathway of the microtubule-dynein ATPase and the structure of dynein: a comparison with actomyosin. Annu. Rev. Biophys. Biophys. Chem. 1985;14:161–188. doi: 10.1146/annurev.bb.14.060185.001113. [DOI] [PubMed] [Google Scholar]
- 103.Reck-Peterson S.L., Yildiz A., Carter A.P., Gennerich A., Zhang N., Vale R.D. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Burgess S.A., Knight P.J. Is the dynein motor a winch? Curr. Opin. Struct. Biol. 2004;14:138–146. doi: 10.1016/j.sbi.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 105.Ueno H., Yasunaga T., Shingyoji C., Hirose K. Dynein pulls microtubules without rotating its stalk. Proc. Natl Acad. Sci. USA. 2008;105:19702–19707. doi: 10.1073/pnas.0808194105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mizuno N., Narita A., Kon T., Sutoh K., Kikkawa M. Three-dimensional structure of cytoplasmic dynein bound to microtubules. Proc. Natl Acad. Sci. USA. 2007;104:20832–20837. doi: 10.1073/pnas.0710406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Warner F.D., Satir P. The structural basis of ciliary bend formation. Radial spoke positional changes accompanying microtubule sliding. J. Cell. Biol. 1974;63:35–63. doi: 10.1083/jcb.63.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Warner F.D. New observations on flagellar fine structure. The relationship between matrix structure and the microtubule component of the axoneme. J. Cell. Biol. 1970;47:159–182. doi: 10.1083/jcb.47.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Patel-King R.S., Gorbatyuk O., Takebe S., King S.M. Flagellar radial spokes contain a Ca2 +‐stimulated nucleoside diphosphate kinase. Mol. Biol. Cell. 2004;15:3891–3902. doi: 10.1091/mbc.E04-04-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Piperno G., Huang B., Ramanis Z., Luck D.J. Radial spokes of Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J. Cell. Biol. 1981;88:73–79. doi: 10.1083/jcb.88.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang P., Diener D.R., Rosenbaum J.L., Sale W.S. Localization of calmodulin and dynein light chain LC8 in flagellar radial spokes. J. Cell. Biol. 2001;153:1315–1326. doi: 10.1083/jcb.153.6.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yang P., Diener D.R., Yang C., Kohno T., Pazour G.J., Dienes J.M. Radial spoke proteins of Chlamydomonas flagella. J. Cell. Sci. 2006;119:1165–1174. doi: 10.1242/jcs.02811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Curry A.M., Rosenbaum J.L. Flagellar radial spoke: a model molecular genetic system for studying organelle assembly. Cell Motil. Cytoskeleton. 1993;24:224–232. doi: 10.1002/cm.970240403. [DOI] [PubMed] [Google Scholar]
- 114.Huang B., Piperno G., Ramanis Z., Luck D.J. Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J. Cell. Biol. 1981;88:80–88. doi: 10.1083/jcb.88.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Witman G.B., Plummer J., Sander G. Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J. Cell. Biol. 1978;76:729–747. doi: 10.1083/jcb.76.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Goodenough U.W., Heuser J.E. Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J. Cell. Biol. 1985;100:2008–2018. doi: 10.1083/jcb.100.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Qin H., Diener D.R., Geimer S., Cole D.G., Rosenbaum J.L. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell. Biol. 2004;164:255–266. doi: 10.1083/jcb.200308132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Diener D.R., Yang P., Geimer S., Cole D.G., Sale W.S., Rosenbaum J.L. Sequential assembly of flagellar radial spokes. Cytoskeleton (Hoboken) 2011;68:389–400. doi: 10.1002/cm.20520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Warner F.D. Ciliary inter-microtubule bridges. J. Cell. Sci. 1976;20:101–114. doi: 10.1242/jcs.20.1.101. [DOI] [PubMed] [Google Scholar]
- 120.Summers K.E., Gibbons I.R. Effects of trypsin digestion on flagellar structures and their relationship to motility. J. Cell. Biol. 1973;58:618–629. doi: 10.1083/jcb.58.3.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Piperno G., Mead K., Shestak W. The inner dynein arms I2 interact with a “dynein regulatory complex” in Chlamydomonas flagella. J. Cell. Biol. 1992;118:1455–1463. doi: 10.1083/jcb.118.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mastronarde D.N., O'Toole E.T., McDonald K.L., McIntosh J.R., Porter M.E. Arrangement of inner dynein arms in wild-type and mutant flagella of Chlamydomonas. J. Cell. Biol. 1992;118:1145–1162. doi: 10.1083/jcb.118.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gardner L.C., O'Toole E., Perrone C.A., Giddings T., Porter M.E. Components of a “dynein regulatory complex” are located at the junction between the radial spokes and the dynein arms in Chlamydomonas flagella. J. Cell. Biol. 1994;127:1311–1325. doi: 10.1083/jcb.127.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lin J., Tritschler D., Song K., Barber C.F., Cobb J.S., Porter M.E., Nicastro D. Building blocks of the nexin–dynein regulatory complex in Chlamydomonas flagella. J. Biol. Chem. 2011;286:29175–29191. doi: 10.1074/jbc.M111.241760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Piperno G., Mead K., LeDizet M., Moscatelli A. Mutations in the “dynein regulatory complex” alter the ATP-insensitive binding sites for inner arm dyneins in Chlamydomonas axonemes. J. Cell. Biol. 1994;125:1109–1117. doi: 10.1083/jcb.125.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hoops H.J., Witman G.B. Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonas flagella. J. Cell. Biol. 1983;97:902–908. doi: 10.1083/jcb.97.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mitchell D.R., Smith B. Analysis of the central pair microtubule complex in Chlamydomonas reinhardtii. Methods Cell Biol. 2009;92:197–213. doi: 10.1016/S0091-679X(08)92013-6. [DOI] [PubMed] [Google Scholar]
- 128.Bernstein M., Beech P.L., Katz S.G., Rosenbaum J.L. A new kinesin-like protein (Klp1) localized to a single microtubule of the Chlamydomonas flagellum. J. Cell. Biol. 1994;125:1313–1326. doi: 10.1083/jcb.125.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lechtreck K.-F., Witman G.B. Chlamydomonas reinhardtii hydin is a central pair protein required for flagellar motility. J. Cell. Biol. 2007;176:473–482. doi: 10.1083/jcb.200611115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dentler W. Intraflagellar transport (IFT) during assembly and disassembly of Chlamydomonas flagella. J. Cell. Biol. 2005;170:649–659. doi: 10.1083/jcb.200412021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ringo D.L. Flagellar motion and fine structure of the flagellar apparatus in Chlamydomonas. J. Cell. Biol. 1967;33:543–571. doi: 10.1083/jcb.33.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cole D.G., Chinn S.W., Wedaman K.P., Hall K., Vuong T., Scholey J.M. Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature. 1993;366:268–270. doi: 10.1038/366268a0. [DOI] [PubMed] [Google Scholar]
- 133.Pazour G.J., Wilkerson C.G., Witman G.B. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT) J. Cell. Biol. 1998;141:979–992. doi: 10.1083/jcb.141.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Piperno G., Mead K. Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc. Natl Acad. Sci. USA. 1997;94:4457–4462. doi: 10.1073/pnas.94.9.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Piperno G., Siuda E., Henderson S., Segil M., Vaananen H., Sassaroli M. Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. J. Cell. Biol. 1998;143:1591–1601. doi: 10.1083/jcb.143.6.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pedersen L.B., Geimer S., Rosenbaum J.L. Dissecting the molecular mechanisms of intraflagellar transport in Chlamydomonas. Curr. Biol. 2006;16:450–459. doi: 10.1016/j.cub.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 137.Qin H., Wang Z., Diener D., Rosenbaum J. Intraflagellar transport protein 27 is a small G protein involved in cell-cycle control. Curr. Biol. 2007;17:193–202. doi: 10.1016/j.cub.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lechtreck K.-F., Johnson E.C., Sakai T., Cochran D., Ballif B.A., Rush J. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J. Cell. Biol. 2009;187:1117–1132. doi: 10.1083/jcb.200909183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Engel B.D., Ludington W.B., Marshall W.F. Intraflagellar transport particle size scales inversely with flagellar length: revisiting the balance-point length control model. J. Cell. Biol. 2009;187:81–89. doi: 10.1083/jcb.200812084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mueller J., Perrone C.A., Bower R., Cole D.G., Porter M.E. The FLA3 KAP subunit is required for localization of kinesin-2 to the site of flagellar assembly and processive anterograde intraflagellar transport. Mol. Biol. Cell. 2005;16:1341–1354. doi: 10.1091/mbc.E04-10-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lucker B.F., Behal R.H., Qin H., Siron L.C., Taggart W.D., Rosenbaum J.L., Cole D.G. Characterization of the intraflagellar transport complex B core: direct interaction of the IFT81 and IFT74/72 subunits. J. Biol. Chem. 2005;280:27688–27696. doi: 10.1074/jbc.M505062200. [DOI] [PubMed] [Google Scholar]
- 142.Lucker B.F., Miller M.S., Dziedzic S.A., Blackmarr P.T., Cole D.G. Direct interactions of intraflagellar transport complex B proteins IFT88, IFT52, and IFT46. J. Biol. Chem. 2010;285:21508–21518. doi: 10.1074/jbc.M110.106997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fan Z.-C., Behal R.H., Geimer S., Wang Z., Williamson S.M., Zhang H. Chlamydomonas IFT70/CrDYF-1 is a core component of IFT particle complex b and is required for flagellar assembly. Mol. Biol. Cell. 2010;21:2696–2706. doi: 10.1091/mbc.E10-03-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Behal R.H., Miller M.S., Qin H., Lucker B.F., Jones A., Cole D.G. Subunit interactions and organization of the Chlamydomonas reinhardtii intraflagellar transport complex a proteins. J. Biol. Chem. 2012;287:11689–11703. doi: 10.1074/jbc.M111.287102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Taschner M., Bhogaraju S., Vetter M., Morawetz M., Lorentzen E. Biochemical mapping of interactions within the intraflagellar transport (IFT) B core complex: IFT52 binds directly to four other IFT-B subunits. J. Biol. Chem. 2011;286:26344–26352. doi: 10.1074/jbc.M111.254920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Geimer S. Chapter 3—immunogold labeling of flagellar components in situ. Methods Cell Biol. 2010;91:63–80. doi: 10.1016/S0091-679X(08)91003-7. [DOI] [PubMed] [Google Scholar]
- 147.Qin H., Rosenbaum J.L., Barr M.M. An autosomal recessive polycystic kidney disease gene homolog is involved in intraflagellar transport in C. elegans ciliated sensory neurons. Curr. Biol. 2001;11:457–461. doi: 10.1016/s0960-9822(01)00122-1. [DOI] [PubMed] [Google Scholar]
- 148.Pazour G.J., San Agustin J.T., Follit J.A., Rosenbaum J.L., Witman G.B. Polycystin-2 localizes to kidney cilia and the ciliary level is elevated in orpk mice with polycystic kidney disease. Curr. Biol. 2002;12:R378–R380. doi: 10.1016/s0960-9822(02)00877-1. [DOI] [PubMed] [Google Scholar]
- 149.Qin H., Burnette D.T., Bae Y.-K., Forscher P., Barr M.M., Rosenbaum J.L. Intraflagellar transport is required for the vectorial movement of TRPV channels in the ciliary membrane. Curr. Biol. 2005;15:1695–1699. doi: 10.1016/j.cub.2005.08.047. [DOI] [PubMed] [Google Scholar]
- 150.Jenkins P.M., Hurd T.W., Zhang L., McEwen D.P., Brown R.L., Margolis B. Ciliary targeting of olfactory CNG channels requires the CNGB1b subunit and the kinesin-2 motor protein, KIF17. Curr. Biol. 2006;16:1211–1216. doi: 10.1016/j.cub.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 151.Huang K., Diener D.R., Mitchell A., Pazour G.J., Witman G.B., Rosenbaum J.L. Function and dynamics of PKD2 in Chlamydomonas reinhardtii flagella. J. Cell. Biol. 2007;179:501–514. doi: 10.1083/jcb.200704069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Taschner M., Bhogaraju S., Lorentzen E. Architecture and function of IFT complex proteins in ciliogenesis. Differentiation. 2011 [Google Scholar]
- 153.Ramelot T.A., Raman S., Kuzin A.P., Xiao R., Ma L.-C., Acton T.B. Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study. Proteins. 2009;75:147–167. doi: 10.1002/prot.22229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bhogaraju S., Taschner M., Morawetz M., Basquin C., Lorentzen E. Crystal structure of the intraflagellar transport complex 25/27. EMBO J. 2011;30:1907–1918. doi: 10.1038/emboj.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Follit J.A., Xu F., Keady B.T., Pazour G.J. Characterization of mouse IFT complex B. Cell Motil. Cytoskeleton. 2009;66:457–468. doi: 10.1002/cm.20346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang Z., Fan Z.-C., Williamson S.M., Qin H. Intraflagellar transport (IFT) protein IFT25 is a phosphoprotein component of IFT complex B and physically interacts with IFT27 in Chlamydomonas. PLoS One. 2009;4:e5384. doi: 10.1371/journal.pone.0005384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lechtreck K.-F., Luro S., Awata J., Witman G.B. HA-tagging of putative flagellar proteins in Chlamydomonas reinhardtii identifies a novel protein of intraflagellar transport complex B. Cell Motil. Cytoskeleton. 2009;66:469–482. doi: 10.1002/cm.20369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Keady B.T., Samtani R., Tobita K., Tsuchya M., San Agustin J.T., Follit J.A. IFT25 links the signal-dependent movement of hedgehog components to intraflagellar transport. Dev. Cell. 2012;22:940–951. doi: 10.1016/j.devcel.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev., Mol. Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]