Abstract
Progress in live-cell imaging of the cytoskeleton has significantly extended our knowledge about the organization and dynamics of actin filaments near the plasma membrane of plant cells. Noticeably, two populations of filamentous structures can be distinguished. On the one hand, fine actin filaments which exhibit an extremely dynamic behavior basically characterized by fast polymerization and prolific severing events, a process referred to as actin stochastic dynamics. On the other hand, thick actin bundles which are composed of several filaments and which are comparatively more stable although they constantly remodel as well. There is evidence that the actin cytoskeleton plays critical roles in trafficking and signaling at both the cell cortex and organelle periphery but the exact contribution of actin bundles remains unclear. A common view is that actin bundles provide the long-distance tracks used by myosin motors to deliver their cargo to growing regions and accordingly play a particularly important role in cell polarization. However, several studies support that actin bundles are more than simple passive highways and display multiple and dynamic roles in the regulation of many processes, such as cell elongation, polar auxin transport, stomatal and chloroplast movement, and defense against pathogens. The list of identified plant actin-bundling proteins is ever expanding, supporting that plant cells shape structurally and functionally different actin bundles. Here I review the most recently characterized actin-bundling proteins, with a particular focus on those potentially relevant to membrane trafficking and/or signaling.
Keywords: actin bundling, fimbrins, formins, LIM proteins, SCAB1, THRUMIN1, V-ATPases, villins
Introduction
Actin is one of the most abundant, ubiquitous, and conserved proteins in eukaryotes. In the cell, globular actin subunits polymerize into actin filaments which themselves assemble into higher order structures, such as orthogonal networks and parallel bundles (Figure 1). This system, referred to as the actin cytoskeleton, exhibits an extraordinary high degree of plasticity allowing the formation, destruction, and recycling of diverse filamentous structures within a short time scale, and offers countless possibilities to cells. The primary level of the regulation of actin cytoskeleton organization and dynamics consists in various (>100) actin-binding proteins which control, in time and space, actin filament nucleation, elongation, stabilization, capping, severing, and crosslinking (Pollard et al., 2000; Winder and Ayscough, 2005). In animal and yeast cells, cortical actin filaments and the plasma membrane undergo a dynamic interplay (Pollard and Cooper, 2009). For instance, the coordinated polymerization of actin filaments against the membrane provides the force necessary to modify cell shape and promote cell locomotion and division. As a consequence, dysfunctions in the actin polymerization machinery or in its regulation frequently results in diseases, including cancers (Van Troys et al., 2008a). Although related mechanisms are not excluded in plant cells, they are obviously not prevalent. The rigid plant cell wall precludes any modification of the cell boundary by an actin polymerization-based process. However, actin filaments are in close proximity with the cell membrane in plant cells too.
Figure 1.
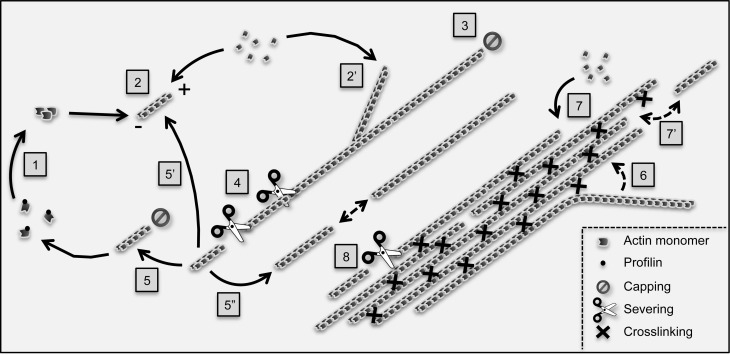
Main reactions controlling actin filament dynamics and organization in plant cells. The G-actin monomer binding protein profilin inhibits spontaneous actin nucleation in the cytoplasm. Nucleation is promoted de novo (1) by nucleating proteins such as formins. In addition, non-processive formins, such as Arabidopsis AtFH1, can also induce nucleation from the side of pre-existing filaments, a process which likely contributes to the initiation of actin bundles (not illustrated; Michelot et al., 2006; Blanchoin et al., 2010). Following nucleation, actin filaments undergo fast polymerization (2) and (2′) before being capped (3). The aging section of actin filaments (which contains ADP-loaded actin subunits, not shown) is fragmented by severing proteins such as actin-depolymerizing factors (4). The resulting fragments can be capped at their barbed end and depolymerize from their pointed (−) end to replenish the pool of monomers (5). Alternatively, they can re-elongate through polymerization (5′) although this process rarely occurs immediately following severing, suggesting intense barbed end capping activity (Staiger et al., 2009). Finally, actin fragments can serve as building blocks to assemble novel filaments by an end-joining mechanism (5″). Actin filaments are crosslinked into bundles by bundling proteins (right part of the cartoon). Both in vitro and live cell TIRFM-based analyses support that actin bundles form by a “catch and zipper” mechanism (6) (Khurana et al., 2010). Actin bundles subsequently grow by elongation of filaments at their ends (7) as well as by end-association of pre-existing filaments (7′), a process which might be facilitated by bundling proteins. Like single filaments, actin bundles are severed although at a lower frequency (Khurana et al., 2010; Smertenko et al., 2010). Current data support that unipolar bundles (here-exemplified) predominate in plant cells. However, the existence of bundles containing actin filaments of mixed polarity is not excluded.
Recent studies combining advanced imaging approaches, such as variable-angle epifluorescence microscopy (VAEM), spinning disc confocal microscopy, and reliable fluorescent actin markers have advanced our understanding of the organization and dynamics of the actin cytoskeleton near the cell cortex (Staiger et al., 2009; Khurana et al., 2010; Smertenko et al., 2010; Henty et al., 2011; Wang et al., 2011; Toth et al., 2012; Figure 1). Cortical filaments arrange into complex networks whose stochastic behavior is largely consistent with the predictions of a biomimetic system (Michelot et al., 2007; Staiger et al., 2009; Blanchoin et al., 2010). Single filaments randomly polymerize at extremely high growth rates (1.7 μm/s in hypocotyl epidermal cells from Arabidopsis seedlings) and exhibit prominent buckling and straightening behavior. Most filaments are short-lived (<30 s), which was demonstrated to be primarily due to prolific severing activity rather than to filament end depolymerization. Beside single filaments, thicker and longer bundles adopt less convoluted configurations and tend to align with the long axis of the cell (Staiger et al., 2009; Smertenko et al., 2010; Henty et al., 2011). In comparison with finer filaments, thick fibers experience slower but qualitatively similar dynamics. Indeed, they elongate, buckle, bundle, and are severed (Staiger et al., 2009; Blanchoin et al., 2010; Smertenko et al., 2010). Although actin filament severing emerges as the leading driver of actin cytoskeleton remodeling, an additional mechanism involving filament bundling, unbundling, and myosin-dependent sliding events was suggested to contribute to the permanent reorganization of the cortical actin network (Smertenko et al., 2010). Using VAEM and quantitative approaches, Henty et al. (2011) provided, for the first time, direct evidence of the contribution of an ABP, namely Arabidopsis actin depolymerizing factor 4 (AtADF4), to actin stochastic dynamics in vitro. In agreement with the biochemical actin severing activity of AtADF4, hypocotyl epidermal cells from adf4 knockout mutants exhibited a 2.5–3-fold decrease in the rate of severing, as well as increased filament lengths and lifetimes. The loss of AtADF4 also led to excessive actin bundling and cell growth in the apical region of the hypocotyl, where active cell expansion takes place.
Actin bundles have been repeatedly reported to play a critical role in cell morphogenesis (Baluska et al., 2001; Smith and Oppenheimer, 2005; Thomas et al., 2009; Higaki et al., 2010a). The relationships between the extent of actin bundling and cell growth are however complex as illustrated by contradictory observations. For instance, the growing epidermal cells from petioles of adf4 mutants also exhibit enhanced bundling but, contrarily to hypocotyl epidermal cells, are shorter than in wildtype plants (Henty et al., 2011). Some studies have established that in specific cells, such as rice coleoptiles, increased actin bundling negatively impacts cell elongation (Nick et al., 2009; Nick, 2010). As an underlying mechanism, it has been proposed that actin bundling prevents efficient delivery of auxin-efflux carriers to their site of action at the plasma membrane, a process which would require a more unbundled actin configuration (Nick, 2010). Actin bundles most likely impact cell growth by different ways. Indeed, actin bundling can alter turgor pressure, the main physical driver of cell expansion (Szymanski and Cosgrove, 2009), by modifying the thickness of the cell wall and the shape of the transvacuolar strands and vacuole (e.g., Staiger et al., 1994; Higaki et al., 2010a,b, 2011). Finally, there is much evidence that actin bundles serve as main tracks used by myosin motors to drive endomembrane compartments over long distances to sites of growth (Smith and Oppenheimer, 2005; Thomas et al., 2009; Higaki et al., 2010a). Interestingly, the organization and dynamics of the actin cytoskeleton significantly differ in isotropically and anisotropically growing cells, and this was confirmed by recent quantitative analyses using VAEM (Smertenko et al., 2010). Therefore, the role of actin bundling in cell growth appears multiple and cell-type dependent.
In animal cells, the assembly of actin bundles is required for the formation and/or function of various specialized cellular structures, such as filopodia, lamellipodia, stress fibers, microvilli, and invadopodia (Stevenson et al., 2012). In most of these structures, actin bundles are in close connection with the cell membrane and, in some cases, with the extracellular environment referred to as the extracellular matrix. For instance, the α-actinin-induced actin bundles that constitute the ventral stress fibers of non-muscle cells are anchored to focal adhesions at each of their extremities. Focal adhesions transmit the force generated by myosin II-dependent stress fiber contraction to the extracellular matrix, allowing to pull the cell body during cell migration (Vicente-Manzanares et al., 2009; Ciobanasu et al., 2012). Key players of focal adhesions are the cell surface membrane receptors integrins around which assemble complex networks made of about 160 proteins that contribute to link the extracellular matrix to the actin cytoskeleton and to create a high-performance environmental sensing system (Geiger et al., 2009). Plants lack most focal adhesion components, including true integrin homologs. However, there is no doubt that plant cells perceive and transduce many external signals from their cell wall to their cytoskeleton (Baluska et al., 2003; Drobak et al., 2004; Humphrey et al., 2007; Fu, 2010; Higaki et al., 2011). Day et al. (2011) recently comprehensively reviewed the potential roles played by the actin cytoskeleton in the organization and activation of host responses to biotic stress. One of the earliest and well-documented responses of plant cells to fungal or oomycete pathogens is a reorganization of the actin cytoskeleton and endomembrane components which both focus at the site of infection (e.g., Kobayashi et al., 1994; Leckie et al., 1995; Xu et al., 1998; Opalski et al., 2005; Takemoto et al., 2006; Day et al., 2011). Such reorganization is thought to culminate in the formation of cell wall appositions rich in antimicrobial compounds (Hardham et al., 2007). During this process actin filaments become more bundled, suggesting an important role for actin-bundling proteins in the dynamic relocalization of organelles during interactions with pathogens. Interestingly, Hardham et al. (2008) could mimic pathogen-induced actin remodeling by applying a gentle and local pressure on the surface of Arabidopsis cotyledon epidermal cells, indicating that the actin cytoskeleton can readily reorganize (3–5 min after stimulation) in response to the physical force exerted by pathogens. Considering the continuous and fast remodeling of the cortical actin array observed in both growing and non-growing epidermal cells, an emerging and seducing idea is that cortical actin plays a sentinel role capable of initiating basal defense against pathogen-induced diseases or abiotic stress, such as mechanical stress, within short time scales (Staiger et al., 2009). How actin filaments and bundles communicate with the cell membrane and cell wall largely remains enigmatic.
Since our last review on actin bundling in plants (Thomas et al., 2009), more than fifteen additional plant actin-bundling proteins were isolated and characterized. Several of those belong to the previously known villin, formin, fimbrin, and LIM protein families, whereas others define novel families. Some of these proteins are likely direct linkers between actin bundles and the cell or organelle membranes. Here we review the last advances in plant actin-bundling proteins with a particular interest for those that further move ahead our comprehension about how actin bundles physically or functionally interact with membranes.
Formins
Over the last years, formins have emerged as a large and major family of plant actin nucleating factors with critical functions in cell growth and division (Blanchoin and Staiger, 2008). Beside their core nucleating activity, plant formins display additional actin regulatory activities including nucleation, capping, severing, and bundling (Staiger and Blanchoin, 2006). The Arabidopsis formin AtFH1 (Banno and Chua, 2000), was the first plant formin reported to promote the formation of actin bundles both in live cells and in vitro (Cheung and Wu, 2004; Michelot et al., 2005). Its overexpression in pollen tubes stimulates the formation of actin bundles from the cell membrane and locally induces membrane deformation, suggesting that a proper density and distribution of actin bundles is critical for membrane assembly and/or maintenance, and that formins play substantial roles in these processes (Cheung and Wu, 2004). Mechanistic studies, employing total internal reflection fluorescence microscopy (TIRFM), revealed that AtFH1 functions as a non-processive formin which moves from the barbed end to the side of an actin filament after the nucleation event, and that this property is involved in AtFH1 actin bundling activity (Michelot et al., 2006). Recently, several other plant formins were shown to promote the formation of actin bundles in an autonomous manner, including Arabidopsis AtFH4, AtFH8, and AtFH14 (Deeks et al., 2010; Li et al., 2010; Xue et al., 2011), and rice OsFH5 (Yang et al., 2011; Zhang et al., 2011a).
Interaction of class I plant formins with a membrane is predicted by the characteristic membrane-targeting domain present in their N-terminal region which consists in a signal peptide followed by a transmembrane domain (Deeks et al., 2002; Blanchoin and Staiger, 2008). Accordingly, most class I formins examined so far were shown to accumulate at the cell periphery or in membrane-rich structures such as the cell plate using immunocytochemistry and/or GFP-fusion strategies (Cheung and Wu, 2004; Favery et al., 2004; Van Damme et al., 2004; Deeks et al., 2005; Ingouff et al., 2005; Cheung et al., 2010). Interestingly, AtFH8-GPF localizes primarily to the nuclear envelope in interphase cells, suggesting functional differences among class I formins (Xue et al., 2011). In addition, biochemical analyses indicate that pollen-specific AtFH3 lacks the ability to generate actin bundles in vitro (Ye et al., 2009). Nevertheless, with or without intrinsic actin-bundling activities, class I formins were convincingly demonstrated to promote actin bundling in vivo (Cheung and Wu, 2004; Ye et al., 2009; Cheung et al., 2010). Indeed, both gain- and loss-of-function genetic studies pointed out a central role of AtFH3 in regulating the long and thick actin bundles running along the pollen tube shank and in controlling the direction and velocity of cytoplasmic streaming (Ye et al., 2009). Therefore, AtFH3 likely cooperates with pollen actin-bundling proteins to assemble the tracks required for long distance actomyosin-dependent movement. Beside AtFH3, the pollen tube tip-enriched formin AtFH5 was found to play more specific functions in the formation of the subapical actin structure often referred to as the cortical actin fringe, and in membrane-targeted vesicular trafficking (Cheung et al., 2010).
It is noteworthy that class I formins exhibit divergent and potentially highly glycosylated extracellular domains, and accordingly represent excellent candidates for mediating extracellular stimuli to the actin cytoskeleton (Cvrckova, 2000; Blanchoin and Staiger, 2008), e.g., during the guidance of pollen tube growth in response to female tissue signals (Cheung and Wu, 2004). In this context, Martiniere et al. (2011) recently provided compelling evidence that the extracellular domain of AtFH1 is anchored to the cell wall and thereby reduces the lateral mobility of AtFH1. Domain analyses highlighted the central role in AtFH1 immobilization of a short, 15 amino acid-long, domain which includes a signature peptide of extensins, a class of cell wall-associated hydroxyproline-rich glycoproteins (Banno and Chua, 2000; Showalter et al., 2010). Although the biochemical nature of formin-cell wall interactions has not been resolved yet, it is tempting to propose that cell wall heterogeneity is responsible for targeting formins to specific plasma membrane subdomains. For instance, the accumulation of AtFH4 in cell-to-cell contact areas and of AtFH5 in the pollen-tube apical dome (Deeks and Hussey, 2005; Cheung et al., 2010) might reflect specificities in cell wall composition at these locations. Interestingly, the extracellular domain responsible for anchoring AtFH1 to the cell wall was required for AtFH1-mediated actin cytoskeleton remodeling in overexpression experiments (Martiniere et al., 2011). Although this remains speculative, anchoring of AtFH1 has been suggested to contribute to the formation and/or stabilization of AtFH1 functional dimers. Together these data support that AFH1, and most likely other class I formins, provide stable anchor points for the actin cytoskeleton at the cell membrane and can induce actin remodeling upon external signal perception. It is noteworthy that time lapse imaging analyses suggested that some of the AtFH5-nucleated and membrane-anchored actin filaments in the subapical region of pollen tubes are fragmented and released to the cytoplasm, providing precursors of some long actin bundles in the core cytoplasm (Cheung et al., 2010). Membrane-associated formins might therefore also indirectly contribute to the formation of more internal actin structures.
In addition to their function as interface between cell membrane and actin cytoskeleton, plant formins recently emerged as central links between actin filaments and microtubules. For instance, class I AtFH4, class II AtFH14, and the closely related rice OsFH5 bind to and bundle both actin filaments and microtubules and are accordingly expected to functionally coordinate the corresponding cytoskeletons (Deeks et al., 2010; Li et al., 2010; Yang et al., 2011; Zhang et al., 2011a). There is accumulating evidence that such coordination is crucial for many developmental processes such as intracellular transport, directional cell growth, and cell division (e.g., Fu et al., 2005; Collings, 2008; Wightman and Turner, 2008; Crowell et al., 2009; Petrasek and Schwarzerova, 2009). A recent quantitative study using VAEM revealed that microtubule depolymerization induces faster elongation and shortening of actin filaments, suggesting that actin dynamics at the cell cortex are modulated by microtubules (Smertenko et al., 2010). Although the underlying mechanism remains unknown, it seems reasonable to speculate that some formin family members are involved. Interestingly, endogenous OsFH5 localizes to specific regions at the chloroplast surface (Zhang et al., 2011a). Like other class II formins, OsFH5 possesses an N-terminal phosphate tensin (PTEN)-like domain instead of the typical transmembrane domain of most class I formins. Transient expression experiments indicate that the PTEN-like domain of FH5 is sufficient to target a fluorescent protein reporter to the chloroplast outer surface of tobacco cells, suggesting that it is responsible for the anchoring of OsFH5 to chloroplasts. Therefore OsFH5 emerges as a potential linker between actin filaments/bundles, microtubules, and chloroplasts, and might accordingly contribute to chloroplast motility, a process that has been proposed to rely on both cytoskeletons at least in some species (e.g., Chuong et al., 2006).
Thrumin1
Chloroplasts change their subcellular location in response to light. They move toward weak light to optimize light capture for photosynthesis and away from intense light to minimize photodamage, the latter process being referred to as the avoidance response (Kasahara et al., 2002; Suetsugu and Wada, 2007). In plants, organelle movement primarily relies on class XI myosins which are predicted to transport their cargos along cytosplasmic actin bundles (Avisar et al., 2008; Peremyslov et al., 2008; Sparkes et al., 2008). Although myosin inhibitor studies support that chloroplast movement also depends on myosin activity to some extent (e.g., Paves and Truve, 2007), recent data indicate that chloroplasts primarily use another type of actin-based mechanism to rapidly change their direction in response to light. A population of so-called chloroplast actin filaments (cp-actin filaments) was shown to anchor chloroplasts to the plasma membrane suggesting that they are involved in light-induced chloroplast repositioning (Kadota et al., 2009; Suetsugu et al., 2010a,b). In this context, THRUMIN1 was recently identified as a novel actin-bundling protein with a potential critical role in linking phototropin photoreceptor activity at the plasma membrane and actin-dependent chloroplast movements (Whippo et al., 2011).
Compared to wild-type plants, thrumin1 mutants exhibit slower and more randomized chloroplast movements in response to light stimuli. In vitro biochemical analyses indicate that THRUMIN1 binds to actin filaments in a direct manner and promotes the formation of actin bundles. Consistent with these data and the previously reported association of THRUMIN1 with the plasma membrane (Alexandersson et al., 2004), YFP-fused THRUMIN1 (THRUMIN1-YFP) extensively decorates the filamentous actin cytoskeleton along the plasma membrane, and in association with chloroplasts (Whippo et al., 2011). Upon stimulation of the chloroplast avoidance response by a localized blue-light irradiation, THRUMIN1-YFP further accumulates along actin filaments and apparently increases actin-bundling locally. The underlying mechanism was proven to be dependent on the phototropin blue-light photoreceptors PHOT1 and PHOT2. Indeed, no elevation of THRUMIN1-YFP fluorescence occurred in phot1phot2 double mutants upon blue light stimulation. Together these data support that THRUMIN1 promotes the formation of actin bundles from the plasma membrane in response to light and in a phototropin-dependent manner. However, the exact role of such actin bundles in chloroplast movement remains to be established. In addition, how THRUMIN1 cooperates with CHUP1, a chloroplast outer envelope ABP involved in cp-actin filament formation (Oikawa et al., 2003, 2008; Schmidt Von Braun and Schleiff, 2008a,b; Kadota et al., 2009), to remodel the actin cytoskeleton and drive chloroplast movement upon light perception by PHOT1 and PHOT2 are central questions that should be addressed in future studies. As already stated in the previous section, class II formins represent additional potential linkers between chloroplasts and actin bundles (Zhang et al., 2011a).
Vacuolar H+-ATPases B subunits
Vacuolar H+-ATPases (V-ATPases) are evolutionary-conserved multisubunit complexes that consist in a cytosolic ATP-hydrolyzing V1 subcomplex and a membrane-associated proton-translocating V0 subcomplex (Nishi and Forgac, 2002; Nelson, 2003; Ma et al., 2011). They mediate ATP-dependent transport of protons across plasma and intracellular membranes and thereby contribute to (1) the acidification of the lumen of various organelles such as vacuoles, secretory vesicles, endosomes, Golgi apparatus, and lysosomes and (2) the production of the energy required for various coupled transport processes. Accordingly, V-ATPases are involved in a wide range of critical processes including membrane trafficking and fusion, and cell expansion (Schumacher et al., 1999; Padmanaban et al., 2004; Dettmer et al., 2006; Brux et al., 2008). In mammals and yeast, both B and C subunits of the V1 subcomplex were previously reported to directly bind to F-actin with high affinity (Lee et al., 1999; Holliday et al., 2000; Vitavska et al., 2003, 2005; Chen et al., 2004; Zuo et al., 2008).
Functional studies support that the actin binding activity of V-ATPase B and C subunits is not involved in the regulation of V-ATPase assembly or activity. However, under stress conditions, it provides a significant survival advantage in yeast, supporting that it is biologically relevant (Xu and Forgac, 2001; Zuo et al., 2008). In addition, several studies have highlighted that the targeting of V-ATPases to specific sites relies on their interaction with the actin cytoskeleton (Lee et al., 1999; Adams et al., 2006; Zuo et al., 2006). Carnell et al. (2011) recently suggested a novel and elegant model in which nucleation-promoting factor WASH-dependent actin polymerization on mature lysosomes from Dictyostelium would sort V-ATPases to recycling vesicles, leading to subsequent lysosome neutralization and exocytosis. In the absence of WASH, no polymerization would occur and V-ATPases would remain on the lysosome, which in turn would remain acidic and unable to exocytose. Such a model assumes that the actin-binding activity of V-ATPases functions as tags for actin-mediated sorting.
A similar mechanism in plant cells is plausible since homologs of V-ATPases (Zimniak et al., 1988; Krebs et al., 2010), ARP2/3 complex and associated nucleation-promoting factors have been identified (Deeks and Hussey, 2005; Szymanski, 2005). In addition, the three Arabidopsis V-ATPase B subunits (AtAVB1, AtVAB2, and AtVAB3) were recently shown to display direct actin binding and bundling activities in vitro (Ma et al., 2012). Therefore, the multiple actin-binding sites responsible for the in vitro actin bundling activity of AtAVB1-3 may confer these proteins an increased affinity for actin filaments/bundles and trigger their clustering and/or recycling upon actin polymerization. Such a scenario of an actin-mediated sorting mechanism in plants remains however highly hypothetical. As an alternative, plant V-ATPases might serve as more passive points for anchoring organelles to actin bundles. A last possibility is that AtAVB1-3 function in a complex dissociated form in the cytoplasm, and therefore contribute to increase actin-bundling upon V-ATPase complex dissociation. Obviously, much work is required to examine each of these possibilities. Nevertheless, V-ATPases emerge as potential additional links between the actin cytoskeleton and membrane trafficking.
SCAB1
Stomatal movement is driven by modifications in turgor pressure of the guard cells. Stomata open when the guard cell volume increases, and they close when the guard cell volume decreases. It is well established that stomatal closure and opening involves reorganization of the actin cytoskeleton at the cell cortex and that such reorganization plays a key role in stomatal movement (e.g., Kim et al., 1995; Eun and Lee, 1997; Liu and Luan, 1998; Hwang and Lee, 2001; Lemichez et al., 2001; Macrobbie and Kurup, 2007; Choi et al., 2008; Gao et al., 2008). Recently, Higaki et al. (2010b) developed a novel quantitative image analysis method allowing a more detailed and reliable characterization of the changes in actin configurations during the diurnal cycles of Arabidopsis guard cells. Data confirmed previous observations that actin filaments adopt a well-organized and radial orientation in open stomata and a more longitudinal orientation in closed stomata. They also provide clear evidence that actin-bundling transiently increases during stomatal opening, and drastically reduces once this process is completed. Interestingly, the abnormally thick and long-lasting actin bundles induced by the expression of a mouse talin-derived actin reporter compromised stomatal opening. This is in good agreement with previous pharmacological and genetic studies indicating that changes in actin dynamics control stomatal movement (Kim et al., 1995; Liu and Luan, 1998; Dong et al., 2001; Lemichez et al., 2001; Macrobbie and Kurup, 2007).
Higaki et al. (2010a,b) suggest that unbundling of actin bundles (rather than their complete depolymerization; Liu and Luan, 1998) stimulates membrane trafficking and increases the number of activated potassium channels in the plasma membrane, which in turn promotes an increase of turgor pressure. In this context, a novel plant-specific actin-bundling protein, termed STOMATAL CLOSURE-RELATED ACTIN BINDING PROTEIN 1 (SCAB1), was isolated from a genetic screen aimed at identifying Arabidopsis mutants defective in stomatal movement (Zhao et al., 2011). In vitro biochemical data revealed that SCAB1 is a simple actin bundling protein unable to promote actin nucleation or capping. Depletion of SCAB1 reduces actin filament stability, delays the switch from a radial to a longitudinal actin filament configuration in guard cells during stomatal closure, and reduces stomatal closure sensitivity to abscisic acid, H2O2, and CaCl2. In contrast, the overexpression of SCAB1 increases actin filament stability and promotes excessive bundling. Both SCAB1 knockout and overexpressing lines exhibit a retardation of stomatal closure, suggesting that proper levels of SCAB1 and actin bundling are required for normal stomatal movements.
Structural and domain analyses indicate that SCAB1 functions as a single actin-binding domain protein that dimerizes through its central coiled coils to achieve the bivalent organization required for actin filament crosslinking (Zhang et al., 2012). Contrary to some other ABPs, SCAB1 activities are insensitive to pH and Ca2+ in vitro (Zhao et al., 2011). Nevertheless, the SCAB1 C-terminal pleckstrin homology domain was shown to weakly bind to inositol phosphates, suggesting a possible SCAB1 regulation by phosphoinositides at the cell membrane (Zhang et al., 2012). In addition to its potential impact on potassium channel density at the membrane of guard cells, actin bundling might also play a structural role in the control of the vacuolar shape and volume. Indeed, the radial actin filament configuration in open stomata allows the vacuole to occupy a maximal volume. In contrast, the long and heavy bundles spanning along the longitudinal axis of the guard cells of closed stomata might contribute to reduce the vacuole volume. Although the exact roles of actin bundles and the newly discovered actin bundling protein SCAB1 in stomatal movement remain to be established, there is accumulating evidence that they are central players.
Villins
Plant villins define a class of multifunctional ABPs which can combine several actin regulatory activities, including actin filament severing, barbed-end capping, and bundling activities. The Arabidopsis genome contains five villin genes (AtVLN1-5), each of which being highly expressed in a wide range of tissues (Klahre et al., 2000; Huang et al., 2005). Whereas atypical AtVLN1 was reported to function as a simple and calcium-insensitive bundling protein (Huang et al., 2005; Khurana et al., 2010), recent biochemical work supports that the rest of the family, including AtVLN2-5, retains the full set of typical villin activities and is calcium-responsive (Khurana et al., 2010; Zhang et al., 2010, 2011b; Bao et al., 2012; van der Honing et al., 2012). The analysis of AtVLN4 and AtVLN5 loss-of-function mutants (Zhang et al., 2010, 2011b) confirmed the predicted roles of villins in the formation and/or stabilization of the long actin bundles running along the shank of pollen tubes and root hairs (e.g., Yokota et al., 1998, 2003; Tominaga et al., 2000; Ketelaar et al., 2002). These studies also further validated the primary role of such actin bundles in the intracellular transport of organelles and vesicles in tip-growing cells (e.g., Miller et al., 1999; Sheahan et al., 2004; Lovy-Wheeler et al., 2005; Ye et al., 2009). Interestingly, the fact that beside their bundling activity, AtVLN4 and AtVLN5 possess calcium-dependent actin severing and capping activities suggests that they also actively contribute to assembling and disassembling the typical short actin bundle-based structures observed in the subapical region of pollen tubes and root hairs (Zhang et al., 2010, 2011b). As these structures remain at a constant distance from the growing cell tip, they inevitably undergo continuous cycles of disassembly/reassembly, a process which is thought to be primarily regulated by changes in the concentration of ions including [Ca+] and [H+], and reactive oxygen species (Holdaway-Clarke and Hepler, 2003; Knight, 2007; Cheung and Wu, 2008).
The analysis of vln2vln3 double T-DNA insertion mutants supports that AtVLN2 and AtVLN3 together play a major role in the generation of thick actin bundles in tissues other than pollen and root hairs, and that such bundles are involved in the regulation of directional organ growth (van der Honing et al., 2012). Indeed, unlike single vln2 and vln3 mutants, double mutants exhibit much thinner actin bundles as compared to wild type plants, and develop twisted leaves, stems, siliques, and roots. Only full-length AtVLN3, but not a truncated version lacking the headpiece region which is required for actin bundling in vitro, could rescue both actin and developmental phenotypes of vln2vln3 double mutants, supporting that villin-induced thick bundles are required for proper regulation of coordinated cell expansion. It is noteworthy that cell shape and size and plant growth rates are similar in control and double mutant plants, indicating that cell expansion itself is unaffected. Surprisingly, another recent study, which also focused on vln2vln3 double T-DNA insertion mutants (Bao et al., 2012), reported a morphological phenotype differing from the one described by van der Honing et al. (2012). In this study, the inflorescence stem of vln2vln3 seedlings developed a pendent phenotype which was correlated to defects in sclerenchyma development (Bao et al., 2012). Although petioles were modestly twisted, this malformation was obviously milder than the prominent twisted phenotype exhibited by various organs of van der Honing's double mutants. Both the pendent and faint twisted phenotypes of Bao's double mutants could be rescued by the expression of either VLN2 or VLN3. In addition, quantitative analyses indicate that xylem fiber cells of double mutant inflorescence stems contain abnormally fine actin bundles, supporting van der Honing's conclusions that VLN2 and VLN3 work as effective and functionally redundant actin-bundling proteins in vitro. The morphological differences between the Bao and van der Honing phenotypes remain intriguing and might reflect the presence of truncated forms of VLN3 (the same vln2 mutant is used as a parental line in both studies) and/or the use of dissimilar plant growth conditions.
LIM proteins
Plant LIM proteins or LIMs (the acronym LIM derived from the first letter of the three first identified LIM domain-containing proteins, namely LIN-11, ISl1, and MEC-3; Way and Chalfie, 1988; Freyd et al., 1990; Karlsson et al., 1990) define a ubiquitous family of actin-bundling proteins. Since our first report on tobacco NtWLIM1 describing the promotion of actin-bundle formation both in vitro and in live cells (Thomas et al., 2006, 2007), several additional LIMs, including the six Arabidopsis AtLIMs (Papuga et al., 2010; Ye and Xu, 2012), lilium LlLIM1 (Wang et al., 2008), and tobacco NtWLIM2 (Moes et al., 2012), were biochemically characterized and recognized as actin-bundling proteins. Contrary to formins and villins, no other actin-regulatory activity has been attributed to any plant LIM protein so far, supporting that they function as simple actin bundlers. In Papuga et al. (2010), we showed that the actin bundling activity of the three pollen-enriched Arabidopsis LIMs (AtPLIM2a–c) is regulated by pH and calcium (AtPLIM2c), whereas that of the three widely-expressed LIMs (AtWLIM1, AtWLIM2a, and AtWLIM2b) is not. These data are particularly relevant considering the central roles previously proposed for pH and calcium gradients/oscillations in the regulation of ABPs activities and cytoskeletal organization during pollen tube elongation (Staiger et al., 2010). Using an Arabidopsis cell suspension-based system, we could demonstrate that the interaction of PLIMs with the actin cytoskeleton can be specifically and reversibly inhibited by a controlled increase of the intracellular pH (Papuga et al., 2010). Overexpression of LlLIM1 modifies the actin cytoskeleton architecture in growing pollen tubes of lily, disturbs endomembrane trafficking, including the Golgi apparatus and endo/exocytic vesicles, and impairs normal targeting of signaling molecules, including phosphatidylinositol-4,5-bisphosphate, phospholipase C, and diacyl glycerol (Wang et al., 2008). As an additional proof of the biological functions of LIMs in pollen, the partial co-suppression of the three AtPLIMs by an RNAi approach was recently shown to provoke important defects in pollen development and tube growth (Ye and Xu, 2012).
Beside their cytosplasmic functions, plant LIMs were repeatedly reported to enter the nucleus, although their roles in this compartment have been comparatively less studied (Mundel et al., 2000; Kawaoka and Ebinuma, 2001; Briere et al., 2003; Thomas et al., 2006; Papuga et al., 2010). One of the first hints of the nuclear roles of plant LIMs was the identification of tobacco NtWLIM1 as a trans factor binding to a PAL-box motif of the horseradish C2 peroxidase (prxC2) gene whose product is involved in phenylpropanoid biosynthesis (Kawaoka et al., 1992, 2000). Supporting the biological relevance of this finding, transgenic tobacco plants with an antisense NtWLIM1 exhibited abnormally low levels of transcripts of several key phenylpropanoid pathway genes as well as a 27% reduction in lignin content (Kawaoka et al., 2000; Kaothien et al., 2002). We recently evaluated the nuclear functions of the tobacco NtWLIM2 and found that NtWLIM2 can specifically and directly bind to the conserved octamer cis-element of the histone AtH4A748 promoter and activate the corresponding promoter in live cell reporter-based experiments (Moes et al., 2012). Similar activities were also shown for the Arabidopsis homolog of NtWLIM2, namely AtWLIM2a, whereas the more distant NtWLIM1 and AtWLIM1 proteins were unable to bind to and to activate the AtH4A748 promoter, suggesting a specialization of LIM protein subfamilies in their nuclear targets.
Like all the other plant LIMs previously characterized, NtWLIM2 decorates the actin cytoskeleton in live cells, and binds to and bundles actin filaments in vitro (Moes et al., 2012). Interestingly, we observed that the NtWLIM2 nuclear fraction readily increases after cell treatment with the F-actin disrupting drug latrunculin B, suggesting that the compartmentalization of NtWLIM2 is modulated by the cytoskeletal status of the cell. It is noteworthy that the mammalian counterparts of plant LIMs, namely the cysteine-rich proteins (CRP1-3) were also reported to shuttle between the cytoplasm and the nucleus where they function as co-activators of genes involved in muscle differentiation (Arber et al., 1994; Arber and Caroni, 1996; Kong et al., 1997; Chang et al., 2003, 2007). In addition, some data support that CRP3 translocates to the nucleus in response to mechanical cues (Boateng et al., 2007, 2009) and that both CRP2 and CRP3 are involved in the stretch response and the regulation of the cell contractile force through their interaction with actin stress fibers (Knoll et al., 2002; Kim-Kaneyama et al., 2005). It is therefore tempting to propose that plant LIMs function as sensors able to perceive mechanical signals and to regulate in turn the mechanical properties of the cell by regulating gene expression (Kawaoka et al., 2000; Kaothien et al., 2002) and remodeling the actin cytoskeleton. In addition, recent expression analyses have highlighted that a subset of poplar LIMs is up-regulated in tension wood (Arnaud et al., 2012), further indicating a connection between plant LIMs and mechanical stress. Such a hypothesis is currently tested in our lab.
Fimbrins
Fimbrins (also known as plastins in humans) define an evolutionary-conserved family of actin bundling proteins whose activities, biological functions, and roles in diseases have been extensively analyzed in animals/humans and yeast (e.g., Bretscher, 1981; Samstag and Klemke, 2007; Al Tanoury et al., 2010; Skau et al., 2011; Morley, 2012; Shinomiya, 2012). The Arabidopsis genome encodes five fimbrins (AtFIM1-5; Staiger and Hussey, 2004). Although the structural bases underlying the actin binding and crosslinking activities of AtFIM1 were characterized in detail, only few studies have directly addressed the biological functions of plant fimbrins (Kovar et al., 2000, 2001; Klein et al., 2004; Wang et al., 2004). Wu et al. (2010) recently provided evidence that pollen-enriched AtFIM5 is required for the proper organization of the actin cytoskeleton in pollen grains and growing pollen tubes. The loss of AtFIM5 disorganizes the typical longitudinal configuration of actin bundles in the shank of the pollen tube and causes some bundles to invade the extreme tip. Such aberrant cytoskeletal organization in turn alters the pattern and velocity of cytoplasmic streaming. Biochemical data revealed that AtFIM5 is a calcium-insensitive actin bundling factor. Although the mechanism by which the loss-of-function of an actin-bundling protein leads to an increase in actin bundles a the tip of pollen tubes remains obscure, together these data highlight an important role for FIM5 in maintaining the normal actin organization and/or dynamics in pollen tubes.
SB401
SB401 is a pollen-specific protein from Solanum berthaultii (Liu et al., 1997) which was previously reported to bind to and bundle both microtubules and actin filaments and proposed to function as a linker between microtubule and actin cytoskeletons (Huang et al., 2007). In agreement with its higher in vitro affinity for microtubules, SB401 was observed to preferentially interact with the microtubule cytoskeleton in immunolabeled pollen tubes. However, recent in vitro biochemical analyses support that phosphorylation of SB401 by casein kinase II specifically inhibits SB401 microtubule regulatory activities, suggesting that phosphorylation can switch the protein toward its actin regulatory function(s) (Liu et al., 2009). Future work should validate SB401 cytoskeleton regulatory activities in a live cell context and provide an insight into its biological function(s) in potato pollen tubes.
AtADF9
Members of the ADF/cofilin family are well-established ABPs able to bind both actin monomers and filaments and whose main function is to increase actin dynamics (Staiger and Blanchoin, 2006; Ono, 2007; Van Troys et al., 2008b; Bernstein and Bamburg, 2010). Whereas vertebrates typically possess three ADFs/cofilins, plant ADF families are particularly large. Indeed, Arabidopsis expresses 11 functional ADFs (AtADF1-11) which can be divided into 5 subclasses according to their tissular expression and phylogeny (Ruzicka et al., 2007). Recently, time-lapse TIRFM analyses provided direct evidence that subclass I AtADF1 and AtADF4 sever actin filaments in vitro (Khurana et al., 2010; Henty et al., 2011), an activity displayed by most animal, protozoa, and yeast ADFs/cofilins (e.g., Andrianantoandro and Pollard, 2006; Chan et al., 2009). In agreement with these data and the role predicted for actin severing in the stochastic dynamics of plant actin filaments (Michelot et al., 2007; Blanchoin et al., 2010; Staiger et al., 2010), Henty et al. (2011) established that Arabidopsis adf4 knockout mutants exhibit a 2.5-fold reduced severing frequency as well as other characteristics of reduced actin dynamics in the cortical region of hypocotyl epidermal cells.
We recently compared the biochemical activities of Arabidopsis ADFs from different subclasses (unpublished data). We found that, contrary to other ADFs, subclass III AtADF9 is unable to enhance actin depolymerization in vitro (Tholl et al., 2011). Instead, AtADF9 stabilizes and crosslinks actin filaments into large bundles. By transiently expressing GFP-tagged and untagged AtADF9 recombinant proteins in tobacco BY2 cells, we confirmed the actin-bundling activity of AtADF9 in a live cell context. Indeed, contrary to AtADF1 which induced many breaks in the actin cytoskeleton, AtADF9 reduced the density and increased the thickness of actin fibers. Interestingly, similar data were obtained with AtADF5 (unpublished data), the other member of Arabidopsis ADF subclass III. Future work should identify the structural features responsible for the unconventional activities of subclass III ADFs, and compare the developmental and actin cytoskeleton phenotypes of adf5 and adf9 mutants to those recently reported for the knockout mutant of the conventional ADF AtADF4 (Henty et al., 2011).
Table 1 lists the actin bundling proteins cited in the present article and emphasizes some of their important features.
Table 1.
List of the actin bundling promoting proteins cited in this article.
| Name | Remarkable features | Reported subcellular locations | Identified or suggested biological functions | Key references |
|---|---|---|---|---|
| FORMINS | ||||
| AtFH1 | Non-processive formin; anchors in the cell wall | Cell membrane | Pollen tube growth, cell expansion | Cheung and Wu, 2004; Michelot et al., 2005, 2006; Martiniere et al., 2011 |
| AtFH3 | Lacks in vitro actin bundling activity | Nuclear envelope; cell plate | Pollen tube growth polarity | Ye et al., 2009 |
| AtFH4 | Bundles both AFs and MTs; AtFH4-GFP co-aligns the ER and MTs | Cell membrane at cell-to-cell contacts; ER membrane | Cell expansion | Deeks et al., 2005, 2010 |
| AtFH5 | In vitro actin bundling activity not reported so far | Growing cell plate; cell membrane in the pollen tube tip | Cell cytokinesis; pollen tube growth | Ingouff et al., 2005; Cheung et al., 2010 |
| AtFH8 | AtFH8(FH1FH2) induces stellar structures in vitro | Cell membrane at cell-to-cell contacts; nuclear envelope | Primary root growth; lateral root initiation; cell expansion and division | Deeks et al., 2005; Yi et al., 2005; Xue et al., 2011 |
| AtFH14 | Bundles both AFs and MTs; crosslinks AFs and MTs together | Preprophase band; phragmoplast | Cell division | Li et al., 2010 |
| OsFH5 | Bundles both AFs and MTs | Chloroplast surface | Cell expansion | Yang et al., 2011; Zhang et al., 2011a |
| AtTHRUMIN1 | Light-dependent actin bundling activity | Cell membrane | Chloroplast movement | Whippo et al., 2011 |
| AtAVB1-3 | Part of the V-ATPase multimeric complex | Endomembrane system | – | Ma et al., 2012 |
| AtSCAB1 | Dimerizes; likely regulated by phosphoinositides | Cytoplasm | Stomatal movement | Zhao et al., 2011; Zhang et al., 2012 |
| VILLINS | ||||
| LlP-135-ABP and LlP-115-ABP | Ca2+ sensitive; bundle AFs with uniform polarity | Cytoplasm | Direction of cytoplasmic streaming in pollen tubes and root hair cells | Yokota et al., 1998, 2000, 2003, 2005; Yokota and Shimmen, 1999; Tominaga et al., 2000 |
| AtVLN1 | Ca2+ insensitive; lacks severing and capping activities | – | – | Huang et al., 2005; Khurana et al., 2010 |
| AtVLN2 | Ca2+ sensitive; has severing and capping activities | Cytoplasm | Directional organ growth; Sclerenchyma development | Bao et al., 2012; van der Honing et al., 2012 |
| AtVLN3 | Ca2+ sensitive; has severing and capping activities; can sever AtVLN1-induced bundles in vitro | Cytoplasm | Directional organ growth; Sclerenchyma development | Khurana et al., 2010; Bao et al., 2012; van der Honing et al., 2012 |
| AtVLN4 | Ca2+ sensitive; has severing and capping activities | Cytoplasm | Root hair growth and cytoplasmic streaming | Zhang et al., 2011b |
| AtVLN5 | Ca2+ sensitive; has severing and capping activities | Cytoplasm | Pollen tube growth | Zhang et al., 2010 |
| LIM PROTEINS | ||||
| NtWLIM1 | Interacts directly with DNA | Cytoplasm; nucleus | Gene expression (lignin biosynthesis) | Kawaoka et al., 2000; Kaothien et al., 2002; Thomas et al., 2006, 2007 |
| NtWLIM2 | Interacts directly with DNA; dimerizes | Cytoplasm; nucleus | Gene expression (Histones) | Moes et al., 2012 |
| AtWLIM1, 2a and b | Ca2+ and pH insensitive | Cytoplasm; nucleus | – | Papuga et al., 2010 |
| AtPLIM2a and b | Only pH sensitive | Cytoplasm; nucleus | Pollen tube growth | Papuga et al., 2010; Ye and Xu, 2012 |
| AtPLIM2c | Ca2+ and pH sensitive | Cytoplasm; nucleus | Pollen tube growth | Papuga et al., 2010; Ye and Xu, 2012 |
| LlLIM1 | Ca2+ and pH sensitive | Cytoplasm; nucleus | Pollen tube growth | Wang et al., 2008 |
| FIMBRINS | ||||
| AtFIM1 | Ca2+ insensitive | Cytoplasm | Cytoplasmic streaming | Kovar et al., 2000, 2001 |
| AtFIM5 | Ca2+ insensitive | Cytoplasm | Pollen tube germination and growth | Wu et al., 2010 |
| OTHER ABP FAMILIES | ||||
| Sb401 | Bundles both AFs and MTs; activity possibly switched toward actin bundling by phosphorylation; genus-specific protein | Cytoplasm; cell cortex | – | Huang et al., 2007; Liu et al., 2009 |
| AtADF9 | Expression induced by hormones; lacks conventional ADF AF severing activity | Cytoplasm; nucleus | Gene expression (repression of flowering); development | Burgos-Rivera et al., 2008; Tholl et al., 2011 |
In the column “Reported subcellular locations,” the term “cytoplasm” means no association with any specific organelle. Note that, in some cases, the “Identified or suggested biological functions” is not directly related to the actin bundling activity of the protein, e.g., nuclear functions. AFs, actin filaments; ER, endoplasmic reticulum; MT, microtubules.
Conclusions
The growing number and diversity of actin-bundling proteins identified in plants indicate that, like animals, plants elaborate various types of actin-bundles with specific structural features and distinct functions (Table 1). This implies that the functions of actin bundles extend beyond the traditional definition of stable tracks for long distance intracellular transport. The characterization of novel types of actin-bundling proteins points out potential functions for actin bundles in stomatal movement, ion channel trafficking and/or activities, and chloroplast movement. In addition, actin bundles most likely play an important role in the regulation of hormone carriers cycling between plasma membrane and intracellular compartments (Nick, 2010). The specific actin bundling proteins involved in this process as well as their mode of regulation however remain to be identified.
Precise actin filament dynamics and organization near the plant cell cortex have been resolved only recently, and the elucidation of the roles of actin filaments and bundles at this location will keep researchers busy during the years to come. Future work should establish why epidermal plant cells keep their cortical actin network so dynamic and whether the other cell types do the same. A number of actin-bundling proteins reviewed in this article support the existence of a physical linkage between actin bundles and membranes. Among those, formins emerge as key multifunctional ABPs able to initiate polymerization and bundling of filaments from diverse types of subcellular locations including the cell membrane. Noticeably, the recent work by Martiniere et al. (2011) provides compelling evidence that the Arabidopsis formin AtAFH1 is anchored by its predicted extracellular domain within the cell wall and bridges the latter to the actin cytoskeleton. A next important step consists in identifying the external signals that target the extracellular domain of class I formins and in characterizing how such signals modulate the intracellular activities of formins. In addition, one can expect that, following the pioneering work on AtADF4 by Henty et al. (2011), the exact contribution of formins to actin nucleation and bundling near the cell cortex, and more generally to the actin stochastic dynamics, will be soon characterized. During the reviewing process of the present article, a publication by Deeks et al. (2012) reporting the identification of a novel and plant-specific superfamily of ABPs termed Networked (NET) was released. Localization analyses strongly suggest that the Arabidopsis NET proteins function as linkers between the actin cytoskeleton and diverse types of membranes, including specific subdomains of the plasma membrane, the tonoplast and the nuclear membrane. There is no doubt that such an exciting discovery will boost the field and contribute to a better understanding of how AFs and actin bundles are coupled to membranes in plant cells.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Work in the author's lab is supported by the Ministry of Culture, Higher Education, and Research and the National Research Fund from Luxembourg (project: HUMCRP, C10/BM/784171).
References
- Adams D. S., Robinson K. R., Fukumoto T., Yuan S., Albertson R. C., Yelick P., Kuo L., McSweeney M., Levin M. (2006). Early, H+-V-ATPase-dependent proton flux is necessary for consistent left-right patterning of non-mammalian vertebrates. Development 133, 1657–1671 10.1242/dev.02341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandersson E., Saalbach G., Larsson C., Kjellbom P. (2004). Arabidopsis plasma membrane proteomics identifies components of transport, signal transduction and membrane trafficking. Plant Cell Physiol. 45, 1543–1556 10.1093/pcp/pch209 [DOI] [PubMed] [Google Scholar]
- Al Tanoury Z., Schaffner-Reckinger E., Halavatyi A., Hoffmann C., Moes M., Hadzic E., Catillon M., Yatskou M., Friederich E. (2010). Quantitative kinetic study of the actin-bundling protein L-plastin and of its impact on actin turn-over. PLoS ONE 5:e9210 10.1371/journal.pone.0009210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianantoandro E., Pollard T. D. (2006). Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24, 13–23 10.1016/j.molcel.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Arber S., Caroni P. (1996). Specificity of single LIM motifs in targeting and LIM/LIM interactions in situ. Genes Dev. 10, 289–300 10.1101/gad.10.3.289 [DOI] [PubMed] [Google Scholar]
- Arber S., Halder G., Caroni P. (1994). Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell 79, 221–231 10.1016/0092-8674(94)90192-9 [DOI] [PubMed] [Google Scholar]
- Arnaud D., Dejardin A., Leple J. C., Lesage-Descauses M. C., Boizot N., Villar M., Benedetti H., Pilate G. (2012). Expression analysis of LIM gene family in poplar, toward an updated phylogenetic classification. BMC Res. Notes 5, 102 10.1186/1756-0500-5-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D., Prokhnevsky A. I., Makarova K. S., Koonin E. V., Dolja V. V. (2008). Myosin XI-K is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol. 146, 1098–1108 10.1104/pp.107.113647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluska F., Jasik J., Edelmann H. G., Salajova T., Volkmann D. (2001). Latrunculin B-induced plant dwarfism: plant cell elongation is F-actin-dependent. Dev. Biol. 231, 113–124 10.1006/dbio.2000.0115 [DOI] [PubMed] [Google Scholar]
- Baluska F., Samaj J., Wojtaszek P., Volkmann D., Menzel D. (2003). Cytoskeleton-plasma membrane-cell wall continuum in plants. Emerging links revisited. Plant Physiol. 133, 482–491 10.1104/pp.103.027250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banno H., Chua N. H. (2000). Characterization of the arabidopsis formin-like protein AFH1 and its interacting protein. Plant Cell Physiol. 41, 617–626 [DOI] [PubMed] [Google Scholar]
- Bao C., Wang J., Zhang R., Zhang B., Zhang H., Zhou Y., Huang S. (2012). Arabidopsis VILLIN2 and VILLIN3 act redundantly in sclerenchyma development via bundling of actin filaments. Plant J. [Epub ahead of print]. 10.1111/j.1365-313X.2012.05044.x [DOI] [PubMed] [Google Scholar]
- Bernstein B. W., Bamburg J. R. (2010). ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 20, 187–195 10.1016/j.tcb.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L., Boujemaa-Paterski R., Henty J. L., Khurana P., Staiger C. J. (2010). Actin dynamics in plant cells: a team effort from multiple proteins orchestrates this very fast-paced game. Curr. Opin. Plant Biol. 13, 714–723 10.1016/j.pbi.2010.09.013 [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Staiger C. J. (2008). Plant formins: diverse isoforms and unique molecular mechanism. Biochim. Biophys. Acta. 1803, 201–206 10.1016/j.bbamcr.2008.09.015 [DOI] [PubMed] [Google Scholar]
- Boateng S. Y., Belin R. J., Geenen D. L., Margulies K. B., Martin J. L., Hoshijima M., De Tombe P. P., Russell B. (2007). Cardiac dysfunction and heart failure are associated with abnormalities in the subcellular distribution and amounts of oligomeric muscle LIM protein. Am. J. Physiol. Heart Circ. Physiol. 292, H259–H269 10.1152/ajpheart.00766.2006 [DOI] [PubMed] [Google Scholar]
- Boateng S. Y., Senyo S. E., Qi L., Goldspink P. H., Russell B. (2009). Myocyte remodeling in response to hypertrophic stimuli requires nucleocytoplasmic shuttling of muscle LIM protein. J. Mol. Cell. Cardiol. 47, 426–435 10.1016/j.yjmcc.2009.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. (1981). Fimbrin is a cytoskeletal protein that crosslinks F-actin in vitro. Proc. Natl. Acad. Sci. U.S.A. 78, 6849–6853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briere C., Bordel A. C., Barthou H., Jauneau A., Steinmetz A., Alibert G., Petitprez M. (2003). Is the LIM-domain protein HaWLIM1 associated with cortical microtubules in sunflower protoplasts? Plant Cell Physiol. 44, 1055–1063 10.1093/pcp/pcg126 [DOI] [PubMed] [Google Scholar]
- Brux A., Liu T. Y., Krebs M., Stierhof Y. D., Lohmann J. U., Miersch O., Wasternack C., Schumacher K. (2008). Reduced V-ATPase activity in the trans-Golgi network causes oxylipin-dependent hypocotyl growth Inhibition in Arabidopsis. Plant Cell 20, 1088–1100 10.1105/tpc.108.058362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos-Rivera B., Ruzicka D. R., Deal R. B., McKinney E. C., King-Reid L., Meagher R. B. (2008). ACTIN DEPOLYMERIZING FACTOR9 controls development and gene expression in Arabidopsis. Plant Mol. Biol. 68, 619–632 10.1007/s11103-008-9398-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnell M., Zech T., Calaminus S. D., Ura S., Hagedorn M., Johnston S. A., May R. C., Soldati T., Machesky L. M., Insall R. H. (2011). Actin polymerization driven by WASH causes V-ATPase retrieval and vesicle neutralization before exocytosis. J. Cell Biol. 193, 831–839 10.1083/jcb.201009119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C., Beltzner C. C., Pollard T. D. (2009). Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr. Biol. 19, 537–545 10.1016/j.cub.2009.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. F., Belaguli N. S., Chang J., Schwartz R. J. (2007). LIM-only protein, CRP2, switched on smooth muscle gene activity in adult cardiac myocytes. Proc. Natl. Acad. Sci. U.S.A. 104, 157–162 10.1073/pnas.0605635103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. F., Belaguli N. S., Iyer D., Roberts W. B., Wu S. P., Dong X. R., Marx J. G., Moore M. S., Beckerle M. C., Majesky M. W., Schwartz R. J. (2003). Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev. Cell 4, 107–118 10.1016/S1534-5807(02)00396-9 [DOI] [PubMed] [Google Scholar]
- Chen S. H., Bubb M. R., Yarmola E. G., Zuo J., Jiang J., Lee B. S., Lu M., Gluck S. L., Hurst I. R., Holliday L. S. (2004). Vacuolar H+-ATPase binding to microfilaments: regulation in response to phosphatidylinositol 3-kinase activity and detailed characterization of the actin-binding site in subunit B. J. Biol. Chem. 279, 7988–7998 10.1074/jbc.M305351200 [DOI] [PubMed] [Google Scholar]
- Cheung A. Y., Niroomand S., Zou Y., Wu H. M. (2010). A transmembrane formin nucleates subapical actin assembly and controls tip-focused growth in pollen tubes. Proc. Natl. Acad. Sci. U.S.A. 107, 16390–16395 10.1073/pnas.1008527107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A. Y., Wu H. M. (2004). Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell 16, 257–269 10.1105/tpc.016550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A. Y., Wu H. M. (2008). Structural and signaling networks for the polar cell growth machinery in pollen tubes. Annu. Rev. Plant Biol. 59, 547–572 10.1146/annurev.arplant.59.032607.092921 [DOI] [PubMed] [Google Scholar]
- Choi Y., Lee Y., Jeon B. W., Staiger C. J. (2008). Phosphatidylinositol 3- and 4-phosphate modulate actin filament reorganization in guard cells of day flower. Plant Cell Environ. 31, 366–377 10.1111/j.1365-3040.2007.01769.x [DOI] [PubMed] [Google Scholar]
- Chuong S. D., Franceschi V. R., Edwards G. E. (2006). The cytoskeleton maintains organelle partitioning required for single-cell C4 photosynthesis in Chenopodiaceae species. Plant Cell 18, 2207–2223 10.1105/tpc.105.036186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciobanasu C., Faivre B., Le Clainche C. (2012). Actin dynamics associated with focal adhesions. Int. J. Cell Biol. 2012, 941292 10.1155/2012/941292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collings D. (2008). Crossed-wires: interactions and cross-talk between the microtubule and microfilament networks in plants, in Plant Microtubules, Plant Cell Monographs, ed N. Peter (Berlin: Springer; ), 47–79 [Google Scholar]
- Crowell E. F., Bischoff V., Desprez T., Rolland A., Stierhof Y. D., Schumacher K., Gonneau M., Hofte H., Vernhettes S. (2009). Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21, 1141–1154 10.1105/tpc.108.065334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrckova F. (2000). Are plant formins integral membrane proteins? Genome Biol. 1, RESEARCH001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day B., Henty J. L., Porter K. J., Staiger C. J. (2011). The pathogen-actin connection: a platform for defense signaling in plants. Annu. Rev. Phytopathol. 49, 483–506 10.1146/annurev-phyto-072910-095426 [DOI] [PubMed] [Google Scholar]
- Deeks M. J., Calcutt J. R., Ingle E. K., Hawkins T. J., Chapman S., Richardson A. C., Mentlak D. A., Dixon M. R., Cartwright F., Smertenko A. P., Oparka K., Hussey P. J. (2012). A Superfamily of Actin-Binding Proteins at the Actin-Membrane Nexus of Higher Plants. Curr. Biol. [Epub ahead of print]. 10.1016/j.cub.2012.06.041 [DOI] [PubMed] [Google Scholar]
- Deeks M. J., Cvrckova F., Machesky L. M., Mikitova V., Ketelaar T., Zarsky V., Davies B., Hussey P. J. (2005). Arabidopsis group Ie formins localize to specific cell membrane domains, interact with actin-binding proteins and cause defects in cell expansion upon aberrant expression. New Phytol. 168, 529–540 10.1111/j.1469-8137.2005.01582.x [DOI] [PubMed] [Google Scholar]
- Deeks M. J., Fendrych M., Smertenko A., Bell K. S., Oparka K., Cvrckova F., Zarsky V., Hussey P. J. (2010). The plant formin AtFH4 interacts with both actin and microtubules, and contains a newly identified microtubule-binding domain. J. Cell Sci. 123, 1209–1215 10.1242/jcs.065557 [DOI] [PubMed] [Google Scholar]
- Deeks M. J., Hussey P. J. (2005). Arp2/3 and SCAR: plants move to the fore. Nat. Rev. Mol. Cell Biol. 6, 954–964 10.1038/nrm1765 [DOI] [PubMed] [Google Scholar]
- Deeks M. J., Hussey P. J., Davies B. (2002). Formins: intermediates in signal-transduction cascades that affect cytoskeletal reorganization. Trends Plant Sci. 7, 492–498 10.1016/S1360-1385(02)02341-5 [DOI] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y. D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18, 715–730 10.1105/tpc.105.037978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C. H., Xia G. X., Hong Y., Ramachandran S., Kost B., Chua N. H. (2001). ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 13, 1333–1346 10.1105/TPC.010051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobak B. K., Franklin-Tong V. E., Staiger C. J. (2004). The role of the actin cytoskeleton in plant cell signaling. New Phytol. 163, 13–30 [DOI] [PubMed] [Google Scholar]
- Eun S. O., Lee Y. (1997). Actin filaments of guard cells are reorganized in response to light and abscisic acid. Plant Physiol. 115, 1491–1498 10.1104/pp.115.4.1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery B., Chelysheva L. A., Lebris M., Jammes F., Marmagne A., De Almeida-Engler J., Lecomte P., Vaury C., Arkowitz R. A., Abad P. (2004). Arabidopsis formin AtFH6 is a plasma membrane-associated protein upregulated in giant cells induced by parasitic nematodes. Plant Cell 16, 2529–2540 10.1105/tpc.104.024372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyd G., Kim S. K., Horvitz H. R. (1990). Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature 344, 876–879 10.1038/344876a0 [DOI] [PubMed] [Google Scholar]
- Fu Y. (2010). The actin cytoskeleton and signaling network during pollen tube tip growth. J. Integr. Plant Biol. 52, 131–137 10.1111/j.1744-7909.2010.00922.x [DOI] [PubMed] [Google Scholar]
- Fu Y., Gu Y., Zheng Z., Wasteneys G., Yang Z. (2005). Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120, 687–700 10.1016/j.cell.2004.12.026 [DOI] [PubMed] [Google Scholar]
- Gao X. Q., Chen J., Wei P. C., Ren F., Wang X. C. (2008). Array and distribution of actin filaments in guard cells contribute to the determination of stomatal aperture. Plant Cell Rep. 27, 1655–1665 10.1007/s00299-008-0581-2 [DOI] [PubMed] [Google Scholar]
- Geiger B., Spatz J. P., Bershadsky A. D. (2009). Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 10, 21–33 10.1038/nrm2593 [DOI] [PubMed] [Google Scholar]
- Hardham A. R., Jones D. A., Takemoto D. (2007). Cytoskeleton and cell wall function in penetration resistance. Curr. Opin. Plant Biol. 10, 342–348 10.1016/j.pbi.2007.05.001 [DOI] [PubMed] [Google Scholar]
- Hardham A. R., Takemoto D., White R. G. (2008). Rapid and dynamic subcellular reorganization following mechanical stimulation of Arabidopsis epidermal cells mimics responses to fungal and oomycete attack. BMC Plant Biol. 8, 63 10.1186/1471-2229-8-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henty J. L., Bledsoe S. W., Khurana P., Meagher R. B., Day B., Blanchoin L., Staiger C. J. (2011). Arabidopsis actin depolymerizing factor4 modulates the stochastic dynamic behavior of actin filaments in the cortical array of epidermal cells. Plant Cell 23, 3711–3726 10.1105/tpc.111.090670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki T., Kojo K. H., Hasezawa S. (2010a). Critical role of actin bundling in plant cell morphogenesis. Plant Signal. Behav. 5, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki T., Kutsuna N., Sano T., Kondo N., Hasezawa S. (2010b). Quantification and cluster analysis of actin cytoskeletal structures in plant cells: role of actin bundling in stomatal movement during diurnal cycles in Arabidopsis guard cells. Plant J. 61, 156–165 10.1111/j.1365-313X.2009.04032.x [DOI] [PubMed] [Google Scholar]
- Higaki T., Kurusu T., Hasezawa S., Kuchitsu K. (2011). Dynamic intracellular reorganization of cytoskeletons and the vacuole in defense responses and hypersensitive cell death in plants. J. Plant Res. 124, 315–324 10.1007/s10265-011-0408-z [DOI] [PubMed] [Google Scholar]
- Holdaway-Clarke T. L., Hepler P. K. (2003). Control of pollen tube growth: role of ion gradients and fluxes. New Phytol. 159, 539–563 [DOI] [PubMed] [Google Scholar]
- Holliday L. S., Lu M., Lee B. S., Nelson R. D., Solivan S., Zhang L., Gluck S. L. (2000). The amino-terminal domain of the B subunit of vacuolar H+-ATPase contains a filamentous actin binding site. J. Biol. Chem. 275, 32331–32337 10.1074/jbc.M004795200 [DOI] [PubMed] [Google Scholar]
- Huang S., Jin L., Du J., Li H., Zhao Q., Ou G., Ao G., Yuan M. (2007). SB401, a pollen-specific protein from Solanum berthaultii, binds to and bundles microtubules and F-actin. Plant J. 51, 406–418 10.1111/j.1365-313X.2007.03153.x [DOI] [PubMed] [Google Scholar]
- Huang S., Robinson R. C., Gao L. Y., Matsumoto T., Brunet A., Blanchoin L., Staiger C. J. (2005). Arabidopsis VILLIN1 generates actin filament cables that are resistant to depolymerization. Plant Cell 17, 486–501 10.1105/tpc.104.028555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey T. V., Bonetta D. T., Goring D. R. (2007). Sentinels at the wall: cell wall receptors and sensors. New Phytol. 176, 7–21 10.1111/j.1469-8137.2007.02192.x [DOI] [PubMed] [Google Scholar]
- Hwang J. U., Lee Y. (2001). Abscisic acid-induced actin reorganization in guard cells of dayflower is mediated by cytosolic calcium levels and by protein kinase and protein phosphatase activities. Plant Physiol. 125, 2120–2128 10.1104/pp.125.4.2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M., Fitz Gerald J. N., Guerin C., Robert H., Sorensen M. B., Van Damme D., Geelen D., Blanchoin L., Berger F. (2005). Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat. Cell Biol. 7, 374–380 10.1038/ncb1238 [DOI] [PubMed] [Google Scholar]
- Kadota A., Yamada N., Suetsugu N., Hirose M., Saito C., Shoda K., Ichikawa S., Kagawa T., Nakano A., Wada M. (2009). Short actin-based mechanism for light-directed chloroplast movement in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 13106–13111 10.1073/pnas.0906250106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaothien P., Kawaoka A., Ebinuma H., Yoshida K., Shinmyo A. (2002). Ntlim1, a PAL-box binding factor, controls promoter activity of the horseradish wound-inducible peroxidase gene. Plant Mol. Biol. 49, 591–599 10.1023/A:1015504515492 [DOI] [PubMed] [Google Scholar]
- Karlsson O., Thor S., Norberg T., Ohlsson H., Edlund T. (1990). Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys-His domain. Nature 344, 879–882 10.1038/344879a0 [DOI] [PubMed] [Google Scholar]
- Kasahara M., Kagawa T., Oikawa K., Suetsugu N., Miyao M., Wada M. (2002). Chloroplast avoidance movement reduces photodamage in plants. Nature 420, 829–832 10.1038/nature01213 [DOI] [PubMed] [Google Scholar]
- Kawaoka A., Ebinuma H. (2001). Transcriptional control of lignin biosynthesis by tobacco LIM protein. Phytochemistry 57, 1149–1157 10.1016/S0031-9422(01)00054-1 [DOI] [PubMed] [Google Scholar]
- Kawaoka A., Kaothien P., Yoshida K., Endo S., Yamada K., Ebinuma H. (2000). Functional analysis of tobacco LIM protein Ntlim1 involved in lignin biosynthesis. Plant J. 22, 289–301 10.1046/j.1365-313x.2000.00737.x [DOI] [PubMed] [Google Scholar]
- Kawaoka A., Sato S., Nakahara K., Matsushima N., Okada N., Sekine M., Shinmyo A., Takano M. (1992). Expression and promoter activity of genes for isozymes of horseradish peroxidase. Plant Cell Physiol. 33, 1143–1150 [Google Scholar]
- Ketelaar T., Faivre-Moskalenko C., Esseling J. J., De Ruijter N. C., Grierson C. S., Dogterom M., Emons A. M. (2002). Positioning of nuclei in Arabidopsis root hairs: an actin-regulated process of tip growth. Plant Cell 14, 2941–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana P., Henty J. L., Huang S., Staiger A. M., Blanchoin L., Staiger C. J. (2010). Arabidopsis VILLIN1 and VILLIN3 have overlapping and distinct activities in actin bundle formation and turnover. Plant Cell 22, 2727–2748 10.1105/tpc.110.076240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Hepler P. K., Eun S. O., Ha K. S., Lee Y. (1995). Actin filaments in mature guard cells are radially distributed and involved in stomatal movement. Plant Physiol. 109, 1077–1084 10.1104/pp.109.3.1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Kaneyama J. R., Suzuki W., Ichikawa K., Ohki T., Kohno Y., Sata M., Nose K., Shibanuma M. (2005). Uni-axial stretching regulates intracellular localization of Hic-5 expressed in smooth-muscle cells in vitro. J. Cell Sci. 118, 937–949 10.1242/jcs.01683 [DOI] [PubMed] [Google Scholar]
- Klahre U., Friederich E., Kost B., Louvard D., Chua N. H. (2000). Villin-like actin-binding proteins are expressed ubiquitously in Arabidopsis. Plant Physiol. 122, 35–48 10.1104/pp.122.1.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M. G., Shi W., Ramagopal U., Tseng Y., Wirtz D., Kovar D. R., Staiger C. J., Almo S. C. (2004). Structure of the actin crosslinking core of fimbrin. Structure (Camb.) 12, 999–1013 10.1016/j.str.2004.04.010 [DOI] [PubMed] [Google Scholar]
- Knight M. R. (2007). New ideas on root hair growth appear from the flanks. Proc. Natl. Acad. Sci. U.S.A. 104, 20649–20650 10.1073/pnas.0710632105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll R., Hoshijima M., Hoffman H. M., Person V., Lorenzen-Schmidt I., Bang M. L., Hayashi T., Shiga N., Yasukawa H., Schaper W., McKenna W., Yokoyama M., Schork N. J., Omens J. H., McCulloch A. D., Kimura A., Gregorio C. C., Poller W., Schaper J., Schultheiss H. P., Chien K. R. (2002). The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell 111, 943–955 10.1016/S0092-8674(02)01226-6 [DOI] [PubMed] [Google Scholar]
- Kobayashi I., Kobayashi Y., Hardham A. R. (1994). Dynamic reorganization of microtubules and microfilaments in flax cells during the resistance response to flax rust infection. Planta 195, 237–247 [Google Scholar]
- Kong Y., Flick M. J., Kudla A. J., Konieczny S. F. (1997). Muscle LIM protein promotes myogenesis by enhancing the activity of MyoD. Mol. Cell. Biol. 17, 4750–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D. R., Gibbon B. C., McCurdy D. W., Staiger C. J. (2001). Fluorescently-labeled fimbrin decorates a dynamic actin filament network in live plant cells. Planta 213, 390–395 10.1007/s004250000494 [DOI] [PubMed] [Google Scholar]
- Kovar D. R., Staiger C. J., Weaver E. A., McCurdy D. W. (2000). AtFim1 is an actin filament crosslinking protein from Arabidopsis thaliana. Plant J. 24, 625–636 10.1046/j.1365-313x.2000.00907.x [DOI] [PubMed] [Google Scholar]
- Krebs M., Beyhl D., Gorlich E., Al-Rasheid K. A., Marten I., Stierhof Y. D., Hedrich R., Schumacher K. (2010). Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc. Natl. Acad. Sci. U.S.A. 107, 3251–3256 10.1073/pnas.0913035107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leckie C. P., Callow J. A., Green J. R. (1995). Reorganization of the endoplasmic reticulum in pea leaf epidermal cells infected by the powdery mildew fungus Erysiphe pisi. New Phytol. 131, 211–221 [Google Scholar]
- Lee B. S., Gluck S. L., Holliday L. S. (1999). Interaction between vacuolar H(+)-ATPase and microfilaments during osteoclast activation. J. Biol. Chem. 274, 29164–29171 10.1074/jbc.274.41.29164 [DOI] [PubMed] [Google Scholar]
- Lemichez E., Wu Y., Sanchez J. P., Mettouchi A., Mathur J., Chua N. H. (2001). Inactivation of AtRac1 by abscisic acid is essential for stomatal closure. Genes Dev. 15, 1808–1816 10.1101/gad.900401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Shen Y., Cai C., Zhong C., Zhu L., Yuan M., Ren H. (2010). The type II Arabidopsis formin14 interacts with microtubules and microfilaments to regulate cell division. Plant Cell 22, 2710–2726 10.1105/tpc.110.075507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B. Q., Jin L., Zhu L., Li J., Huang S., Yuan M. (2009). Phosphorylation of microtubule-associated protein SB401 from Solanum berthaultii regulates its effect on microtubules. J. Integr. Plant Biol. 51, 235–242 10.1111/j.1744-7909.2008.00797.x [DOI] [PubMed] [Google Scholar]
- Liu J., Seul U., Thompson R. (1997). Cloning and characterization of a pollen-specific cDNA encoding a glutamic-acid-rich protein (GARP) from potato Solanum berthaultii. Plant. Mol. Biol. 33, 291–300 10.1023/A:1005746713759 [DOI] [PubMed] [Google Scholar]
- Liu K., Luan S. (1998). Voltage-dependent K+ channels as targets of osmosensing in guard cells. Plant Cell 10, 1957–1970 10.1105/tpc.10.11.1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovy-Wheeler A., Wilsen K. L., Baskin T. I., Hepler P. K. (2005). Enhanced fixation reveals the apical cortical fringe of actin filaments as a consistent feature of the pollen tube. Planta 221, 95–104 10.1007/s00425-004-1423-2 [DOI] [PubMed] [Google Scholar]
- Ma B., Qian D., Nan Q., Tan C., An L., Xiang Y. (2012). Arabidopsis vacuolar H+-ATPase (V-ATPase) B subunits are involved in actin cytoskeleton remodeling via binding to, bundling, and stabilizing F-actin. J. Biol. Chem. 287, 19008–19017 10.1074/jbc.M111.281873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B., Xiang Y., An L. (2011). Structural bases of physiological functions and roles of the vacuolar H(+)-ATPase. Cell. Signal. 23, 1244–1256 10.1016/j.cellsig.2011.03.003 [DOI] [PubMed] [Google Scholar]
- Macrobbie E. A., Kurup S. (2007). Signalling mechanisms in the regulation of vacuolar ion release in guard cells. New Phytol. 175, 630–640 10.1111/j.1469-8137.2007.02131.x [DOI] [PubMed] [Google Scholar]
- Martiniere A., Gayral P., Hawes C., Runions J. (2011). Building bridges: formin1 of Arabidopsis forms a connection between the cell wall and the actin cytoskeleton. Plant J. 66, 354–365 10.1111/j.1365-313X.2011.04497.x [DOI] [PubMed] [Google Scholar]
- Michelot A., Berro J., Guerin C., Boujemaa-Paterski R., Staiger C. J., Martiel J. L., Blanchoin L. (2007). Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr. Biol. 17, 825–833 10.1016/j.cub.2007.04.037 [DOI] [PubMed] [Google Scholar]
- Michelot A., Derivery E., Paterski-Boujemaa R., Guerin C., Huang S., Parcy F., Staiger C. J., Blanchoin L. (2006). A novel mechanism for the formation of actin-filament bundles by a nonprocessive formin. Curr. Biol. 16, 1924–1930 10.1016/j.cub.2006.07.054 [DOI] [PubMed] [Google Scholar]
- Michelot A., Guerin C., Huang S., Ingouff M., Richard S., Rodiuc N., Staiger C. J., Blanchoin L. (2005). The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell 17, 2296–2313 10.1105/tpc.105.030908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. D., De Ruijter N. C., Bisseling T., Emons A. M. (1999). The role of actin in root hair morphogenesis: studies with lipochito-oligosaccharide as a growth stimulator and cytochalasin as an actin perturbing drug. Plant J. 17, 141–154 [Google Scholar]
- Moes D., Gatti S., Hoffmann C., Dieterle M., Moreau F., Neumann K., Schumacher M., Diederich M., Grill E., Shen W. H., Steinmetz A., Thomas C. (2012). A LIM domain protein from tobacco involved in actinbundling and histone gene transcription. Mol. Plant. [Epub ahead of print]. 10.1093/mp/SSS075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley S. C. (2012). The actin-bundling protein L-plastin: a critical regulator of immune cell function. Int. J. Cell Biol. 2012, Article ID: 935173. 10.1155/2012/935173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundel C., Baltz R., Eliasson A., Bronner R., Grass N., Krauter R., Evrard J. L., Steinmetz A. (2000). A LIM-domain protein from sunflower is localized to the cytoplasm and/or nucleus in a wide variety of tissues and is associated with the phragmoplast in dividing cells. Plant Mol. Biol. 42, 291–302 10.1023/A:1006333611189 [DOI] [PubMed] [Google Scholar]
- Nelson N. (2003). A journey from mammals to yeast with vacuolar H+-ATPase (V-ATPase). J. Bioenerg. Biomembr. 35, 281–289 10.1023/A:1025768529677 [DOI] [PubMed] [Google Scholar]
- Nick P. (2010). Probing the actin-auxin oscillator. Plant Signal. Behav. 5, 94–98 10.4161/psb.5.2.10337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick P., Han M. J., An G. (2009). Auxin stimulates its own transport by shaping actin filaments. Plant Physiol. 151, 155–167 10.1104/pp.109.140111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi T., Forgac M. (2002). The vacuolar (H+)-ATPases–nature's most versatile proton pumps. Nat. Rev. Mol. Cell Biol. 3, 94–103 10.1038/nrm729 [DOI] [PubMed] [Google Scholar]
- Oikawa K., Kasahara M., Kiyosue T., Kagawa T., Suetsugu N., Takahashi F., Kanegae T., Niwa Y., Kadota A., Wada M. (2003). Chloroplast unusual positioning1 is essential for proper chloroplast positioning. Plant Cell 15, 2805–2815 10.1105/tpc.016428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa K., Yamasato A., Kong S. G., Kasahara M., Nakai M., Takahashi F., Ogura Y., Kagawa T., Wada M. (2008). Chloroplast outer envelope protein CHUP1 is essential for chloroplast anchorage to the plasma membrane and chloroplast movement. Plant Physiol. 148, 829–842 10.1104/pp.108.123075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S. (2007). Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int. Rev. Cytol. 258, 1–82 10.1016/S0074-7696(07)58001-0 [DOI] [PubMed] [Google Scholar]
- Opalski K. S., Schultheiss H., Kogel K. H., Huckelhoven R. (2005). The receptor-like MLO protein and the RAC/ROP family G-protein RACB modulate actin reorganization in barley attacked by the biotrophic powdery mildew fungus Blumeria graminis f.sp. hordei. Plant J. 41, 291–303 10.1111/j.1365-313X.2004.02292.x [DOI] [PubMed] [Google Scholar]
- Padmanaban S., Lin X., Perera I., Kawamura Y., Sze H. (2004). Differential expression of vacuolar H+-ATPase subunit c genes in tissues active in membrane trafficking and their roles in plant growth as revealed by RNAi. Plant Physiol. 134, 1514–1526 10.1104/pp.103.034025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papuga J., Hoffmann C., Dieterle M., Moes D., Moreau F., Tholl S., Steinmetz A., Thomas C. (2010). Arabidopsis LIM proteins: a family of actin bundlers with distinct expression patterns and modes of regulation. Plant Cell 22, 3034–3052 10.1105/tpc.110.075960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paves H., Truve E. (2007). Myosin inhibitors block accumulation movement of chloroplasts in Arabidopsis thaliana leaf cells. Protoplasma 230, 165–169 10.1007/s00709-006-0230-y [DOI] [PubMed] [Google Scholar]
- Peremyslov V. V., Prokhnevsky A. I., Avisar D., Dolja V. V. (2008). Two class XI myosins function in organelle trafficking and root hair development in Arabidopsis. Plant Physiol. 146, 1109–1116 10.1104/pp.107.113654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasek J., Schwarzerova K. (2009). Actin and microtubule cytoskeleton interactions. Curr. Opin. Plant Biol. 12, 728–734 10.1016/j.pbi.2009.09.010 [DOI] [PubMed] [Google Scholar]
- Pollard T. D., Blanchoin L., Mullins R. D. (2000). Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29, 545–576 10.1146/annurev.biophys.29.1.545 [DOI] [PubMed] [Google Scholar]
- Pollard T. D., Cooper J. A. (2009). Actin, a central player in cell shape and movement. Science 326, 1208–1212 10.1126/science.1175862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka D. R., Kandasamy M. K., McKinney E. C., Burgos-Rivera B., Meagher R. B. (2007). The ancient subclasses of Arabidopsis actin depolymerizing factor genes exhibit novel and differential expression. Plant J. 52, 460–472 10.1111/j.1365-313X.2007.03257.x [DOI] [PubMed] [Google Scholar]
- Samstag Y., Klemke M. (2007). Ectopic expression of L-plastin in human tumor cells: diagnostic and therapeutic implications. Adv. Enzyme Regul. 47, 118–126 10.1016/j.advenzreg.2006.12.008 [DOI] [PubMed] [Google Scholar]
- Schmidt Von Braun S., Schleiff E. (2008a). The chloroplast outer membrane protein CHUP1 interacts with actin and profilin. Planta 227, 1151–1159 10.1007/s00425-007-0688-7 [DOI] [PubMed] [Google Scholar]
- Schmidt Von Braun S., Schleiff E. (2008b). Moving the green: CHUP1 and chloroplast movement-an obvious relationship? Plant Signal. Behav. 3, 488–489 10.4161/psb.3.7.5683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher K., Vafeados D., McCarthy M., Sze H., Wilkins T., Chory J. (1999). The Arabidopsis det3 mutant reveals a central role for the vacuolar H(+)-ATPase in plant growth and development. Genes Dev. 13, 3259–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan M. B., Staiger C. J., Rose R. J., McCurdy D. W. (2004). A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol. 136, 3968–3978 10.1104/pp.104.049411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomiya H. (2012). Plastin family of actin-bundling proteins: its functions in leukocytes, neurons, intestines, and cancer. Int. J. Cell Biol. 2012, 213492 10.1155/2012/213492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter A. M., Keppler B., Lichtenberg J., Gu D., Welch L. R. (2010). A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 153, 485–513 10.1104/pp.110.156554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skau C. T., Courson D. S., Bestul A. J., Winkelman J. D., Rock R. S., Sirotkin V., Kovar D. R. (2011). Actin filament bundling by fimbrin is important for endocytosis, cytokinesis, and polarization in fission yeast. J. Biol. Chem. 286, 26964–26977 10.1074/jbc.M111.239004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko A. P., Deeks M. J., Hussey P. J. (2010). Strategies of actin reorganisation in plant cells. J. Cell Sci. 123, 3019–3028 10.1242/jcs.071126 [DOI] [PubMed] [Google Scholar]
- Smith L. G., Oppenheimer D. G. (2005). Spatial control of cell expansion by the plant cytoskeleton. Annu. Rev. Cell Dev. Biol. 21, 271–295 10.1146/annurev.cellbio.21.122303.114901 [DOI] [PubMed] [Google Scholar]
- Sparkes I. A., Teanby N. A., Hawes C. (2008). Truncated myosin XI tail fusions inhibit peroxisome, golgi, and mitochondrial movement in tobacco leaf epidermal cells: a genetic tool for the next generation. J. Exp. Bot. 59, 2499–2512 10.1093/jxb/ern114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger C. J., Blanchoin L. (2006). Actin dynamics: old friends with new stories. Curr. Opin. Plant Biol. 9, 554–562 10.1016/j.pbi.2006.09.013 [DOI] [PubMed] [Google Scholar]
- Staiger C. J., Hussey P. J. (2004). Actin and actin-modulating proteins, in The Plant Cytoskeleton in Cell Differentiation and Development, ed P. J. Hussey (Oxford, UK: Blackwell Publishing; ), 32–80 [Google Scholar]
- Staiger C. J., Poulter N. S., Henty J. L., Franklin-Tong V. E., Blanchoin L. (2010). Regulation of actin dynamics by actin-binding proteins in pollen. J. Exp. Bot. 61, 1969–1986 10.1093/jxb/erq012 [DOI] [PubMed] [Google Scholar]
- Staiger C. J., Sheahan M. B., Khurana P., Wang X., McCurdy D. W., Blanchoin L. (2009). Actin filament dynamics are dominated by rapid growth and severing activity in the Arabidopsis cortical array. J. Cell Biol. 184, 269–280 10.1111/j.1365-313X.2004.02292.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger C. J., Yuan M., Valenta R., Shaw P. J., Warn R. M., Lloyd C. W. (1994). Microinjected profilin affects cytoplasmic streaming in plant cells by rapidly depolymerizing actin microfilaments. Curr. Biol. 4, 215–219 10.1016/S0960-9822(00)00050-6 [DOI] [PubMed] [Google Scholar]
- Stevenson R. P., Veltman D., Machesky L. M. (2012). Actin-bundling proteins in cancer progression at a glance. J. Cell Sci. 125, 1073–1079 10.1242/jcs.093799 [DOI] [PubMed] [Google Scholar]
- Suetsugu N., Yamada N., Kagawa T., Yonekura H., Uyeda T. Q., Kadota A., Wada M. (2010a). Two kinesin-like proteins mediate actin-based chloroplast movement in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 8860–8865 10.1073/pnas.0912773107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu N., Dolja V. V., Wada M. (2010b). Why have chloroplasts developed a unique motility system? Plant Signal. Behav. 5, 1190–1196 10.4161/psb.5.10.12802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu N., Wada M. (2007). Chloroplast photorelocation movement mediated by phototropin family proteins in green plants. Biol. Chem. 388, 927–935 10.1515/BC.2007.118 [DOI] [PubMed] [Google Scholar]
- Szymanski D. B. (2005). Breaking the WAVE complex: the point of Arabidopsis trichomes. Curr. Opin. Plant Biol. 8, 103–112 10.1016/j.pbi.2004.11.004 [DOI] [PubMed] [Google Scholar]
- Szymanski D. B., Cosgrove D. J. (2009). Dynamic coordination of cytoskeletal and cell wall systems during plant cell morphogenesis. Curr. Biol. 19, R800– R811 10.1016/j.cub.2009.07.056 [DOI] [PubMed] [Google Scholar]
- Takemoto D., Jones D. A., Hardham A. R. (2006). Re-organization of the cytoskeleton and endoplasmic reticulum in the Arabidopsis pen1-1 mutant inoculated with the non-adapted powdery mildew pathogen, Blumeria graminis f. sp. hordei. Mol. Plant Pathol. 7, 553–563 10.1111/j.1364-3703.2006.00360.x [DOI] [PubMed] [Google Scholar]
- Tholl S., Moreau F., Hoffmann C., Arumugam K., Dieterle M., Moes D., Neumann K., Steinmetz A., Thomas C. (2011). Arabidopsis actin-depolymerizing factors (ADFs) 1 and 9 display antagonist activities. FEBS Lett. 585, 1821–1827 10.1016/j.febslet.2011.05.019 [DOI] [PubMed] [Google Scholar]
- Thomas C., Hoffmann C., Dieterle M., Van Troys M., Ampe C., Steinmetz A. (2006). Tobacco WLIM1 is a novel F-actin binding protein involved in actin cytoskeleton remodeling. Plant Cell 18, 2194–2206 10.1105/tpc.106.040956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C., Moreau F., Dieterle M., Hoffmann C., Gatti S., Hofmann C., Van Troys M., Ampe C., Steinmetz A. (2007). The LIM domains of WLIM1 define a new class of actin bundling modules. J. Biol. Chem. 282, 33599–33608 10.1074/jbc.M703691200 [DOI] [PubMed] [Google Scholar]
- Thomas C., Tholl S., Moes D., Dieterle M., Papuga J., Moreau F., Steinmetz A. (2009). Actin bundling in plants. Cell. Motil. Cytoskeleton 66, 940–957 10.1002/cm.20389 [DOI] [PubMed] [Google Scholar]
- Tominaga M., Yokota E., Vidali L., Sonobe S., Hepler P. K., Shimmen T. (2000). The role of plant villin in the organization of the actin cytoskeleton, cytoplasmic streaming and the architecture of the transvacuolar strand in root hair cells of Hydrocharis. Planta 210, 836–843 10.1007/s004250050687 [DOI] [PubMed] [Google Scholar]
- Toth R., Gerding-Reimers C., Deeks M. J., Menninger S., Gallegos R. M., Tonaco I. A., Hubel K., Hussey P. J., Waldmann H., Coupland G. (2012). Prieurianin/endosidin 1 is an actin-stabilizing small molecule identified from a chemical genetic screen for circadian clock effectors in Arabidopsis thaliana. Plant J. 71, 338–352 10.1111/j.1365-313X.2012.04991.x [DOI] [PubMed] [Google Scholar]
- Van Damme D., Bouget F. Y., Van Poucke K., Inze D., Geelen D. (2004). Molecular dissection of plant cytokinesis and phragmoplast structure: a survey of GFP-tagged proteins. Plant J. 40, 386–398 10.1111/j.1365-313X.2004.02222.x [DOI] [PubMed] [Google Scholar]
- van der Honing H. S., Kieft H., Emons A. M., Ketelaar T. (2012). Arabidopsis VILLIN2 and VILLIN3 are required for the generation of thick actin filament bundles and for directional organ growth. Plant Physiol. 158, 1426–1438 10.1104/pp.111.192385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Troys M., Vandekerckhove J., Ampe C. (2008a). Actin and actin-binding proteins in cancer progression and metastasis, in Actin-Binding Proteins and Disease, Protein Reviews, eds Dos Remedios C. G., Chhabra D. (New York, NY: Springer; ), 229–277 [Google Scholar]
- Van Troys M., Huyck L., Leyman S., Dhaese S., Vandekerkhove J., Ampe C. (2008b). Ins and outs of ADF/cofilin activity and regulation. Eur. J. Cell Biol. 87, 649–667 10.1016/j.ejcb.2008.04.001 [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Ma X., Adelstein R. S., Horwitz A. R. (2009). Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10, 778–790 10.1038/nrm2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitavska O., Merzendorfer H., Wieczorek H. (2005). The V-ATPase subunit C binds to polymeric F-actin as well as to monomeric G-actin and induces cross-linking of actin filaments. J. Biol. Chem. 280, 1070–1076 10.1074/jbc.M406797200 [DOI] [PubMed] [Google Scholar]
- Vitavska O., Wieczorek H., Merzendorfer H. (2003). A novel role for subunit C in mediating binding of the H+-V-ATPase to the actin cytoskeleton. J. Biol. Chem. 278, 18499–18505 10.1074/jbc.M212844200 [DOI] [PubMed] [Google Scholar]
- Wang H. J., Wan A. R., Jauh G. Y. (2008). An actin binding protein, LlLIM1, mediates Ca and H regulation of actin dynamics in pollen tubes. Plant Physiol. 47, 1619–1636 10.1104/pp.108.118604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. L., Gao X. Q., Wang X. C. (2011). Stochastic dynamics of actin filaments in guard cells regulating chloroplast localization during stomatal movement. Plant Cell Environ. 34, 1248–1257 10.1111/j.1365-3040.2011.02325.x [DOI] [PubMed] [Google Scholar]
- Wang Y. S., Motes C. M., Mohamalawari D. R., Blancaflor E. B. (2004). Green fluorescent protein fusions to Arabidopsis fimbrin 1 for spatio-temporal imaging of F-actin dynamics in roots. Cell. Motil. Cytoskeleton 59, 79–93 10.1002/cm.20024 [DOI] [PubMed] [Google Scholar]
- Way J. C., Chalfie M. (1988). mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans Cell 54, 5–16 10.1016/0092-8674(88)90174-2 [DOI] [PubMed] [Google Scholar]
- Whippo C. W., Khurana P., Davis P. A., Deblasio S. L., Desloover D., Staiger C. J., Hangarter R. P. (2011). THRUMIN1 is a light-regulated actin-bundling protein involved in chloroplast motility. Curr. Biol. 21, 59–64 10.1016/j.cub.2010.11.059 [DOI] [PubMed] [Google Scholar]
- Wightman R., Turner S. R. (2008). The roles of the cytoskeleton during cellulose deposition at the secondary cell wall. Plant J. 54, 794–805 10.1111/j.1365-313X.2008.03444.x [DOI] [PubMed] [Google Scholar]
- Winder S. J., Ayscough K. R. (2005). Actin-binding proteins. J. Cell Sci. 118, 651–654 10.1242/jcs.01670 [DOI] [PubMed] [Google Scholar]
- Wu Y., Yan J., Zhang R., Qu X., Ren S., Chen N., Huang S. (2010). Arabidopsis FIMBRIN5, an actin bundling factor, is required for pollen germination and pollen tube growth. Plant Cell 22, 3745–3763 10.1105/tpc.110.080283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. R., Staiger C. J., Hamer J. E. (1998). Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc. Natl. Acad. Sci. U.S.A. 95, 12713–12718 10.1073/pnas.95.21.12713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Forgac M. (2001). Microtubules are involved in glucose-dependent dissociation of the yeast vacuolar [H+]-ATPase in vitro. J. Biol. Chem. 276, 24855–24861 10.1074/jbc.M100637200 [DOI] [PubMed] [Google Scholar]
- Xue X. H., Guo C. Q., Du F., Lu Q. L., Zhang C. M., Ren H. Y. (2011). AtFH8 is involved in root development under effect of low-dose latrunculin B in dividing cells. Mol. Plant 4, 264–278 10.1093/mp/ssq085 [DOI] [PubMed] [Google Scholar]
- Yang W., Ren S., Zhang X., Gao M., Ye S., Qi Y., Zheng Y., Wang J., Zeng L., Li Q., Huang S., He Z. (2011). BENT UPPERMOST INTERNODE1 encodes the class II formin FH5 crucial for actin organization and rice development. Plant Cell 23, 661–680 10.1105/tpc.110.081802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Xu M. (2012). Actin bundler PLIM2s are involved in the regulation of pollen development and tube growth in Arabidopsis. J. Plant Physiol. 169, 516–522 10.1016/j.jplph.2011.11.015 [DOI] [PubMed] [Google Scholar]
- Ye J., Zheng Y., Yan A., Chen N., Wang Z., Huang S., Yang Z. (2009). Arabidopsis Formin3 directs the formation of actin cables and polarized growth in pollen tubes. Plant Cell 21, 3868–3884 10.1105/tpc.109.068700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K., Guo C., Chen D., Zhao B., Yang B., Ren H. (2005). Cloning and functional characterization of a formin-like protein (AtFH8) from Arabidopsis. Plant Physiol. 138, 1071–1082 10.1104/pp.104.055665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota E., Takahara K., Shimmen Ki T. T. (1998). Actin-bundling protein isolated from pollen tubes of lily. Biochemical and immunocytochemical characterization. Plant Physiol. 116, 1421–1429 10.1104/pp.116.4.1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota E., Shimmen T. (1999). The 135-kDa actin-bundling protein from lily pollen tubes arranges F-actin into bundles with uniform polarity. Planta 209, 264–266 10.1007/s004250050631 [DOI] [PubMed] [Google Scholar]
- Yokota E., Muto S., Shimmen T. (2000). Calcium-calmodulin suppresses the filamentous actin-binding activity of a 135-kilodalton actin-bundling protein isolated from lily pollen tubes. Plant Physiol. 123, 645–654 10.1104/pp.123.2.645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota E., Vidali L., Tominaga M., Tahara H., Orii H., Morizane Y., Hepler P. K., Shimmen T. (2003). Plant 115-kDa actin-filament bundling protein, P-115-ABP, is a homologue of plant villin and is widely distributed in cells. Plant Cell Physiol. 44, 1088–1099 10.1093/pcp/pcg132 [DOI] [PubMed] [Google Scholar]
- Yokota E., Tominaga M., Mabuchi I., Tsuji Y., Staiger C. J., Oiwa K., Shimmen T. (2005). Plant villin, Lily P-135-ABP, possesses G-actin binding activity and accelerates the polymerization and depolymerization of actin in a Ca2+-sensitive manner. Plant Cell Physiol. 46, 1690–1703 10.1093/pcp/pci185 [DOI] [PubMed] [Google Scholar]
- Zhang H., Qu X., Bao C., Khurana P., Wang Q., Xie Y., Zheng Y., Chen N., Blanchoin L., Staiger C. J., Huang S. (2010). Arabidopsis VILLIN5, an actin filament bundling and severing protein, is necessary for normal pollen tube growth. Plant Cell 22, 2749–2767 10.1105/tpc.110.076257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Zhao Y., Guo Y., Ye K. (2012). Plant actin-binding protein SCAB1 is dimeric actin cross-linker with atypical pleckstrin homology domain. J. Biol. Chem. 287, 11981–11990 10.1074/jbc.M111.338525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhang Y., Tan H., Wang Y., Li G., Liang W., Yuan Z., Hu J., Ren H., Zhang D. (2011a). RICE MORPHOLOGY DETERMINANT encodes the type II formin FH5 and regulates rice morphogenesis. Plant Cell 23, 681–700 10.1105/tpc.110.081349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xiao Y., Du F., Cao L., Dong H., Ren H. (2011b). Arabidopsis VILLIN4 is involved in root hair growth through regulating actin organization in a Ca(2+)-dependent manner. New Phytol. 190, 667–682 10.1111/j.1469-8137.2010.03632.x [DOI] [PubMed] [Google Scholar]
- Zhao Y., Zhao S., Mao T., Qu X., Cao W., Zhang L., Zhang W., He L., Li S., Ren S., Zhao J., Zhu G., Huang S., Ye K., Yuan M., Guo Y. (2011). The plant-specific actin binding protein SCAB1 stabilizes actin filaments and regulates stomatal movement in Arabidopsis. Plant Cell 23, 2314–2330 10.1105/tpc.111.086546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimniak L., Dittrich P., Gogarten J. P., Kibak H., Taiz L. (1988). The cDNA sequence of the 69-kDa subunit of the carrot vacuolar H+-ATPase. Homology to the beta-chain of F0F1-ATPases. J. Biol. Chem. 263, 9102–9112 [PubMed] [Google Scholar]
- Zuo J., Jiang J., Chen S. H., Vergara S., Gong Y., Xue J., Huang H., Kaku M., Holliday L. S. (2006). Actin binding activity of subunit B of vacuolar H+-ATPase is involved in its targeting to ruffled membranes of osteoclasts. J. Bone Miner Res. 21, 714–721 10.1359/jbmr.060201 [DOI] [PubMed] [Google Scholar]
- Zuo J., Vergara S., Kohno S., Holliday L. S. (2008). Biochemical and functional characterization of the actin-binding activity of the B subunit of yeast vacuolar H+-ATPase. J. Exp. Biol. 211, 1102–1108 10.1242/jeb.013672 [DOI] [PubMed] [Google Scholar]