Abstract
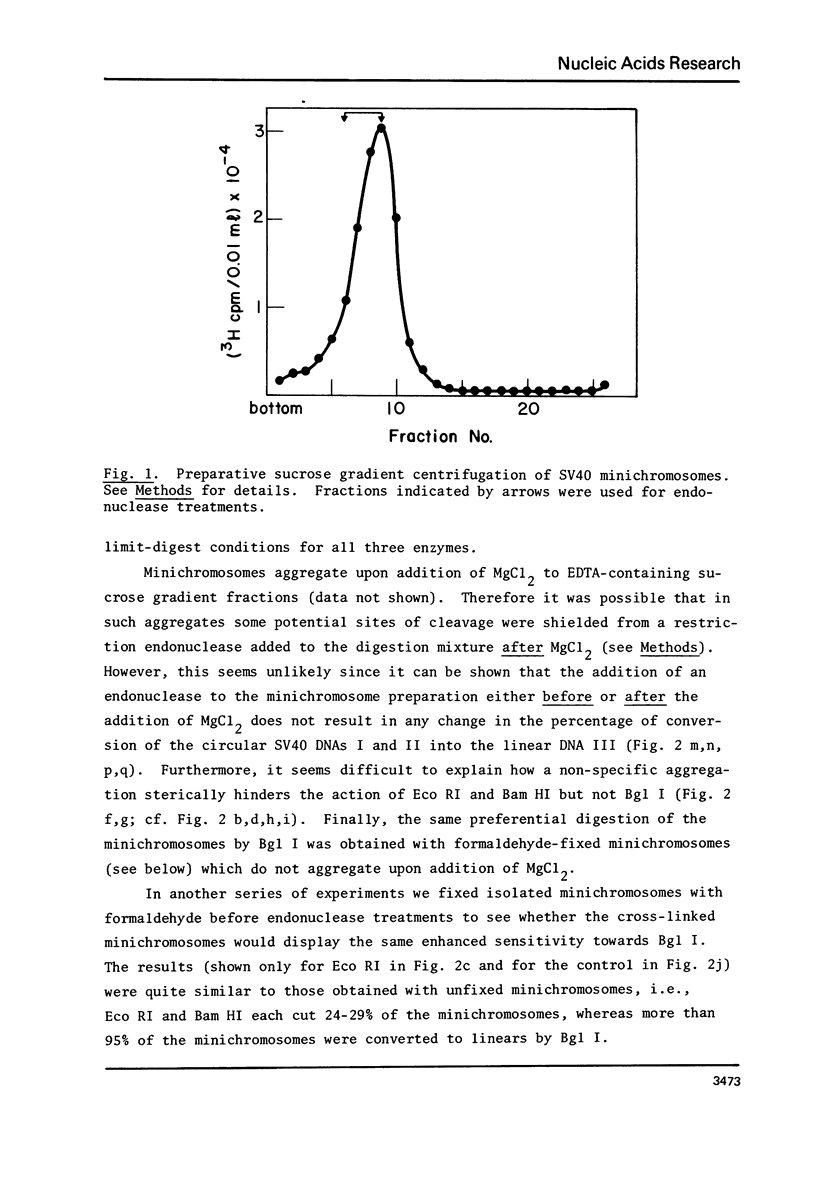
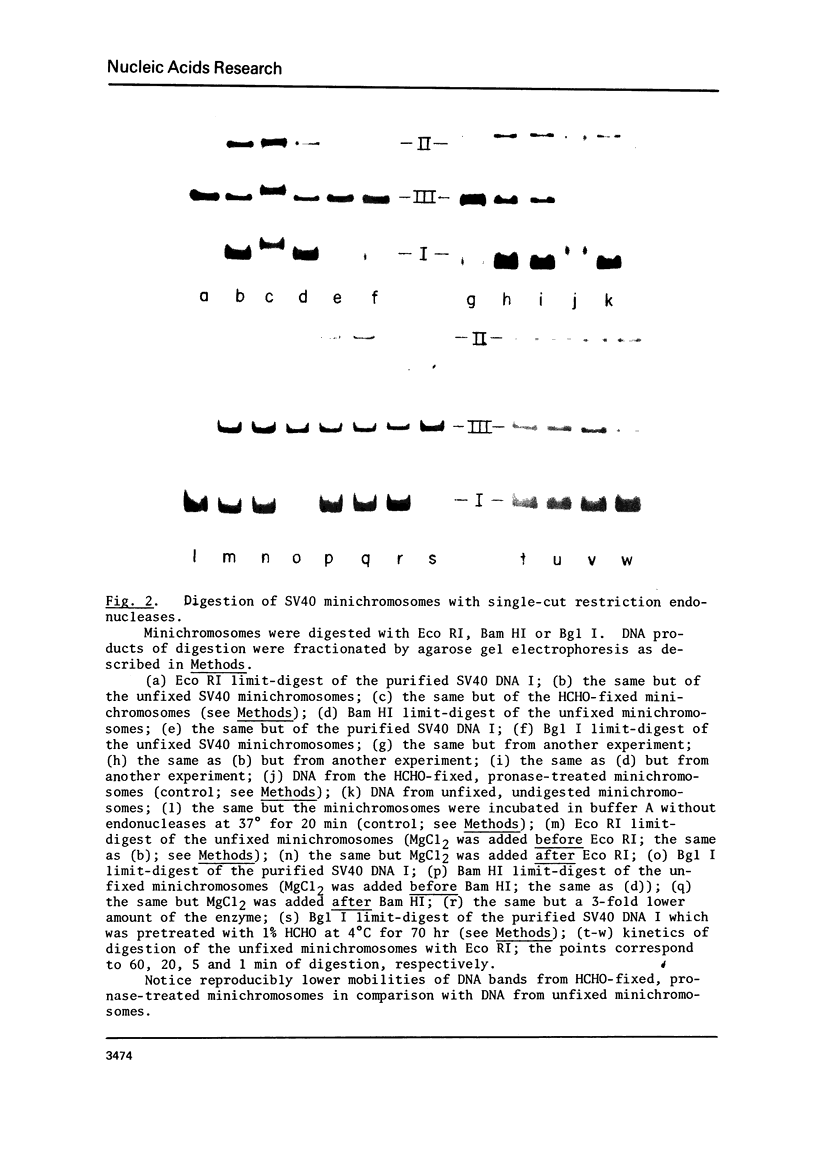
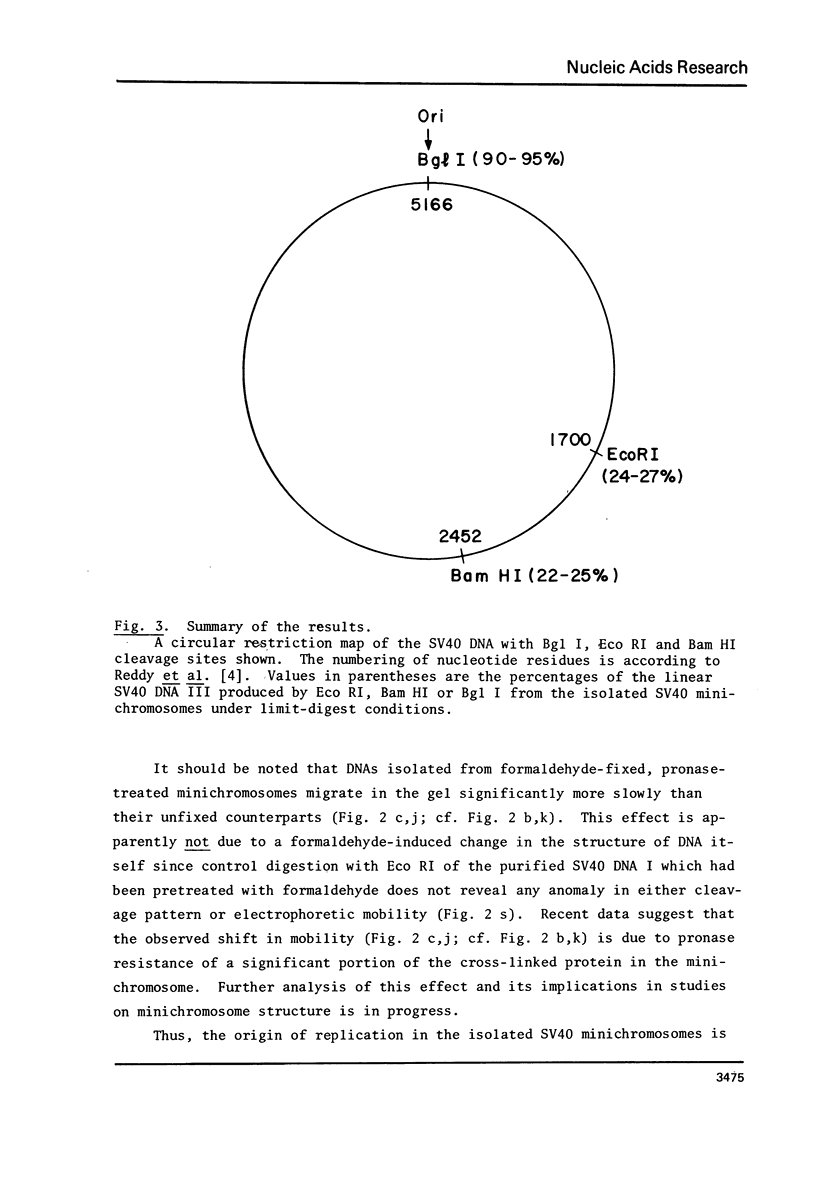
Isolated SV40 minichromosomes [1-3] were treated with different single-cut restriction endonucleases to probe the arrangement of nucleosomes in relation to the SV70 DNA sequence. While Eco RI and Bam HI each cut 22-27% of the SV40 minichromosomes under limit-digest conditions, Bgl I, which cuts SV40 DNA at or very near the origin of replication [4,5], cleaves 90-95% of the minichromosomes in a preparation. Similar results were obtained with minichromosomes which had been fixed with formaldehyde before endonuclease treatment. One possible interpretation of these findings is that the arrangement of nucleosomes in the compact SV40 minichromosomes is nonrandom at least with regard to sequences near the origin of DNA replication.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bellard M., Oudet P., Germond J. E., Chambon P. Subunit structure of simian-virus-40 minichromosome. Eur J Biochem. 1976 Nov 15;70(2):543–553. doi: 10.1111/j.1432-1033.1976.tb11046.x. [DOI] [PubMed] [Google Scholar]
- Christiansen G., Griffith J. Salt and divalent cations affect the flexible nature of the natural beaded chromatin structure. Nucleic Acids Res. 1977 Jun;4(6):1837–1851. doi: 10.1093/nar/4.6.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremisi C., Pignatti P. F., Croissant O., Yaniv M. Chromatin-like structures in polyoma virus and simian virus 10 lytic cycle. J Virol. 1975 Jan;17(1):204–211. doi: 10.1128/jvi.17.1.204-211.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg H. J., Waqar M. A., Huberman J. A. DNA synthesis by partially purified replicating simian virus 40 chromosomes. Nucleic Acids Res. 1977 Sep;4(9):3083–3095. doi: 10.1093/nar/4.9.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin. Nature. 1978 Jan 12;271(5641):115–122. doi: 10.1038/271115a0. [DOI] [PubMed] [Google Scholar]
- Fey G., Hirt B. Fingerprints of polyoma virus proteins and mouse histones. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):235–241. doi: 10.1101/sqb.1974.039.01.030. [DOI] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Haegemann G., Rogiers R., Van de Voorde A., Van Heuverswyn H., Van Herreweghe J., Volckaert G., Ysebaert M. Complete nucleotide sequence of SV40 DNA. Nature. 1978 May 11;273(5658):113–120. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- Frearson P. M., Crawford L. V. Polyoma virus basic proteins. J Gen Virol. 1972 Feb;14(2):141–155. doi: 10.1099/0022-1317-14-2-141. [DOI] [PubMed] [Google Scholar]
- Green M. H., Brooks T. L. Recently replicated simian virus 40 DNA is a preferential template for transcription and replication. J Virol. 1978 May;26(2):325–334. doi: 10.1128/jvi.26.2.325-334.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J. D. Chromatin structure: deduced from a minichromosome. Science. 1975 Mar 28;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- James G. T. Inactivation of the protease inhibitor phenylmethylsulfonyl fluoride in buffers. Anal Biochem. 1978 Jun 1;86(2):574–579. doi: 10.1016/0003-2697(78)90784-4. [DOI] [PubMed] [Google Scholar]
- Kay A. C., Singer M. F. Histone synthesis during infection of monkey kidney cells with Simian Virus 40. Nucleic Acids Res. 1977 Oct;4(10):3371–3386. doi: 10.1093/nar/4.10.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Lake R. S., Barban S., Salzman N. P. Resolutions and identification of the core deoxynucleoproteins of the simian virus 40. Biochem Biophys Res Commun. 1973 Sep 18;54(2):640–647. doi: 10.1016/0006-291x(73)91471-x. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975 Aug 15;56(2):335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- Musich P. R., Maio J. J., Brown F. L. Subunit structure of chromatin and the organization of eukaryotic highly repetitive DNA: indications of a phase relation between restriction sites and chromatin subunits in African green monkey and calf nuclei. J Mol Biol. 1977 Dec 15;117(3):657–677. doi: 10.1016/0022-2836(77)90063-8. [DOI] [PubMed] [Google Scholar]
- Müller U., Zentgraf H., Eicken I., Keller W. Higher order structure of simian virus 40 chromatin. Science. 1978 Aug 4;201(4354):406–415. doi: 10.1126/science.208155. [DOI] [PubMed] [Google Scholar]
- Persico-DiLauro M., Martin R. G., Livingston D. M. Interaction of Simian Virus 40 chromatin with Simian Virus 40 T-antigen. J Virol. 1977 Nov;24(2):451–460. doi: 10.1128/jvi.24.2.451-460.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polisky B., McCarthy B. Location of histones on simian virus 40 DNA. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2895–2899. doi: 10.1073/pnas.72.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder B. A., Crawford L. V. The arrangement of nucleosomes in nucleoprotein complexes from polyoma virus and SV40. Cell. 1977 May;11(1):35–49. doi: 10.1016/0092-8674(77)90315-4. [DOI] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Schaffhausen B. S., Benjamin T. L. Deficiency in histone acetylation in nontransforming host range mutants of polyoma virus. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1092–1096. doi: 10.1073/pnas.73.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjian R. The binding site on SV40 DNA for a T antigen-related protein. Cell. 1978 Jan;13(1):165–179. doi: 10.1016/0092-8674(78)90147-2. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Bakayev V. V., Chumackov P. M., Georgiev G. P. Minichromosome of simian virus 40: presence of histone HI. Nucleic Acids Res. 1976 Aug;3(8):2101–2113. doi: 10.1093/nar/3.8.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. J., Nedospasov S. A., Schmatchenko V. V., Bakayev V. V., Chumackov P. M., Georgiev G. P. Compact form of SV40 viral minichromosome is resistant to nuclease: possible implications for chromatin structure. Nucleic Acids Res. 1977 Oct;4(10):3303–3325. doi: 10.1093/nar/4.10.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]