Abstract
Microbial mats are self-sustained, functionally complex ecosystems that make good models for the understanding of past and present microbial ecosystems as well as putative extraterrestrial ecosystems. Ecological theory suggests that the composition of these communities might be affected by nutrient availability and disturbance frequency. We characterized two microbial mats from two contrasting environments in the oligotrophic Cuatro Ciénegas Basin: a permanent green pool and a red desiccation pond. We analyzed their taxonomic structure and composition by means of 16S rRNA clone libraries and metagenomics and inferred their metabolic role by the analysis of functional traits in the most abundant organisms. Both mats showed a high diversity with metabolically diverse members and strongly differed in structure and composition. The green mat had a higher species richness and evenness than the red mat, which was dominated by a lineage of Pseudomonas. Autotrophs were abundant in the green mat, and heterotrophs were abundant in the red mat. When comparing with other mats and stromatolites, we found that taxonomic composition was not shared at species level but at order level, which suggests environmental filtering for phylogenetically conserved functional traits with random selection of particular organisms. The highest diversity and composition similarity was observed among systems from stable environments, which suggests that disturbance regimes might affect diversity more strongly than nutrient availability, since oligotrophy does not appear to prevent the establishment of complex and diverse microbial mat communities. These results are discussed in light of the search for extraterrestrial life. Key Words: Cuatro Ciénegas—Metagenomics—Microbial mats—Oligotrophic—Phosphorus limitation—Stromatolites. Astrobiology 12, 659–673.
1. Introduction
Microbial mats are self-sustained laminated organosedimentary ecosystems that develop on solid/water interfaces and are composed of tightly interacting microorganisms. They form colored multilayered biofilms embedded in a matrix of extrapolymeric substances that bind cells and inorganic substances together, creating steep biogeochemical microgradients as a result of their own metabolism (Jorgensen et al., 1986; van Gemerden, 1993; Stolz, 2000). This compact gradient (2–50 mm) generates a diverse array of microniches in which different functional guilds can thrive and the spatial closeness allows for the development of complex metabolite exchange networks that ensure survival even under extreme conditions (Paerl and Yannarell, 2010).
Although modern microbial mats are geographically widespread (Gerdes, 2010), they are limited in their environmental occurrence to a selected few aquatic environments, both freshwater and marine (Krumbein et al., 1977; Bebout et al., 2002). This contrasts with their former distribution on Earth, as revealed by the abundance of biogenic, laminated reeflike structures in the fossil record from Precambrian shallow marine environments (Awramik, 1984) as early as 3.4 Ga (Allwood et al., 2006). It is believed that both an exclusion of grazing eukaryotes and a lack of competition for light with fast-growing algae are required for a microbial mat to develop, which restricts their current distribution mostly to extreme environments in modern Earth (Cohen, 1989; Bebout et al., 2002). Many current habitats can be considered environmental analogues to those found in Precambrian shallow waters (Foster and Mobberley, 2010). As such, microbial mats are the oldest ecosystems known to date and have proven to be successful ecological assemblages, given that they have survived billions of years of environmental change due to their stable, but adaptable, structural properties (Awramik, 1976; Green and Jahnke, 2010).
Since microbial mats are self-sustained heterogeneous ecosystems, they are suitable experimental models that can be used to test how ecosystems respond to rapid environmental disturbances, which cannot be tested with any other ecosystem (Paerl et al., 2003; Yannarell et al., 2007). They have also allowed for the reconstruction of putative metabolisms and the characterization of biosignatures of early life on Earth (Kasting, 2001; Bebout et al., 2004; Foster and Mobberley, 2010). Without doubt, the study of Earth's most common, simple, and pervasive ecosystems at the molecular level will help us better understand the evolution of life on Earth and, as a consequence, will facilitate the search for life elsewhere (Foster and Mobberley, 2010).
Microbial mats can be found in extreme conditions, including cold (0.4–3.4°C; Bottos et al., 2008; Varin et al., 2010) and high concentrations of iron (Emerson and Revsbech, 1994), sulfur (Elshahed et al., 2003), and hydrocarbons (Mills et al., 2005). The vast majority of the literature, however, has focused on halophilic coastal mats (like those in Guerrero Negro, Mexico, or in Shark Bay, Australia) or hyperthermophilic mats (like those in Yellowstone) (Ward et al., 1998; Spear et al., 2003). Mats from coastal hypersaline environments can harbor diverse and complex ecosystems (Ley et al., 2006; Kunin et al., 2008). In contrast, mats that grow at high temperatures (64–82°C) are characterized by a low species diversity (Inskeep et al., 2010; Meyer-Dombard et al., 2011), most likely because temperature sets a limit for photosynthesis (Ward et al., 1998). This suggests that nutrient availability and primary production will determine the complexity of a microbial mat community, as has been previously proposed for plant communities (Grime, 1973; Tilman, 1990). The ecological characterization of communities from low-nutrient (oligotrophic) environments can aid the search of life on other planets by defining a nutrient availability range for the development of life, so that astrobiological targets can be more precisely defined.
Another ecological factor to be considered in the search for life beyond the confines of Earth is environmental disturbance, since communities will not develop where disturbances are too frequent or too intense, while an intermediate disturbance frequency in many cases will maximize complexity by creating several microniches (Grime, 1973; Connell, 1978; Rainey and Travisano, 1998). It is therefore relevant to study how communities are affected by disturbance regimes to be able to predict their possible existence in other planets.
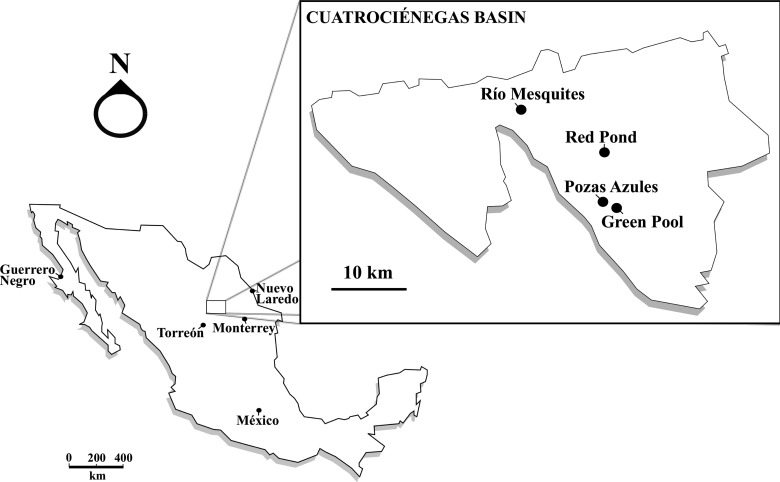
The Cuatro Ciénegas Basin (CCB) is a naturally isolated valley in the Chihuahuan Desert (Coahuila, Mexico) that encompasses hundreds of permanent ponds, marshes, and desiccation pools (Minckley and Cole, 1968) (Fig. 1), which despite their low phosphorus content (Elser et al., 2005; Breitbart et al., 2009, Peimbert et al., 2012 in this issue) harbor varied and diverse microbial communities (Souza et al., 2006; Escalante et al., 2008). The CCB has received the attention of astrobiological research because it is considered a modern analogue of the Precambrian oceans (Elser et al., 2006), which have been characterized as having had low levels of circulating phosphorus (Bjerrum and Canfield, 2002; Planavsky et al., 2010) and a reduced deposition of phosphorite rocks (Papineau, 2010). Moreover, complex microbial communities are known to have developed in Precambrian waters, since abundant phosphate-absorptive calcareous stromatolites have been preserved in the geological record (Grotzinger and Knoll, 1999; Gerdes, 2010). Finally, the CCB has also been identified as an analogue of the martian Olympia Undae gypsum sand dunes (Szynkiewicz et al., 2010) and the dynamic fluvial system of the Gale Crater of early Mars, one of the four landing sites targeted for the Mars Science Laboratory mission later this year (Golombek et al., 2011).
FIG. 1.
Schematic map depicting the geographic origin of the metagenomic data sets analyzed in this study.
All this stresses the need to characterize the microbial communities that inhabit the CCB, while its oligotrophic waters and diversity of aquatic environments offer the opportunity to study the limiting effects of phosphorus availability on microbial community development. In the present study, we taxonomically characterized two aquatic, nonthermophilic, nonhalophilic oligotrophic microbial mats from two environments with contrasting disturbance regimes: a permanent green pool and a red desiccation pond. We analyzed their taxonomic structure and composition by using 16S rRNA gene clone libraries and metagenomics. A low diversity was expected in these oligotrophic environments, with a greater diversity in the mat from the desiccation pond due to intermediate disturbances. We also compared our data with two previously published metagenomes of microbial communities from the CCB (the stromatolites of Río Mesquites and Poza Azul; Breitbart et al., 2009) and with the only other microbial mat metagenome available, the well-studied coastal halophilic microbial mat from the salines of Guerrero Negro (Baja California, Mexico; Spear et al., 2003; Kunin et al., 2008). We expected to find large differences in taxonomic composition, since Guerrero Negro and the CCB are geographically separated by a linear distance of 1200 km.
2. Material and Methods
Two 20×20 cm microbial mat cores were collected in July 2008 from a shallow, seasonal red desiccation pond in the Los Hundidos region (red mat, at 26°52′17″N, 102°01′11.3″W) and a permanent green pool in Pozas Azules Ranch (green mat, at 26°49′24.4″N, 102°00′53.2″W) (Fig. 1). Due to the nature of the desiccation ponds, the temperature disturbance frequency was 5 times higher for the red mat than for the green mat (Table 1). Samples were frozen in liquid nitrogen and transported to the laboratory. DNA extraction was performed by Freeze/Thaw, CTAB, phenol-chloroform extraction as described previously (Zhou et al., 1996; Breitbart et al., 2009). The samples were further purified by electrodialysis as described by Rodríguez-Mejía et al. (2008). Total DNA was amplified with Genomiphi polymerase (GE Healthcare, Piscataway, NJ, USA) according to the manufacturer's instructions. Ten independent reactions were carried out and later pooled before sequencing to reduce amplification bias. From the total DNA, 400 μL of one red mat (551 ng/μL) and 400 μL of the green mat (890 ng/μL) were independently pyrosequenced with 454 FLX (Roche Diagnostics, IN, USA) at CINVESTAV-LANGEBIO, Irapuato, Mexico. Metagenomic reads were noise-corrected with CD-HIT 454 (Li and Godzik, 2006). RNA sequences were identified and masked with rRNA-HMM and tRNA-scan (Huang et al., 2009). Open reading frames calling was performed with GeneMark (Besemer and Borodovsky, 2005) and annotated with RAMMCAP (Li, 2009). Data sets were uploaded to the online server MG-RAST and subjected to its standard quality control pipeline (Meyer et al., 2008). Data is available via the MG-RAST portal (green mat ID is 4441363.3, and red mat ID is 4442466.3).
Table 1.
Physiochemical Properties of the Aquatic Environments from Where the Samples Analyzed in This Study Were Taken
| Tm (°C) | T* (°C) | pH* | Conductivity (μS/cm)* | Stoichiometric N to P ratio** | |
|---|---|---|---|---|---|
| Green | 25 [15–24] | 30.27 | 6.0 | 2.57 | 2:1 |
| Red | 30 [10–60] | 48.26 | 5.5 | 117.6 | 157:1 |
| Red 2 | NA | 47.58 | 7.3 | 83.5 | NA |
At time of sampling.
Numbers of nitrogen atoms per phosphorus atom. For reference, the classic “Redfield” ratio when nutrients are not limiting is 16:1.
Tm=Yearly average temperature mean; minimum and maximum temperatures are shown in brackets.
NA, Data not available.
2.1. 16S rRNA gene clone libraries analyses
A 16S rRNA gene clone library was constructed for each of the two mats and one additional mat from an adjacent red desiccation pool in Los Hundidos (red mat 2). The PCR reaction mixture for amplification of 16S rRNA genes contained 1.5 mM MgCl2, 250 mM of each nucleotide, 4 mM of each primer, 1 U of Taq DNA polymerase (Roche, Mannheim, Germany), and 50 ng of isolated DNA. The bacterial 16S rRNA gene was amplified with the universal primers 27F (5-AGA GTT TGA TCC TGG CTC AG-3) and 1492R (5-GGT TAC CTT GTT ACG ACT T-3). Amplification was performed as follows: a “hot start” (95°C for 5 min) was followed by 25 cycles at 94°C for 40 s, 55°C for 40 s, and 72°C for 90 s with a 10 min extension at 72°C. Amplified products were purified with a Roche PCR purification Kit (Roche, Mannheim, Germany), and PCR products were cloned with a TOPO cloning kit (Invitrogen, Karlsruhe, Germany) following the instructions of the manufacturer. Plasmids were sequenced on one end by using dye terminator chemistry on an automated DNA sequencer (ABI3700, Applied Biosystems).
Raw sequences were chimera-checked with Mallard (Ashelford et al., 2006), trimmed of vector sequences, and checked for errors and low quality with BioEdit (Hall, 1999). Sequences were aligned and analyzed with ARB (Ludwig et al., 2004), assigned to a taxonomic category with the GreenGenes Classifier (DeSantis et al., 2006), and assigned to a phylogenetic position in reference to the SILVA database in ARB. Clustering of operational taxonomic units (OTUs) at 97% identity, alpha-diversity indexes, and rarefaction analyses were performed with MOTHUR (Schloss et al., 2009). Phylogenetic trees were constructed with PhyML software as implemented in ARB (GTR+I model, 1000 bootstraps), and edited with iTOL (Letunic and Bork, 2011).
2.2. Metagenomic protein gene phylogenetic marker analyses
The entire metagenomic data set of stromatolites from Poza Azul (ID 4440067.3) and Río Mesquites (ID 4440060.4) were downloaded from MG-RAST (Meyer et al., 2008) and subjected to the exact same quality control and annotation analyses as described above. The 31 universally conserved, single-copy phylogenetic molecular markers identified with AMPHORA (Wu and Eisen, 2008) were searched for and assigned to taxonomic categories with MEGAN (Huson et al., 2007). Analyses were performed at genus level. Taxa-abundance matrices were built to calculate ecological distance matrices by using the Bray-Curtis distance and were subjected to a principal component analysis. Mantel's test was performed to evaluate correlation between geographical and ecological distance. Community structure for the four data sets was evaluated with Renyi's entropy profiles (Bent and Forney, 2008). All ecological analyses were performed with R (R Development Core Team, 2006). A list of the diversity metrics can be found in Supplementary Material S4b (Supplementary Data are available online at www.liebertonline.com/ast).
2.3. Metagenomic all-reads complement analyses
The resulting metagenomic data sets of the green and red microbial mats were uploaded and annotated with the MG-RAST automatic pipeline, where each single read in a metagenome can be assigned to a taxonomic category by comparing it with the available annotated databases in the SEED platform (Meyer et al., 2008). Hits with an e-value lower than 1×10−5 were used to build taxonomic profiles for the metagenomes. The same analyses were performed with the pyrosequenced metagenomes of the Poza Azul and Río Mesquites stromatolites (Breitbart et al., 2009) and the shotgun capillarily sequenced metagenomes from Guerrero Negro (Kunin et al., 2008), which were all downloaded from the MG-RAST portal (identification numbers 4440060.4, 4440067.3, and 4440963.3 to 4440972.3). The profiles of the 10 data sets from Guerrero Negro were pooled together since the referred study analyzed each layer of the microbial mat separately. These profiles allowed us to compare a larger number of reads from each metagenome by assigning them to different taxonomic categories at different taxonomic levels. We considered abundant taxa as those comprising 75% of the total annotated reads in each metagenome and compared these across metagenomes. The fragment recruitment analyses were calculated in MG-RAST with a maximum e-value cutoff of 1e-05. The presence of specific genes in the genomes that recruited the largest number of reads was analyzed in the Integrated Microbial Genomes portal (Markowitz et al., 2009).
3. Results
We evaluated diversity of two microbial mats, using three different approaches: 16S rRNA gene clone library construction, protein-coding phylogenetic marker genes from metagenomes, and all-reads analysis from metagenomes. These approaches are complementary, as each one analyzed different aspects of community structure and composition. For example, the abundance of minor phyla (Armatimonadetes, Caldisericia, Nitrospira, OD1, Lentisphaera, and Deferribacteres) is only apparent with the clone library approach, while the protein marker approach gives a more precise structure estimation and allows the comparison between previously published metagenomes for which no clone libraries are available. The large proportion of cyanobacterial genomes is revealed by comparing the two previous approaches with the all-reads taxonomic composition.
3.1. 16S rRNA gene clone libraries analyses
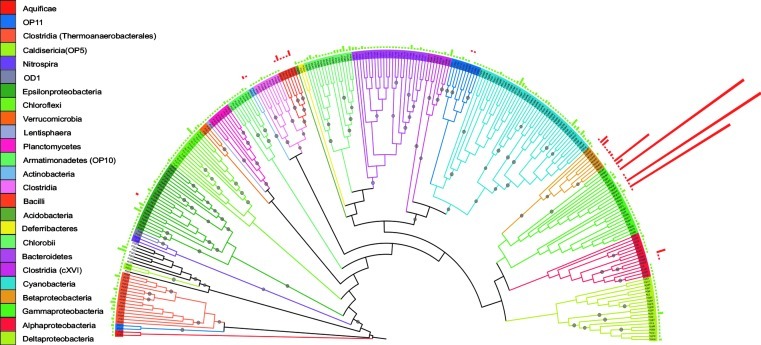
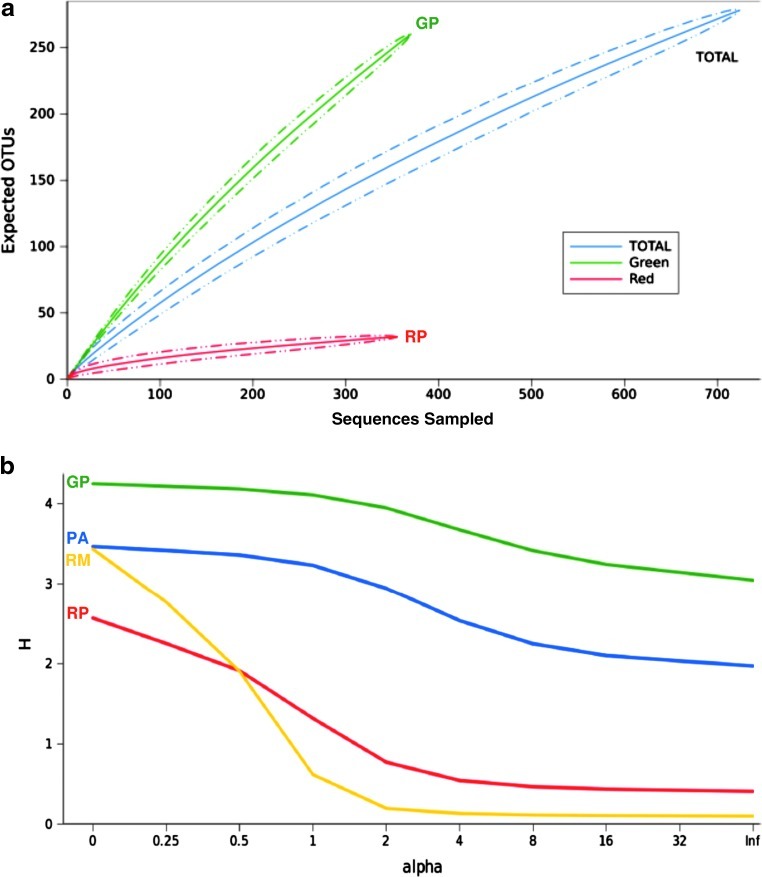
Highly structured microbial mats were sampled from two different kinds of oligotrophic pools in the CCB: a green mat from a stable permanent pool and two red mats from two desiccation ponds. A total of 516 unique phylotypes were recovered from 724 sequences. Clustering at 97% identity produced 32 OTUs for the red mat and 260 for the green mat, comprising 23 phyla. The red mat had low evenness and was dominated by a few OTUs from the genus Pseudomonas (Shannon=2.003, Simpson=0.216), while the green mat showed a very high evenness with no dominant OTU (Shannon=5.389, Simpson=0.003), though the phyla Cyanobacteria, Clostridia, Gammaproteobacteria, Epsilonproteobacteria, and Deltaproteobacteria were abundant (Fig. 2). The Pseudomonas sequences belonged to the P. fluorescens (P. fluorescens and P. corrugata/P. brassicacearum) subgroups and the P. pachastrellae lineages (Mulet et al., 2010). The nonparametric richness estimator Chao estimated 1221 OTUs (C.I.=904–1704) for the green mat and 80 OTUs (C.I.=42–221) for the red mat. A total regional richness of 1198 OTUs (C.I.=902–1643) is expected in microbial mats of the CCB, calculated from a composite pool of the individual libraries. The two microbial mat communities were phylogenetically and statistically different, both in terms of composition (Sorensen=0.095, UniFrac U=0.78) and structure (Morisita-Horn=0.042, Weighted UniFrac W=0.91). Both communities shared only 8 OTUs, with a total of 21 OTUs expected to be shared by the Chao richness estimator.
FIG. 2.
Consensus phylogenetic tree of 16S rRNA gene clone libraries from the green and red microbial mats reconstructed by Maximum Likelihood with 1000 bootstraps. The outer rings represent the relative abundance of each OTU clustered at 97% of the green (green bars) and red (red bars) mats. Black points on the branches indicate clades present in >80% of the phylogenies. Color key for bacterial phyla are given clockwise from top to bottom.
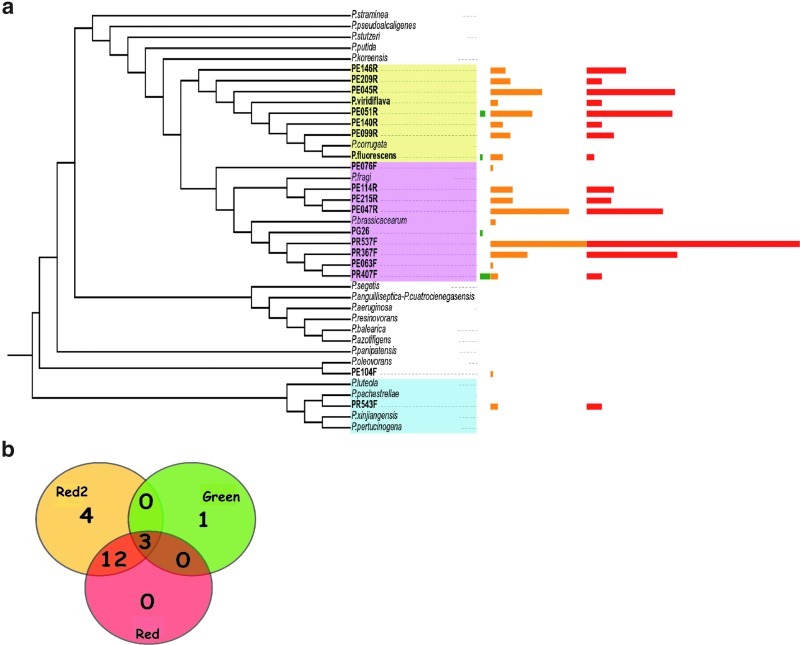
The 16S rRNA gene clone library of a second red mat (red mat 2) from a second desiccation pool was produced for comparison. Forty-two operational taxonomic units were recovered at 97% identity from 218 sequences. This red mat 2 was also dominated by Pseudomonas, although it was slightly more diverse than the first red mat (Shannon=3.546, Simpson=0.022, Chao=122 [73-246]). The red mat 2 was more similar to the first red mat (Sorensen=0.253, Morisita-Horn=0.193) and shared 15 OTUs, with a total of 77 shared OTUs expected (Fig. 3).
FIG. 3.
(a) Phylogenetic tree of the Pseudomonas subclade reconstructed by maximum likelihood. OTUs containing sequences from this study are typed in boldface, and reference sequences are in italics. The abundance bars represent the number of sequences contained in each OTU at 98% identity. Bars are colored as follows: green mat (green), red mat (red), red mat 2 (orange). (b) Venn diagram depicting the number of shared and unique OTUs between libraries. Color images available online at www.liebertpub.com/ast
3.2. Metagenomic protein gene phylogenetic marker analyses
The metagenomes of both mats were sequenced, which generated a total of 150,381,320 bp and 709,799 reads. The green mat metagenome consisted of 427,366 reads with an average length of 202.54 bp and a maximum length of 390 bp. The average GC content was 39.7%. The red mat metagenome consisted of 282,433 reads with an average length of 225.98 bp and a maximum length of 366 bp. The average GC content was 52.8%.
A total of 1066 sequences from 31 protein marker genes were retrieved from the data sets with AMPHORA (Wu and Eisen, 2008): 831 from the green mat and 235 from the red mat. In agreement with the clone library, the red mat was less diverse (Shannon=1.315, Simpson=0.464, Chao=23.5±31.108 SE) and dominated by Pseudomonas. Genera Sphingopixis (Sphingomonadales), Chitinophaga (Sphingobacteria), and Microcoleus (Cyanobacteria) were also abundant. In contrast, the green mat was highly diverse (Shannon=4.108, Simpson=0.019, Chao=148±33.618), with no dominant genus. Among the abundant genera were the phototrophic Synechococcus (Cyanobacteria) and Chloroherpeton (Chlorobi); the heterotrophic Legionella (Gammaproteobacteria), Algoriphagus and Bacteroides (Bacteroidetes), and the deltaproteobacterial Desulfococcus and Synthrophobacter (Fig. 4).
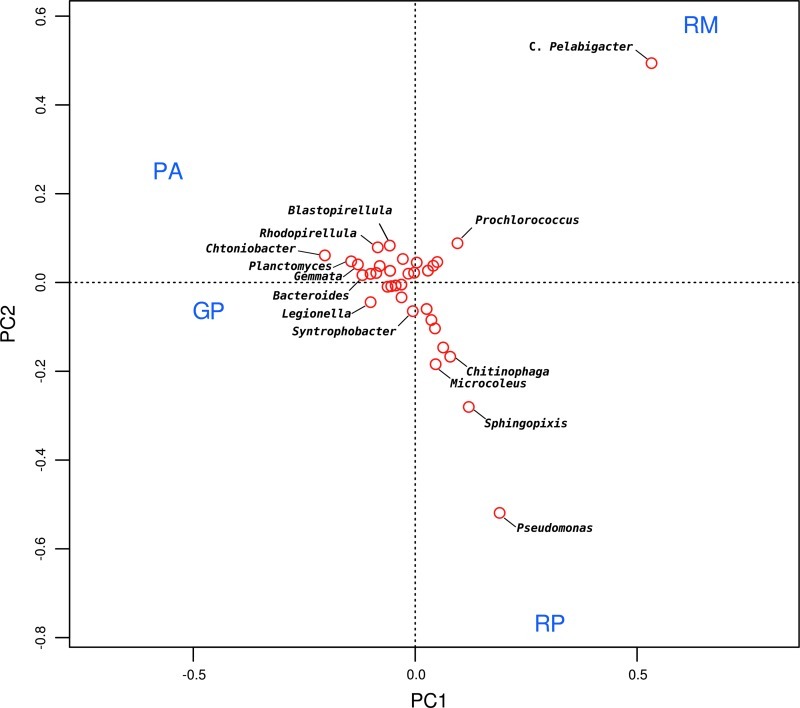
FIG. 4.
Principal component analysis ordination plot of the Cuatro Ciénegas metagenomes using the taxon-abundance matrix constructed with deduced protein markers in the green (GP) and red (RP) microbial mats and Pozas Azules (PA) and Río Mesquites (RM) stromatolites. The names of the top 14 genera most greatly contributing to the principal components are indicated. Color images available online at www.liebertonline.com/ast
The microbial mat metagenomes were compared with two other metagenomes from the CCB: a stromatolite from the permanent pool Pozas Azules and a riverine oncolite from Río Mesquites (Breitbart et al., 2009). The analysis of the same 31 marker genes retrieved 785 sequences from Río Mesquites and 238 from Pozas Azules. The most abundant genera in Pozas Azules all belonged to the superphylum Verrucomicrobia–Planctomycetes (Chthoniobacter, Verrucomicrobium, Rhodopirellula, Akkermansia, Gemmata, Planctomyces) and exhibited a high diversity (Shannon=3.230, Simpson=0.053, Chao=145.2±127.552). Río Mesquites showed far less diversity (Shannon=0.614, Simpson=0.824, Chao=132±147.344), with most sequences belonging to the oligotrophic genus Pelagibacter (formerly known as the SAR11 cluster, Giovannoni et al., 2005). These findings are in agreement with recent clone libraries from DNA obtained from similar structures in the same system (Nitti et al., 2012).
The principal component analysis (Fig. 4) showed a larger similarity between the green mat and the Pozas Azules stromatolite because these samples aggregated together, though independently from the red mat and Río Mesquites stromatolite. Ecological indexes also showed a larger similarity between the green mat and the Pozas Azules stromatolite (Sorensen=0.194, Morisita-Horn=0.2693) than between any other pair of samples (<0.05).
Renyi's community profiles revealed that the green mat was the most diverse sample, while the red mat was the least rich and Río Mesquites showed the largest dominance. Although the two stromatolite samples showed a similar level of richness, they differed in dominance (Fig. 5). Mantel's test was not significant and showed no correlation between ecological and geographical distances (r=0.2821, p=0.704). A guide to the interpretation of Renyi's profiles can be found in Supplementary Material S4c.
FIG. 5.
(a) Estimated rarefaction curves for the 16S rRNA gene clone libraries, with OTUs clustered at 97% identity. (b) Renyi's entropy profile for the four Cuatro Ciénegas metagenomes. Profiles were calculated with the taxon-abundance matrix constructed with protein marker analyses. Green mat (GP), red mat (RP), Río Mesquites (RM), and Pozas Azules (PA). Color images available online at www.liebertonline.com/ast
3.3. Metagenomic all-reads complement analyses
The green mat and the Pozas Azules stromatolite samples displayed the highest evenness: 28 different orders composed 75% of the classified reads in the green mat and 22 orders in Pozas Azules. Among the most abundant taxa in the green mat were photosynthetic taxa (Cyanobacteria, Chloroflexales, Chlorobiales, Chromatiales, Rhodobacterales) and known heterotrophic taxa (Clostridiales, Bacillales, Burkholderiales). In Pozas Azules, the most abundant taxa were the Planctomycetes/Verrucomicrobia complex (Planctomycetales, Verrucomicrobiales, Spartobacteria) and the Cyanobacteria (Chroococcales, Nostocales, Oscillatoriales). The red mat was dominated by reads from heterotrophic orders, with Pseudomonadales, Burkholderiales, and Bacillales comprising 50% of the total reads. Photosynthetic orders represented 15% of total reads. The Río Mesquites stromatolite was dominated by reads from Cyanobacteria, with Nostocales, Chroococcales, and Oscillatoriales comprising 78% of the total reads.
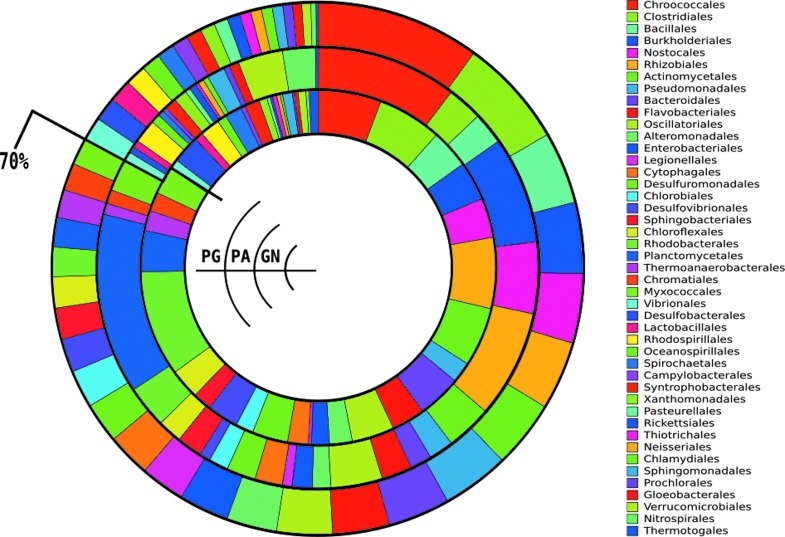
To include another nonthermophilic microbial mat in the study, we analyzed the pooled taxonomic sequence profile from the coastal hypersaline mat metagenome from Guerrero Negro (Kunin et al., 2008). It also displayed a high evenness, with 75% of the reads distributed among 27 orders. Abundant taxa were Rhodobacterales, Rhizobiales, Clostridiales, Chroococcales, Actinomycetales, and Bacteroidales. We found a remarkable conservation in the relative proportion of the analyzed taxa across the green mat, Pozas Azules, and Guerrero Negro (Fig. 6).
FIG. 6.
Relative abundance distributions of the most abundant orders by means of the all-reads metagenomic complement approach. Abundant taxa are considered as those comprising 75% of the total annotated reads in each metagenome. Green mat, outer circle; Pozas Azules, middle circle; Guerrero Negro, inner circle. The black line indicates where the cumulative proportion of the three samples reaches 70%, just after Myxococcales.
Fragment recruitment revealed that reference genomes from members of the Nostocales recruited most reads from the green mat (1140 recruited by Anabaena variabilis and 820 from Nostoc punctiforme), but they did so with low similarity values (mode>1e-10). The genomes of unicellular diazotrophic Cyanothece species (Chroococcales: Aphanotecoideae) recruited a large amount of reads from the green mat data set (1017 reads), while reference genomes from different strains of Pseudomonas fluorescens recruited most reads from the red mat (3996–5626 reads) with high similarity values (mode<1e-30). The cyanobacteria Microcoleus chthonoplastes was the next reference genome that recruited most reads from the red mat (4847 reads). Given the cosmopolitan distribution and high similarity between M. chthonoplastes populations (Garcia-Pichel et al., 1996) and the fact that the recruited fragments from the red mat had very high similarity values (mode<1e-30), the organisms present in the CCB red mat are very likely non-heterocystous filamentous cyanobacteria from M. chthonoplastes. Fragment recruitment plots can be found in Supplementary Material S3.
4. Discussion
4.1. On the dominant organisms in microbial mats
Cyanobacteria are important and often dominant components of microbial mats (Ward et al., 1998), and are responsible for the formation of the mat tissue with interlaced filaments. This study is no exception, since they were abundant in both mats, and order Chroococcales was the most abundant and diverse cyanobacterial order in both mats. Although no single OTU was dominant within Chroococcales, several species from genus Synechococcus were found. Organisms from this order would have had no role in the formation of the mat tissue, since it comprises only unicellular species. Order Oscillatoriales contains mat-forming species, and although it was among the most abundant groups in the green mat, no sequences from the well-known mat-forming cosmopolitan Microcoleus chthonoplastes (Garcia-Pichel et al., 1996) were found in GP. Order Nostocales was the most abundant group that contained known mat-forming Cyanobacteria. Abundant filamentous Nostocales from family Rivulariaceae (Calothrix and Tolypothrix) have been observed in other CCB microbial mats and stromatolites (Garcia-Pichel et al., 2002; Domínguez-Escobar et al., 2011), and this group has also been reported to grow better under low-phosphorus conditions (Berrendero et al., 2008). Unfortunately, family Rivulariaceae cannot be accurately determined from protein sequences, since no reference genomes are available yet, which makes cultured members of this group good targets for future sequencing projects. In consequence, the mat-forming filamentous cyanobacteria in the green mat could be uncharacterized species from order Oscillatoriales, Nostocales, or both.
In contrast, most sequences from Oscillatoriales in the red mat show a remarkable similarity to M. chthonoplastes (BLAST hits below 1e-20), despite the fact that they comprise only 4% of the total metagenome. Nevertheless, only one 16S rRNA sequence from the clone library and only two protein-coding markers from the red mat displayed affinity to this organism. This is what would be expected from organisms with large genomes (M. chthonoplastes has a genome of 7.36 Mb), and this discrepancy has also been noted in other microbial mats (Sørensen et al., 2005; Dillon et al., 2009). All approaches agreed in that the red mat was dominated by Pseudomonas. Although unexpected, this is not surprising in that the existence of mats dominated by noncyanobacterial organisms like Thioploca (Oschmann, 2000), Beggiatoa (Mussmann et al., 2007), and even fungi mycelia (Verrecchia, 2000) have also been reported. Moreover, Pseudomonas species are recognized for their metabolic versatility, and many are acknowledged to form biofilms (Silby et al., 2009). More remarkable is the finding of several closely related but distinct lineages with affinity to P. fluorescens. Since the metagenomes described here represent only a snapshot of the red mat community evolution in time, we can infer that the composition observed in the red mats is actually a radiative burst of Pseudomonas and other heterotrophs after a desiccation disturbance. Even though it has already been shown that environmental heterogeneity promotes diversification in P. fluorescens populations (Rainey and Travisano, 1998), the evolutionary consequences of these continuous disturbances are unknown. Our results suggest that this could lead to an adaptive radiation burst over the long term, but the observed pattern could also be caused simply by the metabolic plasticity of Pseudomonas. The similar composition of a second desiccation pond (red mat 2) suggests that the blooming of Pseudomonas could be an effect of the seasonal cycles common to all desiccation ponds in this area.
4.2. Taxonomic composition and functional trait analysis of the most abundant organisms
Although most organisms in these mats are uncharacterized, we can rely on the abundant literature on related species and genome projects to infer the general metabolic traits of the organisms that are abundant, according to the three methods, and have known and well-characterized relatives. In the green mat, despite the large amount of organisms represented, the main primary producers are likely to be Cyanobacteria, more specifically the abundant picooxygenic Synechococcus (Chroococcales). Moreover, the sequences found reveal high affinity to species OS-A and OS-B′ found in thermophilic mats in Yellowstone (Ward et al., 1998). Interestingly, these are the only Synechococcus genomes to carry nitrogen-fixing genes, as well as other genes that allow them to grow in oligotrophic environments such as genes for urea cycle, sulfolipid production, nitrate, phosphate and phosphonate utilization, and the biosynthesis and degradation of cyanophycin, a reserve polymer rich in nitrogen (Bhaya et al., 2007). This suggests that the most abundant phototrophs in the mat are not filamentous bacteria but efficient unicellular oligotrophic cyanobacteria and potentially important contributors to primary productivity and inorganic nutrient incorporation. Nevertheless, large filamentous cyanobacteria might still be the dominant organisms when considering biomass contribution (S.J. Green et al., 2008).
The organic compounds synthesized by these primary producers in the green mat are likely recycled by the abundant and efficient oxygenic heterotrophs like Legionella, which are common in oligotrophic water bodies, even though they are auxotrophic for seven amino acids, including L-cysteine, and require iron salts for growth (Declerck, 2010). They satisfy their nutrient requirements from living organisms (such as amoeboid hosts and microbial biofilms) or decaying organic matter (Declerck, 2010). Therefore, the green microbial mat appears to provide physicochemical protection from the environment, while it concentrates large amounts of nutrients. Algoriphagus species are aerobic heterotrophs characterized by the degradation of high-molecular-weight polysaccharides, with Algoriphagus sp. PR1 genome coding for 145 polysaccharide degradation enzymes (Alegado et al., 2011). In the anoxic part of microbial mats, Desulfococcus species are able to completely degrade short- and long-chain fatty acids and aromatic compounds coupled to the reduction of sulfate to hydrogen sulfide (Muyzer and Stams, 2008) and can fix CO2 via the Wood-Ljungdahl pathway (Platen et al., 1990). In contrast, the green-sulfur bacteria Chloroherpethon utilizes sulfide ions as electron donors for carbon assimilation (Bryant and Frigaard, 2006), while it contributes with carbon and nitrogen fixation to primary productivity and the production of phosphate granules via polyphosphate kinases. Even though Chlorobii are not properly classified as oligotrophs, they specialize in low light use and have been shown to dominate in environments with low organic matter input (Gonzalez et al., 2011).
The red mat, in contrast, is dominated by Pseudomonas, a metabolically versatile, ubiquitous heterotroph with broad catabolic and transport capabilities (Moore et al., 2006). We corroborated the Pseudomonas dominance with another clone library built from a second red mat, located in a similar desiccation pond that was adjacent to the first red mat pond (Fig. 3). Pseudomonas fluorescens species are aerobes; but some strains can use nitrate as electron acceptor instead of oxygen, and their genomes code for a large number of high-specificity nutrient transporters, as well as a large array of efflux systems for metal, organic solvent, and antibiotic detoxification (Silby et al., 2009). Their large genomes, high-affinity transporters, and broad sensing capabilities position Pseudomonas as copiotrophs (organisms adapted to grow in nutrient-rich environments) (Lauro et al., 2009). Nevertheless, P. fluorescens can use several carbon sources at very low concentrations, with a marked preference for amino acids (van der Kooij et al., 1982), and can form biofilms under starvation conditions to maximize exposure to diluted nutrients (Kroukamp et al., 2010), which suggests that they are at least tolerant to nutrient depletion. More relevant to the red mat environment, several P. fluorescens strains can efficiently solubilize mineralized inorganic phosphates (Fankem et al., 2008; Woo et al., 2010). Acidification increases solubilization by the production of carboxylic acids that have a high affinity for phosphate-bound ions (Khan et al., 2009) and reaches an optimum between a pH of 4.5 and 5.5 (Fankem et al., 2008). Phosphate solubilization is maximized with citrate and malate (Fankem et al., 2008), which are two of the most abundant carboxylic acids produced by P. fluorescens (Vyas and Gulati, 2009). None of the OTUs from the mats belonged to previously reported abundant species in the CCB (P. mendocina, P. otitidis, or P. cuatrocienegasensis, Escalante et al., 2009).
The aerobic heterotroph Chitinophaga is also abundant in the red mat. It is also a copiotroph, with a large genome of 9.1 Mb that contains a large and diverse collection of enzymes for the degradation of sugars (169 glycosyl hydrolases; Del Rio et al., 2010), most notably the degradation of chitin and casein (Sangkhobol and Skerman, 1981). In contrast, other abundant members in the red mat include the oligotrophic ultramicrobia (cell volume<0.1 μm3) Sphingopixis alaskensis (Lauro et al., 2009) and Janthinobacterium sp. Marseille (Minibacterium massiliensis, Audic et al., 2007), whose common traits include very small cells and genomes, preference for amino acids over sugars, active iron scavenging, and a broad array of high-affinity but low-specificity transport systems. In addition, S. alaskensis exhibits degradation of high-energy yielding fatty acids and has a large number of genes for secondary metabolite catabolism and detoxification (Lauro et al., 2009). Surprisingly, the phosphorus metabolism in S. alaskensis is reduced to the nonspecific alkaline phosphatases and a single ATP-dependent transporter, which is the only ABC transporter in the genome (Williams et al., 2009). In contrast, the genome of Janthinobacterium sp. contains ABC transporters for sulfate, sulfonate, thiosulfate, nitrate, most amino acids, phosphate, and phosphonates (Audic et al., 2007).
While Microcoleus chthonoplastes, which is also represented in the red mat, has a large genome (7.36 Mb) and is capable of performing both oxygenic and anoxygenic photosynthesis in the presence of sulfide (Stal, 1991), it cannot utilize nitrate or fix nitrogen (Zimmermann, 1989). Mats dominated by M. chthonoplastes show very low nitrogen fixation (Camacho and de Wit, 2003), and most strains analyzed to date lack the nitrogenase gene (Bolhuis et al., 2009). Moreover, anoxygenic photosynthesis and nitrogen fixation in M. chthonoplastes mats are strongly dependent on phosphate abundance (Zimmermann, 1989; Camacho and de Wit, 2003), so it is not surprising that its genome contains genes for low- and high-affinity transporters (pitA and pstSBCA) and for the synthesis of the reservoir compound polyphosphate (ppk).
4.3. Food or stability? Oligotrophy and disturbance as limiting factors for the development of complex microbial mat communities
The functional trait analysis showed that, in the green mat, oligotrophic organisms are abundant primary producers that can fix nitrogen and optimize phosphorus utilization. Moreover, at least three of the five most abundant organisms are capable of CO2 fixation. The fixed organic matter would then be recycled by non-oligotrophic heterotrophs. In contrast, oligotrophic organisms in the red mat are heterotrophs, while the dominant organisms are versatile copiotrophs. This reveals two very different strategies to cope with an oligotrophic environment, one mainly based on autotrophic primary production and the other on very efficient heterotrophic recycling. The results suggest that oligotrophy is not a limiting factor for the development of complex and functionally diverse microbial communities.
The analysis of diversity revealed that the green mat was by far the most diverse community and that communities in the more stable environments (the green mat and Pozas Azules) harbor higher richness and evenness than those in the more variable environments (the red mat and Río Mesquites). This suggests that disturbance frequency is a determinant factor of community structure in microbial mats, which is consistent with the exclusion of microbial mats by disturbing eukaryotes (Cohen, 1989; Bebout et al., 2002). In the red mat, the disturbance regime is apparently too frequent and may drive the reduction in abundance of several species, most notably autotrophs. These systems, as those growing in hyperthermophilic environments, demonstrate that microbial mats can exist with a simplified diversity, as long as a chemical gradient exists where primary producers are present and their nutrients are recycled by heterotrophs.
4.4. A common high-rank taxonomic composition as a result of trait conservation in microbial mats
Experimental studies on Guerrero Negro microbial mats have reported that, although salinity seems to affect community structure, cyanobacterial communities are only modestly affected by changes in sulfate and salinity concentrations (S.J. Green et al., 2008). Mathematical models of nutrient and population dynamics in microbial mats where complexity is reduced to a few functional groups show a large resemblance to natural mats (Decker et al., 2005). Moreover, functionally similar mats develop under varying oxygen concentrations because oxygenic phototrophs create a similar oxygen gradient in the upper layers (Herman and Kump, 2005). The role of oxygenic phototrophs is stressed not only as a source of nutrient incorporation but also as the generators of this gradient themselves. The potential metabolic differences between the two mats are mainly a result of differences between the proportion of photoautotrophs in the mats, which suggests that primary productivity is determinant to the community structure complexity. Since an unexpected diversity has been found in systems that produce a similar layered pattern with steep biogeochemical gradient (Jorgensen et al., 1986; Stolz, 2000; Kunin et al., 2008), it would appear that microbial mats are completely independent of the taxonomic composition of its conforming species and that similar systems may arise in any place where these gradient conditions are met. Hence, we compared our microbial mat communities with similar structures developed under different environmental conditions, the stromatolites from the CCB, and the hypersaline mat from Guerrero Negro. The recently published literature on microbialite community diversity comprising microbial mats (Ley et al., 2006; Abed et al., 2007; Allen et al., 2009), stromatolites (Burns et al., 2004; Papineau et al., 2005; Baumgartner et al., 2009), and endoevaporites (Sahl et al., 2008) reveals that microbial mat communities have a far larger expected richness than stromatolites and endoevaporites (Table S2b in Supplementary Material). As expected, we found very few shared organisms between the two CCB mats, and no species were shared between the four systems from the CCB. Moreover, only two genomes (M. chthonoplastes and Desulfococcus oleovorans) showed high recruitment in both Guerrero Negro and CCB metagenomes. However, a larger similarity at higher taxonomic levels was observed between the Guerrero Negro microbial mat, the green mat, and the Pozas Azules stromatolite even though these two mats were geographically separated by a linear distance of ca. 1200 km. The pattern appears unexpected, since other clone library–based investigations in which both stromatolites and microbial mats from the same sampling site were analyzed produced very different community compositions (for example Burns et al., 2004; Allen et al., 2009).
The finding of a common phylogenetic pattern at high taxonomic ranks suggests that layered microbial communities at the water-sediment interface assemble in biogeochemical gradients that fill defined ecological niches (photosynthesis, sulfate reduction, heterotrophy) according to their functional traits and independently of their phylogeny. It also supports the theory of the assembly of bacterial communities by functional genes rather than species (Burke et al., 2011) and the existence of general functional traits shared by organisms at deep phylogenetic nodes (J.L. Green et al., 2008; Philippot et al., 2010). Under this model, phylogenetically unrelated species are able to colonize the same niche in an ecosystem as long as they are ecologically equivalent (same trophic level or metabolic function), just as similar ecosystems will have communities with common functional “guilds,” but species within these guilds will be selected at random (Burke et al., 2011). This explains the success of microbial matlike communities because they benefit from a simple set of environmental requirements, namely, a water-sediment interface and a steep physicochemical gradient. Hence, complex communities are likely to be found wherever these conditions are met, with metabolically diverse functional guilds benefiting from the generation of diverse microniches and from the environmental buffering the mat structure provides. This would suggest that the prevalence of remarkably similar, but taxonomically distinct, mat structures in stable environments is a natural consequence of the undisturbed association of metabolically diverse organisms that exploit a locally microdiverse niche. The identification of a conserved taxonomic group core also provides a base system with which to test the effects of different kinds of disturbances on complex microbial ecosystems, while it also sets a start-up for the design of synthetic artificial model microbial communities in mesocosms.
Microbial mats appear to be the most ancient and pervasive ecosystems because they conform biogeochemical structures that are likely to be found wherever a gradient occurs across a sediment-water interface, and they comprise physicochemical structures that protect organisms from environmental extremes (S.J. Green et al., 2008) while concentrating nutrients. This could explain their success even under the low-phosphorus environments of the Precambrian oceans (Papineau, 2010). The fact that their structure depends more on the ecological traits than the taxonomic component of their members suggests that analogues of these kinds of ecosystems are also the most likely structures to be found beyond the confines of Earth. Their organosedimentary nature makes them ideal targets for the detection of evidence of former life in the geological record by means of the identification of stable isotopic signatures and molecules of biological origin.
Currently, a wide array of biosignatures has been proposed with which to narrow the number of astrological bodies that could host life, such as photosynthetic pigments (Seager et al., 2005), sulfur gases (Domagal-Goldman et al., 2011), methane (Hoehler et al., 2001), and ammonia (Des Marais et al., 2002). Theoretically, it would seem more likely to detect a complex community of unknown organisms than a simple one, since a complex community would contain and exude a larger variety of metabolic products, which would increase the probability of detecting one of these as biosignatures. Our results suggest that complex communities can be found in environments where nutrient concentrations are very low, which underscores the importance of considering microhabitats in otherwise nonviable environments. However, an environment with a frequent disturbance regime might significantly reduce diversity, while environments with intense disturbances might not host life at all. This suggests that more complex and, hence, more detectable communities are likely to be found in more stable environments, so that efforts in the search for life should incorporate disturbance intensity and frequency when narrowing the list of planets where search efforts are to be directed.
5. Conclusion
The CCB displays astounding microbial diversity despite being a mixed oligotrophic environment, which makes its ponds desirable astrobiological experimental models. The microbial mats from this study revealed two extremes of diversity, with the green mat showing a large diversity above phylum level and the red mat exhibiting diversity below genus level. Our results widen the ranges within which life may be found, since they show that complex communities can develop in environments with nutrient concentrations below detection levels, even though phosphate is readily mineralized and made biologically unavailable. However, our results also stress the need to incorporate disturbance intensity and frequency models into a more precise definition of astrobiological targets, since high disturbance regimes can lower diversity (with a concomitant lowering of measurable biosignatures) and even prevent the development of life.
Supplementary Material
Acknowledgments
We thank Rodrigo Gonzalez Chauvet for technical logistics and field assistance; E. Lopez, A. Islas, V. Lopez, F. Reverchon, E. Rebollar, M. Avitia, and A. Gutierrez for assistance in sample collection and DNA isolation; and Laura Espinosa from IE/UNAM for laboratory and technical assistance. We thank three anonymous reviewers, whose insightful comments greatly improved this manuscript. We also thank O. Rodriguez and C. Bixler for their inertiatic propulsion. This work was supported by grants CONACyT 057507, SEMARNAT 2006-C01-23459, and WWF-Alianza Carlos Slim L039 to V.S., and CINVESTAV-multidisciplinario to G.O.A. The manuscript was written while V.S. and L.E.E. were on sabbatical at UCI with support from DGAPA to V.S. and UC-Mexus to L.E.E. G.B.R. was supported with Ph.D. scholarship CONACYT 196814 and Programa de Posgrado en Ciencias Biomédicas UNAM.
Abbreviations
CCB, Cuatro Ciénegas Basin; OTUs, operational taxonomic units.
References
- Abed R.M. Kohls K. de Beer D. Effect of salinity changes on the bacterial diversity, photosynthesis and oxygen consumption of cyanobacterial mats from an intertidal flat of the Arabian Gulf. Environ Microbiol. 2007;9:1384–1392. doi: 10.1111/j.1462-2920.2007.01254.x. [DOI] [PubMed] [Google Scholar]
- Alegado R.A. Ferriera S. Nusbaum C. Young S.K. Zeng Q. Imamovic A. Fairclough S.R. King N. Complete genome sequence of Algoriphagus sp. PR1, bacterial prey of a colony-forming choanoflagellate. J Bacteriol. 2011;193:1485–1486. doi: 10.1128/JB.01421-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M.A. Goh F. Burns B.P. Neilan B.A. Bacterial, archaeal and eukaryotic diversity of smooth and pustular microbial mat communities in the hypersaline lagoon of Shark Bay. Geobiology. 2009;7:82–96. doi: 10.1111/j.1472-4669.2008.00187.x. [DOI] [PubMed] [Google Scholar]
- Allwood A.C. Walter M.R. Kamber B.S. Marshall C.P. Burch I.W. Stromatolite reef from the Early Archaean era of Australia. Nature. 2006;441:714–718. doi: 10.1038/nature04764. [DOI] [PubMed] [Google Scholar]
- Ashelford K.E. Chuzhanova N.A. Fry J.C. Jones A.J. Weightman A.J. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol. 2006;72:5734–5741. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic S. Robert C. Campagna B. Parinello H. Claverie J.M. Raoult D. Drancourt M. Genome analysis of Minibacterium massiliensis highlights the convergent evolution of water-living bacteria. PLoS Genet. 2007;3:e138. doi: 10.1371/journal.pgen.0030138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awramik S.A. Ancient stromatolites and microbial mats. In: Cohen Y., editor; Castenholz R.W., editor; Halvorson H.O., editor; Liss Alan R., editor. Microbial Mats: Stromatolites. New York: 1984. pp. 1–22. [Google Scholar]
- Awramik W.S. Gunflint stromatolites: microfossil distribution in relation to stromatolite morphology. In: Walter M.R., editor. Developments in Sedimentology. Vol. 20. Elsevier; Amsterdam: 1976. pp. 311–320. [Google Scholar]
- Baumgartner L.K. Spear J.R. Buckley D.H. Pace N.R. Reid R.P. Dupraz C. Visscher P.T. Microbial diversity in modern marine stromatolites, Highborne Cay, Bahamas. Environ Microbiol. 2009;11:2710–2719. doi: 10.1111/j.1462-2920.2009.01998.x. [DOI] [PubMed] [Google Scholar]
- Bebout B.M. Carpenter S.P. Des Marais D.J. Discipulo M. Embaye T. Garcia-Pichel F. Hoehler T.M. Hogan M. Jahnke L.L. Keller R.M. Miller S.R. Prufert-Bebout L.E. Raleigh C. Rothrock M. Turk K. Long-term manipulations of intact microbial mat communities in a greenhouse collaboratory: simulating Earth's present and past field environments. Astrobiology. 2002;2:383–402. doi: 10.1089/153110702762470491. [DOI] [PubMed] [Google Scholar]
- Bebout B.M. Hoehler T.M. Thamdrup B. Albert D. Carpenter S.P. Hogan M. Turk K. Des Marais D.J. Methane production by microbial mats under low sulphate concentrations. Geobiology. 2004;2:87–96. [Google Scholar]
- Bent S.J. Forney L.J. The tragedy of the uncommon: understanding limitations in the analysis of microbial diversity. ISME J. 2008;2:689–695. doi: 10.1038/ismej.2008.44. [DOI] [PubMed] [Google Scholar]
- Berrendero E. Perona E. Mateo P. Genetic and morphological characterization of Rivularia and Calothrix (Nostocales, Cyanobacteria) from running water. Int J Syst Evol Microbiol. 2008;58:447–460. doi: 10.1099/ijs.0.65273-0. [DOI] [PubMed] [Google Scholar]
- Besemer J. Borodovsky M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005;33:W451–W454. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya D. Grossman A.R. Steunou A.S. Khuri N. Cohan F.M. Hamamura N. Melendrez M.C. Bateson M.M. Ward D.M. Heidelberg J.F. Population level functional diversity in a microbial community revealed by comparative genomic and metagenomic analyses. ISME J. 2007;1:703–713. doi: 10.1038/ismej.2007.46. [DOI] [PubMed] [Google Scholar]
- Bjerrum C.J. Canfield D.E. Ocean productivity before about 1.9 Gyr ago limited by phosphorus adsorption onto iron oxides. Nature. 2002;417:159–162. doi: 10.1038/417159a. [DOI] [PubMed] [Google Scholar]
- Bolhuis H. Severin I. Confurius-Guns V. Wollenzien U.I.A. Stal L.J. Horizontal transfer of the nitrogen fixation gene cluster in the cyanobacterium Microcoleus chthonoplastes. ISME J. 2009;4:121–130. doi: 10.1038/ismej.2009.99. [DOI] [PubMed] [Google Scholar]
- Bottos E.M. Vincent W.F. Greer C.W. Whyte L.G. Prokaryotic diversity of arctic ice shelf microbial mats. Environ Microbiol. 2008;10:950–966. doi: 10.1111/j.1462-2920.2007.01516.x. [DOI] [PubMed] [Google Scholar]
- Breitbart M. Hoare A. Nitti A. Siefert J. Haynes M. Dinsdale E. Edwards R. Souza V. Rohwer F. Hollander D. Metagenomic and stable isotopic analyses of modern freshwater microbialites in Cuatro Ciénegas, Mexico. Environ Microbiol. 2009;11:16–34. doi: 10.1111/j.1462-2920.2008.01725.x. [DOI] [PubMed] [Google Scholar]
- Bryant D.A. Frigaard N.U. Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol. 2006;14:488–496. doi: 10.1016/j.tim.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Burke C. Steinberg P. Rusch D. Kjelleberg S. Thomas T. Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci USA. 2011;108:14288–14293. doi: 10.1073/pnas.1101591108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns B.P. Goh F. Allen M. Neilan B.A. Microbial diversity of extant stromatolites in the hypersaline marine environment of Shark Bay, Australia. Environ Microbiol. 2004;6:1096–1101. doi: 10.1111/j.1462-2920.2004.00651.x. [DOI] [PubMed] [Google Scholar]
- Camacho A. de Wit R. Effect of nitrogen and phosphorus additions on a benthic microbial mat from a hypersaline lake. Aquat Microb Ecol. 2003;32:261–273. [Google Scholar]
- Cohen Y. Photosynthesis in cyanobacterial mats and its relation to the sulfur cycle: a model for microbial sulfur interactions. In: Cohen Y., editor; Rosenberg E., editor. Microbial Mats: Physiological Ecology of Benthic Microbial Communities. American Society for Microbiology; Washington DC: 1989. pp. 22–36. [Google Scholar]
- Connell J.H. Diversity in tropical rain forests and coral reefs. Science. 1978;199:1302–1310. doi: 10.1126/science.199.4335.1302. [DOI] [PubMed] [Google Scholar]
- Decker K. Potter C. Bebout B.M. Des Marais D.J. Carpenter S. Discipulo M. Hoehler T.M. Miller S.R. Thamdrup B. Turk K.A. Visscher P.T. Mathematical simulation of the diel O, S, and C biogeochemistry of a hypersaline microbial mat. FEMS Microbiol Ecol. 2005;52:377–395. doi: 10.1016/j.femsec.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Declerck P. Biofilms: the environmental playground of Legionella pneumophila. Environ Microbiol. 2010;12:557–566. doi: 10.1111/j.1462-2920.2009.02025.x. [DOI] [PubMed] [Google Scholar]
- Del Rio T.G. Abt B. Spring S. Lapidus A. Nolan M. Tice H. Copeland A. Cheng J. Chen F. Bruce D. Goodwin L. Pitluck S. Ivanova N. Mavromatis K. Mikhailova N. Pati A. Chen A. Palaniappan K. Land M. Hauser L. Chang Y. Jeffries C. Chain P. Saunders E. Detter J. Brettin T. Rohde M. Göker M. Bristow J. Eisen J. Markowitz V. Hugenholtz P. Kyrpides N. Klenk H. Lucas S. Complete genome sequence of Chitinophaga pinensis type strain (UQM 2034T) Stand Genomic Sci. 2010;2:87. doi: 10.4056/sigs.661199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais D.J. Harwit M.O. Jucks K.W. Kasting J.F. Lin D.N.C. Lunine J.I. Schneider J. Seager S. Traub W.A. Woolf N.J. Remote sensing of planetary properties and biosignatures on extrasolar terrestrial planets. Astrobiology. 2002;2:153–181. doi: 10.1089/15311070260192246. [DOI] [PubMed] [Google Scholar]
- DeSantis T.Z. Hugenholtz P. Larsen N. Rojas M. Brodie E.L. Keller K. Huber T. Dalevi D. Hu P. Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon J.G. Miller S. Bebout B. Hullar M. Pinel N. Stahl D.A. Spatial and temporal variability in a stratified hypersaline microbial mat community. FEMS Microbiol Ecol. 2009;68:46–58. doi: 10.1111/j.1574-6941.2009.00647.x. [DOI] [PubMed] [Google Scholar]
- Domagal-Goldman S.D. Meadows V.S. Claire M.W. Kasting J.F. Using biogenic sulfur gases as remotely detectable biosignatures on anoxic planets. Astrobiology. 2011;11:419–441. doi: 10.1089/ast.2010.0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Escobar J. Beltrán Y. Bergman B. Díez B. Ininbergs K. Souza V. Falcón L.I. Phylogenetic and molecular clock inferences of cyanobacterial strains within Rivulariaceae from distant environments. FEMS Microbiol Lett. 2011;316:90–99. doi: 10.1111/j.1574-6968.2010.02195.x. [DOI] [PubMed] [Google Scholar]
- Elser J.J. Schampel J.H. Garcia-Pichel F. Wade B.D. Souza V. Eguiarte L. Escalante A. Farmer J.D. Effects of phosphorus enrichment and grazing snails on modern stromatolitic microbial communities. Freshw Biol. 2005;50:1808–1825. [Google Scholar]
- Elser J.J. Watts J. Schampel J.H. Farmer J. Early Cambrian food webs on a trophic knife-edge? A hypothesis and preliminary data from a modern stromatolite-based ecosystem. Ecological Letters. 2006;9:295–303. doi: 10.1111/j.1461-0248.2005.00873.x. [DOI] [PubMed] [Google Scholar]
- Elshahed M.S. Senko J.M. Najar F.Z. Kenton S.M. Roe B.A. Dewers T.A. Spear J.R. Krumholz L.R. Bacterial diversity and sulfur cycling in a mesophilic sulfide-rich spring. Appl Environ Microbiol. 2003;69:5609–5621. doi: 10.1128/AEM.69.9.5609-5621.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson D. Revsbech N.P. Investigation of an iron-oxidizing microbial mat community located near Aarhus, Denmark: field studies. Appl Environ Microbiol. 1994;60:4022–4031. doi: 10.1128/aem.60.11.4022-4031.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante A.E. Eguiarte L.E. Espinosa-Asuar L. Forney L.J. Noguez A.M. Souza-Saldivar V. Diversity of aquatic prokaryotic communities in the Cuatro Cienegas basin. FEMS Microbiol Ecol. 2008;65:50–60. doi: 10.1111/j.1574-6941.2008.00496.x. [DOI] [PubMed] [Google Scholar]
- Escalante A.E. Caballero-Mellado J. Martínez-Aguilar L. Rodríguez-Verdugo A. González-González A. Toribio-Jiménez J. Souza V. Pseudomonas cuatrocienegasensis sp. nov., isolated from an evaporating lagoon in the Cuatro Ciénegas valley in Coahuila, Mexico. Int J Syst Evol Microbiol. 2009;59:1416–1420. doi: 10.1099/ijs.0.006189-0. [DOI] [PubMed] [Google Scholar]
- Fankem H. Ngo Nkot L. Deubel A. Quinn J. Merbach W. Etoa F.-X. Nwaga D. Solubilization of inorganic phosphates and plant growth promotion by strains of Pseudomonas fluorescens isolated from acidic soils of Cameroon. Afr J Microbiol Res. 2008;2:171–178. [Google Scholar]
- Foster J.S. Mobberley J.M. Microbial Mats: Modern and Ancient Microorganisms in Stratified Systems. Springer; Dordrecht: 2010. Past, present, and future: microbial mats as models for astrobiological research; pp. 563–582. [Google Scholar]
- Garcia-Pichel F. Prufert-Bebout L. Muyzer G. Phenotypic and phylogenetic analyses show Microcoleus chthonoplastes to be a cosmopolitan cyanobacterium. Appl Environ Microbiol. 1996;62:3284–3291. doi: 10.1128/aem.62.9.3284-3291.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Pichel F. Wade B.D. Farmer J.D. Jet-suspended, calcite-ballasted cyanobacterial waterwarts in a desert spring. J Phycol. 2002;38:420–428. [Google Scholar]
- Gerdes G. What are microbial mats? In: Seckbach J., editor; Oren A., editor. Microbial Mats: Modern and Ancient Microorganisms in Stratified Systems. Springer; Dordrecht: 2010. pp. 3–25. [Google Scholar]
- Giovannoni S.J. Tripp H.J. Givan S. Podar M. Vergin K.L. Baptista D. Bibbs L. Eads J. Richardson T.H. Noordewier M. Rappé M.S. Short J.M. Carrington J.C. Mathur E.J. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- Golombek M. Grant J. Vasavada A.R. Grotzinger J. Watkins M. Kipp D. Noe Dobrea E. Griffes J. Parker T. Final four landing sites for the Mars Science Laboratory [abstract 1520]. 42nd Lunar and Planetary Science Conference, Lunar and Planetary Institute; Houston. 2011. [Google Scholar]
- Gonzalez B.C. Iliffe T.M. Macalady J.L. Schaperdoth I. Kakuk B. Microbial hotspots in anchialine blue holes: initial discoveries from the Bahamas. Hydrobiologia. 2011;677:149–156. [Google Scholar]
- Green J.L. Bohannan B.J.M. Whitaker R.J. Microbial biogeography: from taxonomy to traits. Science. 2008;320:1039–1043. doi: 10.1126/science.1153475. [DOI] [PubMed] [Google Scholar]
- Green S.J. Jahnke L.L. Molecular investigations and experimental manipulations of microbial mats: a view to paleomicrobial ecosystems. In: Seckbach J., editor; Oren A., editor. Microbial Mats: Modern and Ancient Microorganisms in Stratified Systems. Springer; Dordrecht: 2010. pp. 185–208. [Google Scholar]
- Green S.J. Blackford C. Bucki P. Jahnke L.L. Prufert-Bebout L. A salinity and sulfate manipulation of hypersaline microbial mats reveals stasis in the cyanobacterial community structure. ISME J. 2008;2:457–470. doi: 10.1038/ismej.2008.6. [DOI] [PubMed] [Google Scholar]
- Grime J.P. Competitive exclusion in herbaceous vegetation. Nature. 1973;242:344–347. [Google Scholar]
- Grotzinger J.P. Knoll A.H. Stromatolites in Precambrian carbonates: evolutionary mileposts or environmental dipsticks? Annu Rev Earth Planet Sci. 1999;27:313–358. doi: 10.1146/annurev.earth.27.1.313. [DOI] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Herman E. Kump L. Biogeochemistry of microbial mats under Precambrian environmental conditions: a modelling study. Geobiology. 2005;3:77–92. [Google Scholar]
- Hoehler T.M. Bebout B.M. Des Marais D.J. The role of microbial mats in the production of reduced gases on the early Earth. Nature. 2001;412:324–327. doi: 10.1038/35085554. [DOI] [PubMed] [Google Scholar]
- Huang Y. Gilna P. Li W. Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics. 2009;25:1338–1340. doi: 10.1093/bioinformatics/btp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H. Auch A.F. Qi J. Schuster S.C. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inskeep W.P. Rusch D.B. Jay Z.J. Herrgard M.J. Kozubal M.A. Richardson T.H. Macur R.E. Hamamura N. Jennings R.D. Fouke B.W. Reysenbach A.-L. Roberto F. Young M. Schwartz A. Boyd E.S. Badger J.H. Mathur E.J. Ortmann A.C. Bateson M. Geesey G. Frazier M. Rodriguez-Valera F. Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS One. 2010;5:e9773. doi: 10.1371/journal.pone.0009773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen B.B. Cohen Y. Revsbech N.P. Transition from anoxygenic to oxygenic photosynthesis in a Microcoleus chthonoplastes cyanobacterial mat. Appl Environ Microbiol. 1986;51:408–417. doi: 10.1128/aem.51.2.408-417.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasting J.F. Earth history. The rise of atmospheric oxygen. Science. 2001;293:819–820. doi: 10.1126/science.1063811. [DOI] [PubMed] [Google Scholar]
- Khan A. Jilani G. Akhtar M.S. Naqvi S.M.S. Rasheed M. Phosphorus solubilizing bacteria: occurrence, mechanisms and their role in crop production. J Agric Biol Sci. 2009;1:48–58. [Google Scholar]
- Kroukamp O. Dumitrache R.G. Wolfaardt G.M. Pronounced effect of the nature of the inoculum on biofilm development in flow systems. Appl Environ Microbiol. 2010;76:6025–6031. doi: 10.1128/AEM.00070-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumbein W.E. Cohen Y. Shilo M. Solar Lake (Sinai). 4. Stromatolitic cyanobacterial mats. Limnol Oceanogr. 1977;22:635–656. [Google Scholar]
- Kunin V. Raes J. Harris J.K. Spear J.R. Walker J.J. Ivanova N. von Mering C. Bebout B.M. Pace N.R. Bork P. Hugenholtz P. Millimeter-scale genetic gradients and community-level molecular convergence in a hypersaline microbial mat. Mol Syst Biol. 2008;4:198. doi: 10.1038/msb.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauro F.M. McDougald D. Thomas T. Williams T.J. Egan S. Rice S. DeMaere M.Z. Ting L. Ertan H. Johnson J. Ferriera S. Lapidus A. Anderson I. Kyrpides N. Munk A.C. Detter C. Han C.S. Brown M.V. Robb F.T. Kjelleberg S. Cavicchioli R. The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci USA. 2009;106:15527–15533. doi: 10.1073/pnas.0903507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I. Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R.E. Harris J.K. Wilcox J. Spear J.R. Miller S.R. Bebout B.M. Maresca J.A. Bryant D.A. Sogin M.L. Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl Environ Microbiol. 2006;72:3685–3695. doi: 10.1128/AEM.72.5.3685-3695.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. Analysis and comparison of very large metagenomes with fast clustering and functional annotation. BMC Bioinformatics. 2009;10:359. doi: 10.1186/1471-2105-10-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Ludwig W. Strunk O. Westram R. Richter L. Meier H. Yadhukumar Buchner A. Lai T. Steppi S. Jobb G. Förster W. Brettske I. Gerber S. Ginhart A.W. Gross O. Grumann S. Hermann S. Jost R. König A. Liss T. Lüssmann R. May M. Nonhoff B. Reichel B. Strehlow R. Stamatakis A. Stuckmann N. Vilbig A. Lenke M. Ludwig T. Bode A. Schleifer K.H. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz V.M. Chen I.-M. Palaniappan K. Chu K. Szeto E. Grechkin Y. Ratner A. Anderson I. Lykidis A. Mavromatis K. Ivanova N.N. Kyrpides N.C. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 2009;38:D382–D390. doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F. Paarmann D. D'Souza M. Olson R. Glass E.M. Kubal M. Paczian T. Rodriguez A. Stevens R. Wilke A. Wilkening J. Edwards R.A. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Dombard D.R. Swingley W. Raymond J. Havig J. Shock E.L. Summons R.E. Hydrothermal ecotones and streamer biofilm communities in the Lower Geyser Basin, Yellowstone National Park. Environ Microbiol. 2011;13:2216–2231. doi: 10.1111/j.1462-2920.2011.02476.x. [DOI] [PubMed] [Google Scholar]
- Mills H.J. Martinez R.J. Story S. Sobecky P.A. Characterization of microbial community structure in Gulf of Mexico gas hydrates: comparative analysis of DNA- and RNA-derived clone libraries. Appl Environ Microbiol. 2005;71:3235–3247. doi: 10.1128/AEM.71.6.3235-3247.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minckley W.L. Cole G.A. Preliminary limnologic information on waters of the Cuatro Cienegas basin, Coahuila, Mexico. Southwest Nat. 1968;13:421–431. [Google Scholar]
- Moore E.R.B. Tindall B.J. Martins Dos Santos V. Pieper D.H. Ramos J.L. Palleroni N.J. Nonmedical pseudomonas. In: Dworkin M., editor; Falkow S., editor; Rosenberg E., editor; Schleifer K.-H., editor; Stackebrandt E., editor. The Prokaryotes. Vol. 6. Springer; New York: 2006. pp. 646–703. [Google Scholar]
- Mulet M. Lalucat J. García-Valdés E. DNA sequence-based analysis of the Pseudomonas species. Environ Microbiol. 2010;12:1513–1530. doi: 10.1111/j.1462-2920.2010.02181.x. [DOI] [PubMed] [Google Scholar]
- Mussmann M. Hu F.Z. Richter M. de Beer D. Preisler A. Jørgensen B.B. Huntemann M. Glöckner F.O. Amann R. Koopman W.J. Lasken R.S. Janto B. Hogg J. Stoodley P. Boissy R. Ehrlich G.D. Insights into the genome of large sulfur bacteria revealed by analysis of single filaments. PLoS Biol. 2007;5:e230. doi: 10.1371/journal.pbio.0050230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G. Stams A.J.M. The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol. 2008;6:441–454. doi: 10.1038/nrmicro1892. [DOI] [PubMed] [Google Scholar]
- Nitti A. Daniels C.A. Siefert J. Souza V. Hollander D. Breitbart M. Spatially resolved genomic, stable isotopic, and lipid analyses of a modern freshwater microbialite from Cuatro Ciénegas, Mexico. Astrobiology. 2012;12:685–698. doi: 10.1089/ast.2011.0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oschmann W. Microbes and black shales. In: Riding R.E., editor; Awramik S.M., editor. Microbial Sediments. Springer Verlag; Berlin: 2000. pp. 137–148. [Google Scholar]
- Paerl H.W. Yannarell A.C. Environmental dynamics, community structure and function in a hypersaline microbial mat. In: Seckbach J., editor; Oren A., editor. Microbial Mats: Modern and Ancient Microorganisms in Stratified Systems. Springer; Dordrecht: 2010. pp. 423–444. [Google Scholar]
- Paerl H.W. Steppe T.F. Buchan K.C. Potts M. Hypersaline cyanobacterial mats as indicators of elevated tropical hurricane activity and associated climate change. AMBIO: A Journal of the Human Environment. 2003;32:87–90. doi: 10.1579/0044-7447-32.2.87. [DOI] [PubMed] [Google Scholar]
- Papineau D. Global biogeochemical changes at both ends of the proterozoic: insights from phosphorites. Astrobiology. 2010;10:165–181. doi: 10.1089/ast.2009.0360. [DOI] [PubMed] [Google Scholar]
- Papineau D. Walker J.J. Mojzsis S.J. Pace N.R. Composition and structure of microbial communities from stromatolites of Hamelin Pool in Shark Bay, Western Australia. Appl Environ Microbiol. 2005;71:4822–4832. doi: 10.1128/AEM.71.8.4822-4832.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peimbert M. Alcaraz L.D. Bonilla-Rosso G. Olmedo-Alvarez G. García-Oliva F. Segovia L. Eguiarte L.E. Souza V. Comparative metagenomics of two microbial mats at Cuatro Ciénegas Basin I: ancient lessons on how to cope with an environment under severe nutrient stress. Astrobiology. 2012;12:648–658. doi: 10.1089/ast.2011.0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot L. Andersson S.G.E. Battin T.J. Prosser J.I. Schimel J.P. Whitman W.B. Hallin S. The ecological coherence of high bacterial taxonomic ranks. Nat Rev Microbiol. 2010;8:523–529. doi: 10.1038/nrmicro2367. [DOI] [PubMed] [Google Scholar]
- Planavsky N.J. Rouxel O.J. Bekker A. Lalonde S.V. Konhauser K.O. Reinhard C.T. Lyons T.W. The evolution of the marine phosphate reservoir. Nature. 2010;467:1088–1090. doi: 10.1038/nature09485. [DOI] [PubMed] [Google Scholar]
- Platen H. Temmes A. Schink B. Anaerobic degradation of acetone by Desulfococcus biacutus spec. nov. Arch Microbiol. 1990;154:355–361. doi: 10.1007/BF00276531. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R Foundation for Statistical Computing. Vienna, Austria; 2006. R: a language and environment for statistical computing. [Google Scholar]
- Rainey P.B. Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Mejía J.L. Martinez-Anaya C. Folch-Mallol J.L. Dantán-González E. A two-step electrodialysis method for DNA purification from polluted metallic environmental samples. Electrophoresis. 2008;29:3239–3244. doi: 10.1002/elps.200700829. [DOI] [PubMed] [Google Scholar]
- Sahl J.W. Pace N.R. Spear J.R. Comparative molecular analysis of endoevaporitic microbial communities. Appl Environ Microbiol. 2008;74:6444–6446. doi: 10.1128/AEM.00879-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangkhobol V. Skerman V.B.D. Chitinophaga, a new genus of chitinolytic myxobacteria. Int J Syst Bacteriol. 1981;31:285–293. [Google Scholar]
- Schloss P.D. Westcott S.L. Ryabin T. Hall J.R. Hartmann M. Hollister E.B. Lesniewski R.A. Oakley B.B. Parks D.H. Robinson C.J. Sahl J.W. Stres B. Thallinger G.G. Van Horn D.J. Weber C.F. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seager S. Turner E.L. Schafer J. Ford E.B. Vegetation's red edge: a possible spectroscopic biosignature of extraterrestrial plants. Astrobiology. 2005;5:372–390. doi: 10.1089/ast.2005.5.372. [DOI] [PubMed] [Google Scholar]
- Silby M.W. Cerdeño-Tárraga A.M. Vernikos G.S. Giddens S.R. Jackson R.W. Preston G.M. Zhang X.X. Moon C.D. Gehrig S.M. Godfrey S.A. Knight C.G. Malone J.G. Robinson Z. Spiers A.J. Harris S. Challis G.L. Yaxley A.M. Harris D. Seeger K. Murphy L. Rutter S. Squares R. Quail M.A. Saunders E. Mavromatis K. Brettin T.S. Bentley S.D. Hothersall J. Stephens E. Thomas C.M. Parkhill J. Levy S.B. Rainey P.B. Thomson N.R. Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol. 2009;10:R51. doi: 10.1186/gb-2009-10-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen K.B. Canfield D.E. Teske A.P. Oren A. Community composition of a hypersaline endoevaporitic microbial mat. Appl Environ Microbiol. 2005;71:7352–7365. doi: 10.1128/AEM.71.11.7352-7365.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza V. Espinosa-Asuar L. Escalante A.E. Eguiarte L.E. Farmer J. Forney L. Lloret L. Rodríguez-Martínez J.M. Soberón X. Dirzo R. Elser J.J. An endangered oasis of aquatic microbial biodiversity in the Chihuahuan desert. Proc Natl Acad Sci USA. 2006;103:6565–6570. doi: 10.1073/pnas.0601434103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear J.R. Ley R.E. Berger A.B. Pace N.R. Complexity in natural microbial ecosystems: the Guerrero Negro experience. Biol Bull. 2003;204:168–173. doi: 10.2307/1543553. [DOI] [PubMed] [Google Scholar]
- Stal L.J. The metabolic versatility of the mat-building cyanobacteria Microcoleus chthonoplastes and Oscillatoria limosa and its ecological significance. Algological Studies/Archiv für Hydrobiologie. 1991;64:453–467. [Google Scholar]
- Stolz J.F. Structure of microbial mats and biofilms. In: Riding R.E., editor; Awramik S.M., editor. Microbial Sediments. Springer-Verlag; Berlin: 2000. pp. 1–8. [Google Scholar]
- Szynkiewicz A. Ewing R.C. Moore C.H. Glamoclija M. Bustos D. Pratt L.M. Origin of terrestrial gypsum dunes—implications for martian gypsum-rich dunes of Olympia Undae. Geomorphology. 2010;121:69–83. [Google Scholar]
- Tilman D. Constraints and tradeoffs: toward a predictive theory of competition and succession. Oikos. 1990;58:3–15. [Google Scholar]
- van der Kooij D. Oranje J. Hijnen W. Growth of Pseudomonas aeruginosa in tap water in relation to utilization of substrates at concentrations of a few micrograms per liter. Appl Environ Microbiol. 1982;44:1086–1095. doi: 10.1128/aem.44.5.1086-1095.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gemerden H. Microbial mats: A joint venture. Mar Geol. 1993;113:3–25. [Google Scholar]
- Varin T. Lovejoy C. Jungblut A.D. Vincent W.F. Metagenomic profiling of Arctic microbial mat communities as nutrient scavenging and recycling systems. Limnol Oceanogr. 2010;55:1901–1911. [Google Scholar]
- Verrecchia E.P. Fungi and sediments. In: Riding R.E., editor; Awramik S.M., editor. Microbial Sediments. Springer-Verlag; Berlin: 2000. pp. 69–75. [Google Scholar]
- Vyas P. Gulati A. Organic acid production in vitro and plant growth promotion in maize under controlled environment by phosphate-solubilizing fluorescent Pseudomonas. BMC Microbiol. 2009;9:174. doi: 10.1186/1471-2180-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D.M. Ferris M.J. Nold S.C. Bateson M.M. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol Mol Biol Rev. 1998;62:1353–1370. doi: 10.1128/mmbr.62.4.1353-1370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T.J. Ertan H. Ting L. Cavicchioli R. Carbon and nitrogen substrate utilization in the marine bacterium Sphingopyxis alaskensis strain RB2256. ISME J. 2009;3:1036–1052. doi: 10.1038/ismej.2009.52. [DOI] [PubMed] [Google Scholar]
- Woo S.M. Lee M.K. Hong I.S. Poonguzhali S. Sa T.M. Soil Solutions for a Changing World. Brisbane; Australia: 2010. Isolation and characterization of phosphate solubilizing bacteria from Chinese cabbage; pp. 56–59. [Google Scholar]
- Wu M. Eisen J.A. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 2008;9:R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannarell A.C. Steppe T.F. Paerl H.W. Disturbance and recovery of microbial community structure and function following Hurricane Frances. Environ Microbiol. 2007;9:576–583. doi: 10.1111/j.1462-2920.2006.01173.x. [DOI] [PubMed] [Google Scholar]
- Zhou J. Bruns M.A. Tiedje J.M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann C.F. Nitrogen and phosphorus uptake and release by the blue-green alga Microcoleus lyngbyaceus. Journal of Aquatic Plant Management. 1989;27:49–51. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.