Abstract
Aim
This study compared the perfomance of the RNA-based EndoPredict multigene test on core biopsies and surgical breast cancer specimens and analysed the influence of biopsy-induced tissue injuries on the test result.
Methods
80 formalin-fixed paraffin-embedded samples comprising paired biopsies and surgical specimens from 40 ER-positive, HER2-negative patients were evaluated. Total RNA was extracted and the EndoPredict score was determined.
Results
RNA yield was considerably lower in core biopsies, but sufficient to measure the assay in all samples. The EndoPredict score was highly correlated between paired samples (Pearson r=0.92), with an excellent concordance of classification into a low or high risk of metastasis (overall agreement 95%).
Conclusions
The measurements are comparable between core biopsies and surgical sections, which suggest that the EndoPredict assay can be performed on core biopsy tissue. Inflammatory changes induced by presurgical biopsies had no significant effect on the RNA-based risk assessment in surgical specimens.
Keywords: Breast, breast cancer, breast pathology, cancer, cancer genetics, cancer research, EGFR, endocrine pathology, gynaecological pathology, molecular oncology, molecular pathology, oncology, ovary, statistics, tumour markers
Introduction
Breast cancer is the most frequently diagnosed cancer and the second leading cause of cancer-related death in women.1 The use of validated multigene assays might assist clinical treatment decisions. To this end, several prognostic and predictive tests have been developed to allow a more tailored treatment strategy in breast cancer.2–6 The EndoPredict test has recently been introduced as a novel multigene classifier to predict the likelihood of distant recurrence in ER-positive, HER2-negative breast cancer patients treated with adjuvant endocrine therapy.4 Two large randomised phase III trials (ABCSG6 and ABSCG8) involving endocrine therapy only (n=1702) demonstrated additional prognostic information of the EndoPredict test beyond all common clinicopathological parameters.4
Formalin-fixed, paraffin-embedded (FFPE) tumour tissue sections from surgical specimens are commonly used as the source material for the quantitative PCR-based expression analysis of multigene assays such as the EndoPredict test. However, the comparability of multigene assays between core biopsies and surgical specimens is largely unknown. In addition, the surgical specimens may contain inflammatory changes that are caused by preoperative core biopsy sampling. As a consequence, wound healing and inflammation processes could potentially lead to relevant changes for the established molecular tests. For instance, the protein levels of the prognostic breast cancer markers urokinase-type plasminogen activator (uPA) and the plasminogene activator inhibitor (PAI-1) are considerably altered in the tissue area surrounding the biopsy channels leading to unreliable results of the ELISA test.7 So far it is not clear whether RNA levels of prognostic multigene assays are also altered by the presence of tissue injuries in the tumour sections analysed.
In this study, we compared the EndoPredict score between core biopsies and corresponding surgical specimens of breast cancer patients. Second, we analysed the influence of tissue injuries caused by a preceding biopsy on the quantification of the EndoPredict score in the paired breast cancer samples.
Methods
Study population and histopathological examination
Eighty FFPE tumour blocks comprising 40 paired samples (core biopsies and corresponding surgery specimens) from patients with ER-positive, HER2-negative breast carcinomas were retrospectively evaluated. The cases were selected based on the presence of typical changes of biopsy-induced inflammation, such as inflammatory infiltrates, localised fibroblast proliferation or focal residual haemorrhage in the H&E sections from surgical specimens. There were four cores (median) per FFPE tissue block. The majority of the cores were taken under ultrasound control with 14G needles. The median time between biopsy and resection was 14 days.
Patient characteristics are shown in table 1. Two consecutive 5 μm FFPE tissue sections from each pretherapeutic core biopsy and the corresponding surgical specimen were used for this study, respectively. One section was routinely stained by H&E. The second slide was used for molecular analyses. The hormone receptor and the HER2 status as well as the proliferation activity were extracted from pathology reports. The receptor status was determined during routine diagnosis by immunohistochemistry, in the case of HER2 2+ (Dako score) followed by a silver enhanced in-situ hybridisation.8
Table 1.
Patient characteristics
| Characteristic | No of patients | % |
| All cases* | 40 | 100 |
| Histological type | ||
| Ductal carcinoma | 31 | 77.5 |
| Lobular carcinoma | 5 | 12.5 |
| Other carcinoma | 4 | 10 |
| Tumour size (mm) | ||
| ≤20 | 23 | 57.5 |
| >20 | 17 | 42.5 |
| Nodal status | ||
| pN0 | 27 | 67.5 |
| pN1a | 10 | 25 |
| pN2a | 3 | 7.5 |
| Histological grade | ||
| G1 | 8 | 20 |
| G2 | 26 | 65 |
| G3 | 6 | 15 |
| Tumour proliferation (surgery specimen)† | ||
| MIB-1 <14% | 21 | 52.5 |
| MIB-1≥14% | 16 | 37.5 |
All cases were hormone receptor positive (ER ≥60% positive tumour cells) and HER-2 negative (Dako score 0–2; in the case of Dako score 2+ without amplification using silver-enhanced in-situ hybridisation).
Data were not available for all samples.
RNA extraction and assessment of the EndoPredict score
Total RNA was extracted using a fully automated isolation method, as described previously.9–11 DNA-free total RNA from the FFPE sections was finally eluted with 100 μl elution buffer and stored at −80°C.
The EndoPredict assay was carried out to determine the expression levels of three reference genes and the relative expression levels of eight genes of interest. To obtain reliable results, all genes were assessed in triplicate by quantitative one-step reverse transcription PCR using the SuperScript III PLATINUM (Invitrogen, Karlsruhe, Germany) one-step quantitative reverse transcription PCR system with ROX (final concentration 50 nM). Measurements were carried out with the VERSANT kPCR system (Siemens Healthcare Diagnostics, Tarrytown, USA). Samples were classified as low or high risk of distant metastasis according to the predefined cut-off value of 5.4
Statistical analysis
Absolute expression levels (Ct values) were calculated using VERSANT software. Relative expression levels of the eight prognostic genes (AZGP1, BIRC5, DHCR7, IL6ST, MGP, RBBP8, STC2, UBE2C) were assessed as delta cycle threshold (ΔCt) values with respect to the three reference genes (CALM2, OAZ1 and RPL37A). The EndoPredict score was calculated for the biopsy and tumour sections, as described.4
Results
Forty FFPE tumour sections and 40 corresponding core biopsies were analysed and used for comparison. RNA yield was compared between biopsy and tissue sections by using the mean Ct value of the three reference genes as a surrogate marker. With a median difference of two Ct units, RNA yield was considerably lower in core biopsies. However, the extracted RNA amount was still sufficient to measure the EndoPredict score in all biopsies and tumour tissue areas, respectively.
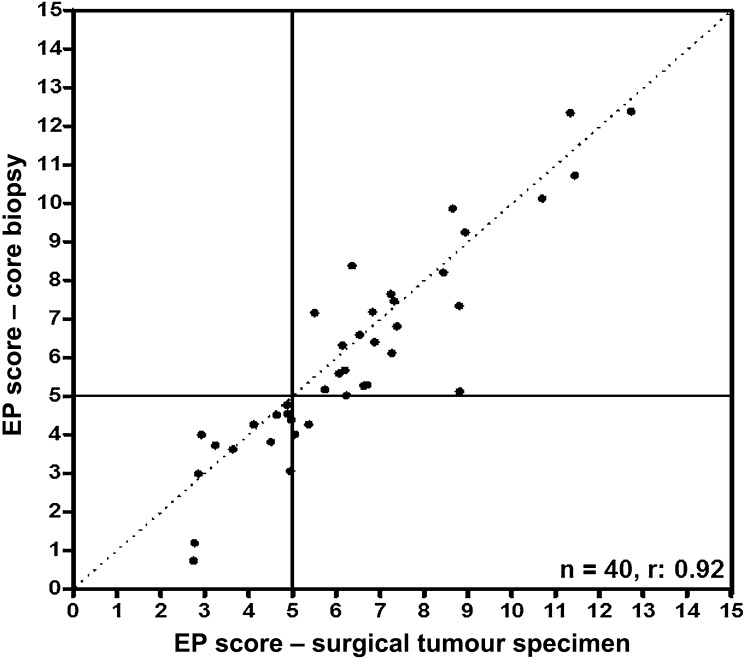
Gene expression levels of the eight EndoPredict genes (AZGP1, BIRC5, DHCR7, IL6ST, MGP, RBBP8, STC2, UBE2C) were compared between the biopsy and the corresponding surgical specimen. Relative RNA levels of each of the eight prognostic genes were significantly correlated between core biopsies and surgical specimens with Pearson correlation coefficients ranging from 0.65 to 0.89 (table 2). The EndoPredict score was also highly correlated between biopsies and tumour sections (r=0.92; table 2, Figure 1), and the concordance for the EndoPredict-based risk classification was excellent (overall agreement 95%, κ=0.89). Only two disagreements were found in terms of risk categorisation; however, both samples exhibited an EndoPredict score that was close to the predefined cut-off level.
Table 2.
Analysed genes and correlation between core biopsy and surgical specimens
| Gene name | Correlation (Pearson r) |
| AZGP1 | 0.82 |
| BIRC5 | 0.72 |
| DHCR7 | 0.77 |
| IL6ST | 0.89 |
| MGP | 0.69 |
| RBBP8 | 0.84 |
| STC2 | 0.65 |
| UBE2C | 0.76 |
| EndoPredict score | 0.92 |
Figure 1.
Correlation between the EndoPredict (EP) scores from biopsies and corresponding surrounding tumour tissue (n=40). Samples with an EndoPredict score below 5 were considered ‘low risk’, whereas samples with an EndoPredict score above 5 were considered ‘high-risk’. Pearson correlation coefficient 0.92.
Discussion
The results indicate that the EndoPredict score between core biopsy and corresponding surgical specimens is identical. In the evaluated samples, a biopsy-induced injury seems to have no influence on the EndoPredict score. This might be due to the selected genes in the EndoPredict assay, which are not directly linked to inflammation and wound repair processes. Haas and colleagues7 recently showed that prognostic markers such as uPA/PAI-1, which are associated with wound healing and inflammation, can be considerably altered by the fibroblastic inflammatory reaction around the biopsy channel. In addition, expression levels of 60 genes were compared between core biopsies and surgical tumour tissue specimens by Zanetti-Dällenbach et al.12 All analysed genes were selected due to their relevance in breast cancer and their association with biological functions such as proliferation, survival, invasiveness and wound healing. Among the genes, uPAR, PAI-1, COX2 and MMP1 showed an increased expression level in surgical tissue compared with the core biopsy. The data also indicate that the expression changes could be influenced by tissue injuries from preoperative core biopsy sampling.12
Despite the marked differences in RNA yield between core biopsies and corresponding surgical specimens, correlations between the EndoPredict scores were very high. The data suggest that the EndoPredict score can be reliably determined in core biopsy specimens. Inflammatory changes induced by presurgical core biopsies had no significant effect on the RNA-based risk assessment in the surgical specimens evaluated in this study. This suggests that tumour areas with biopsy-induced changes might be used for molecular testing.
Take-home messages.
The EndoPredict score was highly correlated between core biopsies and corresponding surgical tissue specimen; core biopsies appear to be a biomaterial to run the EndoPredict.
Tissue injury induced by preoperative biopsy sampling in this cohort seems to have no considerable effect on risk categorization by the EP.
Acknowledgments
The authors would like to thank Britta Beyer and Manuela Averdick for excellent technical assistance and Britta Dahl as well as Martina Eickmann for proofreading.
Footnotes
Contributors: BMM participated in the collection and histological examination of the samples, analysis and interpretation of data and drafting the manuscript. JCB participated in the acquisition of data, drafting of the manuscript, analysis and interpretation of data. HAF participated in the acquisition of data. CD, RK, CP and MD participated in the conception and design, analysis and interpretation of data, critical revision of the manuscript for important intellectual content and supervision. CD further participated in the histological examination of samples. JB and KEW participated in the statistical analysis. JP participated in the collection of samples. All authors read and approved the final paper. The first two authors contributed equally.
Competing interests: KEW, CP, MD, RK and CD are shareholders in Sividon Diagnostics. The remaining authors report no competing interests.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin 2012;62:10–29 [DOI] [PubMed] [Google Scholar]
- 2.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med 2009;360:790–800 [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Klijn JGM, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 2005;365:671–79 [DOI] [PubMed] [Google Scholar]
- 4.Filipits M, Rudas M, Jakesz R, et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin Cancer Res 2011;17:6012–20 [DOI] [PubMed] [Google Scholar]
- 5.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004;351:2817–26 [DOI] [PubMed] [Google Scholar]
- 6.van de Vijver MJ, He YD, van 't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 2002;347:1999–2009 [DOI] [PubMed] [Google Scholar]
- 7.Haas S, Park T-W, Hahne J, et al. Influence of preoperative core biopsies on uPA/PAI-1 expression in breast cancer tissue. Virchows Arch 2008;452:277–83 [DOI] [PubMed] [Google Scholar]
- 8.Dietel M, Ellis IO, Hofler H, et al. Comparison of automated silver enhanced in situ hybridisation (SISH) and fluorescence ISH (FISH) for the validation of HER2 gene status in breast carcinoma according to the guidelines of the American Society of Clinical Oncology and the College of American Pathologists. Virchows Arch 2007;451:19–25 [DOI] [PubMed] [Google Scholar]
- 9.Bohmann K, Hennig G, Rogel U, et al. RNA extraction from archival formalin-fixed paraffin-embedded tissue: a comparison of manual, semiautomated, and fully automated purification methods. Clin Chem 2009;55:1719–27 [DOI] [PubMed] [Google Scholar]
- 10.Hennig G, Gehrmann M, Stropp U, et al. Automated extraction of DNA and RNA from a single formalin-fixed paraffin-embedded tissue section for analysis of both single-nucleotide polymorphisms and mRNA expression. Clin Chem 2010;56:1845–53 [DOI] [PubMed] [Google Scholar]
- 11.Muller BM, Kronenwett R, Hennig G, et al. Quantitative determination of estrogen receptor, progesterone receptor, and HER2 mRNA in formalin-fixed paraffin-embedded tissue – a new option for predictive biomarker assessment in breast cancer. Diagn Mol Pathol 2011;20:1–10 [DOI] [PubMed] [Google Scholar]
- 12.Zanetti-Dallenbach R, Vuaroqueaux V, Wight E, et al. Comparison of gene expression profiles in core biopsies and corresponding surgical breast cancer samples. Breast Cancer Res 2006;8:R51. [DOI] [PMC free article] [PubMed] [Google Scholar]