Microorganisms make a wealth of unusual metabolites that have a secondary role in the organism’s ontogeny, such as self-defense, aggression, or even communication as the need arises. These compounds often have biological activity valuable to humankind (1). Nevertheless, as valuable as antibiotics and other naturally occurring drugs have been over the past 60 years, their future use is threatened by now widespread resistance to antibiotics among human, animal, and agricultural pathogens. Despite the fact that microbial metabolites are being found continually by pharmaceutical companies through targeted screening programs, fewer and fewer fundamentally new types of drugs have been found in this way during the past two decades (2).
Combinatorial biosynthesis seeks to help remedy this situation through genetic engineering, the goal of which is to make new antibiotics by exploiting the microbial genes and enzymes that make these substances, thereby aiming to discover drugs that cannot be found in nature. Combinatorial biosynthesis has been especially successful with bacterial polyketide synthases (PKSs), enzymes that, in essence, polymerize simple fatty acids into a myriad of chemical structures called “polyketides” (3). The polyketides discovered so far number into the thousands and are usually categorized on the basis of their structures: one type resembles branched-chain fatty acids formed into large carbocyclic rings (macrolides), and another type contains two or more aromatic rings fused into polycyclic structures, although nature has invented many variations of these and other structural themes (3). Modular type I PKSs consisting of one or more large multifunctional proteins make the first type, and iterative type II PKSs, complexes of several largely monofunctional proteins, make the second type (4, 5). After the first bacterial PKS genes were cloned between 1989 and 1991, extensive structure–function investigations of both types of PKSs were carried out, and more recently, this has led to many successful attempts to produce new microbial metabolites (4–9). Recently, McDaniel et al. (10) and Shen et al. (11) report major achievements in this direction through the interchange of modules or domains among type I PKSs in one system (10) and the expression of three PKS genes from a type II PKS system (11).
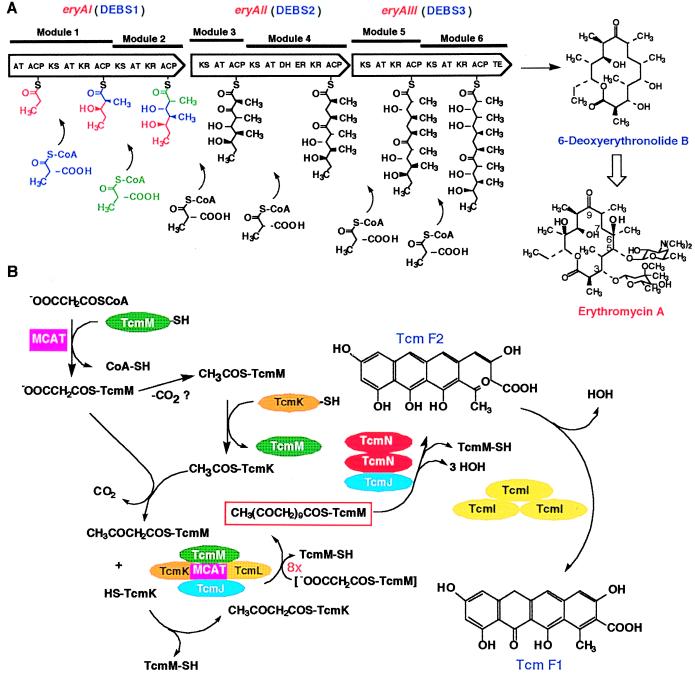
The basis for these seminal developments was laid through studies of the genetics of antibiotic production by the Streptomyces and related filamentous bacteria, begun by David Hopwood and other scientists nearly 20 years ago. The architecture of the type II PKSs was revealed first by analysis of the granaticin (12), tetracenomycin (13), and actinorhodin (14) PKS genes. Enzymological studies of the latter two systems carried out in the past year (15, 16) have established the features of the type II enzyme complex, as illustrated in Fig. 1B for the tetracenomycin PKS encoded by the tcmJKLMN genes. Here, a β-ketoacyl:acyl carrier protein (ACP) synthase (KS) composed of TcmK and TcmL (the latter type of protein is also known as the chain length factor or CLF) interact with a malonyl-CoA:ACP acyltransferase (MCAT), thought to be borrowed from the bacterial fatty acid synthase, and the TcmM ACP. These proteins are called the minimal PKS and select a starter unit (most often acetate, believed to be derived from decarboxylation of enzyme-bound malonate) to use with the malonate chain extender unit to form a poly-β-ketide intermediate by repetitive decarboxylative condensation of acylthioester intermediates bound to the active site of TcmK and the phosphopantetheinylated TcmM. The resulting decaketide is cyclized to Tcm F2 and then to Tcm F1 by the homodimeric TcmN and trimeric TcmI cyclases. In the tetracenomcyin system, the TcmJ protein plays an important but as yet poorly understood role in these processes. The actinorhodin system contains comparable enzymes (except for TcmJ) plus a ketoreductase (KR) that removes one of the aromatic hydroxyls by reduction of a carbonyl group in the poly-β-ketide.
Figure 1.
Structure and function of type I (A) and type II (B) bacterial PKSs.
Work on erythromycin A biosynthesis by Leadlay (17) and Katz (18) and their coworkers led in 1990–1991 to the discovery of the remarkable properties of modular type I PKSs. The 6-deoxyerythronolide B synthase (DEBS) subunits encoded by the eryA genes, oriented 5′ to 3′ as shown by the pointed wedges in Fig. 1A, are large multifunctional proteins containing domains typical of fatty acid synthases: KS, AT (acyltransferase), KR, DH (dehydratase), ER (enoyl reductase), ACP, and TE (thioesterase). These are grouped into six modules that specify how each 3-carbon unit of the final product is assembled, including the choice of the starter unit (governed by the AT and ACP closest to the N terminus of DEBS1, which are known as the loading domains or module) and the chain extender unit (specified by the ATs in each module). Different combinations of KR, DH, and ER domains can create further structural diversity in the product; compare modules 3 and 4 of DEBS2, which are responsible for the C-9 carbonyl and C-7 methylene groups in the product. Colinearity between the order of the modules in the DEBS proteins and the steps of polyketide assembly, as shown in Fig. 1A, is typical of bacterial type I PKSs. Hence, once propionate is loaded onto the ACP of the loading domain and (2S)-2-methylmalonate onto the second ACP of module 1, the system is set up to catalyze a decarboxylative condensation between these two substrates, after the propionate is moved to the KS domain, and then to reduce the β-carbonyl of the first intermediate formed to produce 2-methyl-3-hydroxyvalerate bound to the module 1 ACP. The same set of reactions is repeated, resulting in the formation of a 9-carbon intermediate attached to the ACP of module 2 of DEBS1, and so on, by using the catalytic activities found in each of the modules of DEBS2 and DEBS3, until the final 21-carbon product is released and cyclized (by the TE domain) as the 14-membered macrolide product (Fig. 1A). This is converted to erythromycin A by the tailoring reactions of the pathway, which can tolerate some differences in the structure of 6-deoxyerythronolide B, but not all of them that have been produced by combinatorial biosynthesis.
From the outset, Katz and coworkers (18) recognized that the modular nature of DEBS coupled with the known processivity of PKSs in general possibly meant that the absence of a particular KR, DH, or ER activity would allow the product of that module to be processed by the active sites of DEBS operating subsequently to produce analogs of 6-deoxyerythronolide B. The validity of this concept was first shown by targeted reprogramming of DEBS: deleting the KR domain of eryAIII module 5 (18) or inactivating the ER domain of eryAII module 4 (19) in both cases led to the predicted macrolides, which were formed in reasonable amounts.
The boundaries of this approach to creating novel macrolides by genetic engineering have been extensively explored in several laboratories, by using genetically engineered forms of either the erythromycin producer, Saccharopolyspora erythraea, or Streptomyces coelicolor. Such work was facilitated by Kao et al. (20), who developed an expedient way to express the eryA genes in this heterologous host. Analogs of the erythromycins and 6-deoxyerythronolide B made from starter units other than propionate (21, 22) and ones with varied substitution pattern at the methyl- and oxygen-bearing carbons have been made over the past 5 years, as summarized by McDaniel et al. (10). The principal method used in the earlier work has been replacement of a single domain in the eryA genes with inactivated forms or with their functional homologs from the rapamycin (23) or avermectin (24) PKSs, and included the addition of DH or DH and ER domains to create gain-of-function hybrid PKSs capable of introducing an alkene or alkane group in place of a hydroxyl-bearing carbon (25). The exciting development revealed by McDaniel and his colleagues at Kosan Biosciences, Inc. (10) is the extension of this approach to engineered eryA genes containing multiple changes in a true combinatorial fashion. The most productive single mutations in any module were combined to create double or triple mutants in the same or different modules, and then the effect on the productivity of 6-deoxyerythronolide B analogs was determined. Over 100 new analogs were created, and together they are said to represent about 3% of the total polyketides known (10). More importantly, this number “exceeds the total number of different macrolide ring structures yet discovered [in nature]” (10).
To put this achievement in perspective, four questions have to be asked and answered. First, can this approach be extended to other macrolides as well as to other types of polyketides made by modular PKSs? Surely this will be the case; already the sequence of more than eight sets of PKS genes for macrolide or ansamacrolide biosynthesis, besides those for erythromycin and rapamycin, are in the databases, and several more are being characterized. Although these come from bacterial genera other than the Streptomyces or Saccharopolyspora, it should be possible to express all of them in the currently used Streptomyces host strains where the resulting PKS should function well. The epothilone PKS genes, for instance, are an exciting candidate for genetic engineering as a means to new analogs of this promising anticancer drug (26). Second, will the yields of such analogs be high enough to permit preparation of them or their derivatives, made biologically or chemically, in 1- to 100-g amounts, which would be adequate for extensive biological testing? Since McDaniel et al. (10) state that good yields of single DEBS mutants often presaged the highest yields among the resulting multiple mutants, yield optimization at the former stage should result in acceptable yields of the analogs with more than one structural change. Ideally, DNA shuffling (27) and forms of error-prone PCR will be useful tools for isolating the desired higher yielding PKS mutants. Third, can the numbers of analogs be increased to tens or hundreds of thousands or even millions (the theoretical number approaches 107 for DEBS if all possible combinations of mutations are productive)? This should be done if combinatorial biosynthesis is to compete favorably with combinatorial chemistry methods. The answer would be disheartening if the linear approach to mutant construction illustrated by McDaniel and coworkers (10) were employed. Fortunately, Kosan Biosciences has recently developed an exciting new approach that greatly simplifies the work involved. Xue et al. (Q. Xue, G. Ashley, R. Zierman, M. Betlach, and D. Santi, unpublished work) have found that fully active DEBS is reconstituted in vivo when each eryA gene or its mutant is expressed separately from the same promoter, by using different combinations of episomic or chromosomally integrated plasmid vectors. This technique allows large numbers of combinations of mutations in each PKS gene to be tested against all of the other PKS genes simultaneously. Furthermore, it should be possible to deconvolute the resulting library of analogs by known methods involving direct chemical or biochemical assays of the mixed or individual bacterial cultures. This method is a true combinatorial approach and an expedient way of increasing the diversity of naturally occurring polyketides. Finally, can combinatorial biosynthesis include the addition of further modules to an existing PKS to increase the size of the carbon chain in addition to altering its functionality? Essentially nothing is known about how the carbon chain assembly intermediates are passed between the three DEBS subunits, and only a few experiments have been done to probe the outcome of fusing modules onto these proteins or adding additional modular PKS proteins to the system (28). Yet it is clear that nature has solved this problem because mono- to hexamodular proteins are found among the known modular PKSs.
Returning to the type II PKs system, the questions explored by combinatorial biosynthesis in this arena have mainly involved testing the outcome of substituting the components of one PKS for those of another to find out what determines the choice of starter unit, the length of the carbon chain, and the cyclization pattern (5). Some general rules have evolved from this work (29, 30), which allowed the predictable biosynthesis of certain types of polycyclic compounds (30), but interpretation of the experimental results has been complicated by the well known instability and reactivity of the poly-β-carbonyl intermediates (31). In the absence of the cognate cyclase or ketoreductase enzymes, the polycyclic products of a minimal PKS system were few and led to the belief that the minimal PKS was able to influence the cyclization regiochemistry. Moreover, the compounds made by certain minimal PKS and heterologous cyclase combinations appeared to support this idea (29). Shen et al. (11) now report that the Streptomyces coelicolor whiE minimal PKS, when expressed in an S. coelicolor strain lacking the actinorhodin and whiE genes, produces a multitude of mono- and polycyclic aromatic products made from chain lengths of 14–24 carbons. Among those with 24 carbons (the probable size of the normal whiE spore pigment, whose structure still is unknown, ref. 32), six quite different ring systems were isolated, one of which (TW93h) contains an unusual 2,4-dioxaadamantane moiety, representing a new structural class of polyketides not previously described. Formation of this “unnatural natural product” clearly demonstrates the power of combinatorial biosynthesis for the synthesis of novel chemotypes. The distribution of chain lengths and cyclization regiochemistries observed suggests that the whiE minimal PKS may be less fastidious (“sloppier,” ref. 11) than other systems previously studied, such as the actinorhodin and tetracenomycin minimal PKSs whose two principal products reflected the preference for 16-carbon (act) or 20-carbon (tcm) chain lengths (29). Hence, it now appears that a minimal PKS does not tightly control the chain length or cyclization pattern when assembling the poly-β-carbonyl intermediate. However, like the act and tcm systems (29, 33) and type II PKSs in general, only a single polycyclic product was formed when the cognate whiE cyclase(s) was present (32). This behavior suggests that the cyclases act like chaperones, to direct the behavior of the minimal PKS away from the manifold, spontaneous reactions its product(s) might undergo toward a single outcome set by microbial evolution.
The work of Shen et al. (11) should heighten interest in using type II PKSs to create molecular diversity among such polyketides, even though formation of the polycyclic aromatic products may not be controllable to the same extent as modular PKSs can be directed to produce specific products. Taken together with the advance in manipulating modular PKSs reported by McDaniel et al. (10), these discoveries expand the horizon of combinatorial biosynthesis considerably and will accelerate the rate at which new polyketides can be made.
Footnotes
References
- 1.Lancini G, Parenti F, Gallo G G. Antibiotics A Multidisciplinary Approach. New York: Plenum; 1995. [Google Scholar]
- 2.Strohl W R. In: Biotechnology of Antibiotics. Strohl W R, editor. New York: Marcel Dekker; 1997. pp. 1–48. [Google Scholar]
- 3.O’Hagan D. The Polyketide Metabolites. New York: Ellis Horwood; 1991. [Google Scholar]
- 4.Cane D E, Walsh C T, Khosla C. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 5.Hopwood D A. Chem Rev. 1997;97:2465–2497. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- 6.Katz L. Chem Rev. 1997;97:2557–2575. doi: 10.1021/cr960025+. [DOI] [PubMed] [Google Scholar]
- 7.Khosla C. Chem Rev. 1997;97:2577–2590. doi: 10.1021/cr960027u. [DOI] [PubMed] [Google Scholar]
- 8.Staunton J, Wilkinson B. Chem Rev. 1997;97:2611–2629. doi: 10.1021/cr9600316. [DOI] [PubMed] [Google Scholar]
- 9.Hutchinson C R. Curr Opin Microbiol. 1998;1:319–329. doi: 10.1016/s1369-5274(98)80036-2. [DOI] [PubMed] [Google Scholar]
- 10.McDaniel R M, Thamchaipenet A, Gustafsson C, Fu H, Betlach M, Betlach M, Ashley G. Proc Natl Acad Sci USA. 1999;96:1846–1851. doi: 10.1073/pnas.96.5.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen Y, Yoon P, Yu T-W, Floss H G, Hopwood D, Moore B S. Proc Natl Acad Sci USA. 1999;96:3622–3627. doi: 10.1073/pnas.96.7.3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherman D H, Malpartida F, Bibb M J, Kieser H M, Bibb M J, Hopwood D A. EMBO J. 1989;8:2717–2725. doi: 10.1002/j.1460-2075.1989.tb08413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bibb M J, Biro S, Motamedi H, Collins J F, Hutchinson C R. EMBO J. 1989;8:2727–2736. doi: 10.1002/j.1460-2075.1989.tb08414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez-Moreno M A, Martinez E, Boto L, Hopwood D A, Malpartida F. J Biol Chem. 1992;267:19278–19290. [PubMed] [Google Scholar]
- 15.Carreras C W, Khosla C. Biochemistry. 1998;37:2084–2088. doi: 10.1021/bi972919+. [DOI] [PubMed] [Google Scholar]
- 16.Bao W, Wendt-Pienkowski E, Hutchinson C R. Biochemistry. 1998;37:8132–8138. doi: 10.1021/bi980466i. [DOI] [PubMed] [Google Scholar]
- 17.Cortes J, Haydock S F, Roberts G A, Bevitt D J, Leadlay P F. Nature (London) 1990;346:176–178. doi: 10.1038/348176a0. [DOI] [PubMed] [Google Scholar]
- 18.Donadio S, Staver M J, McAlpine J B, Swanson S J, Katz L. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 19.Donadio S, McAlpine J B, Sheldon P J, Jackson M, Katz L. Proc Natl Acad Sci USA. 1993;90:7119–7123. doi: 10.1073/pnas.90.15.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kao C, Katz L, Khosla C. Science. 1994;265:509–512. doi: 10.1126/science.8036492. [DOI] [PubMed] [Google Scholar]
- 21.Jacobsen J R, Hutchinson C R, Cane D E, Khosla C. Science. 1997;277:367–369. doi: 10.1126/science.277.5324.367. [DOI] [PubMed] [Google Scholar]
- 22.Marsden A F A, Wilkinson B, Cortes J, Dunster N J, Staunton J, Leadlay P F. Science. 1998;279:199–202. doi: 10.1126/science.279.5348.199. [DOI] [PubMed] [Google Scholar]
- 23.Schwecke T, Aparacio J F, Molnar I, Konig A, Khaw L E, Haydock S F, Oliynyk M, Caffrey P, Cortes J, Lester J B, et al. Proc Natl Acad Sci USA. 1995;92:7839–7843. doi: 10.1073/pnas.92.17.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacNeil D J, Occi J L, Gewain K M, MacNeil T. Ann NY Acad Sci. 1994;721:123–132. doi: 10.1111/j.1749-6632.1994.tb47384.x. [DOI] [PubMed] [Google Scholar]
- 25.Kao C M, McPherson M, McDaniel R M, Fu H, Cane D E, Khosla C. J Am Chem Soc. 1997;119:11339–11340. [Google Scholar]
- 26.Nicolaou K C, Roschangar F, Vourloumis D. Angew Chem Int Ed. 1998;37:2014–2045. doi: 10.1002/(SICI)1521-3773(19980817)37:15<2014::AID-ANIE2014>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 27.Patten P A, Howard R J, Stemmer W P C. Curr Opin Biotechnol. 1997;8:724–733. doi: 10.1016/s0958-1669(97)80127-9. [DOI] [PubMed] [Google Scholar]
- 28.McDaniel R M, Kao C M, Hwang S J, Khosla C. Chem Biol. 1997;4:667–674. doi: 10.1016/s1074-5521(97)90222-2. [DOI] [PubMed] [Google Scholar]
- 29.McDaniel R M, Ebert-Khosla S, Fu H, Hopwood D A, Khosla C. Proc Natl Acad Sci USA. 1994;91:11542–11546. doi: 10.1073/pnas.91.24.11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDaniel R M, Ebert-Khosla S, Hopwood D A, Khosla C. Nature (London) 1995;375:549–554. doi: 10.1038/375549a0. [DOI] [PubMed] [Google Scholar]
- 31.Harris T M, Harris C M. Pure Appl Chem. 1986;58:283–294. [Google Scholar]
- 32.Yu T-W, Shen Y, McDaniel R, Floss H G, Khosla C, Hopwood D A, Moore B S. J Am Chem Soc. 1998;120:7749–7759. [Google Scholar]
- 33.Hutchinson C R. Chem Rev. 1997;97:2525–2535. doi: 10.1021/cr960022x. [DOI] [PubMed] [Google Scholar]