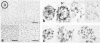Abstract
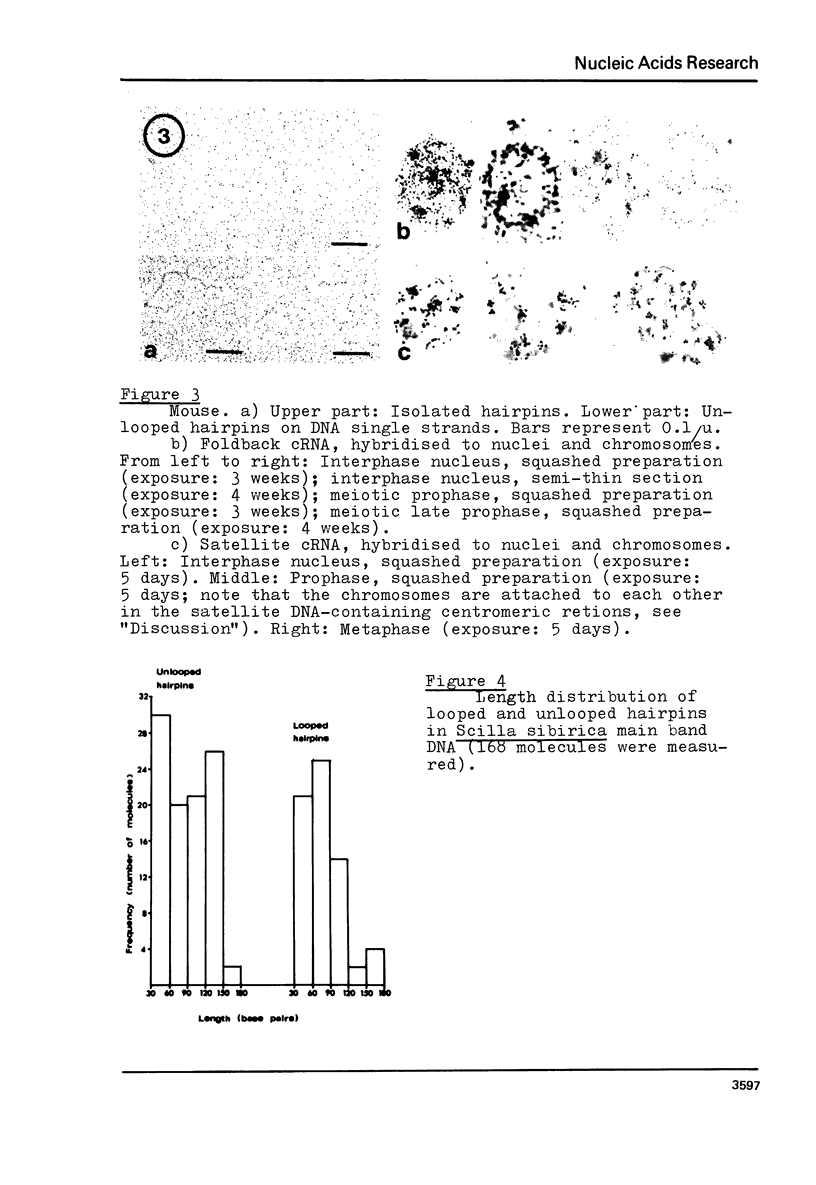
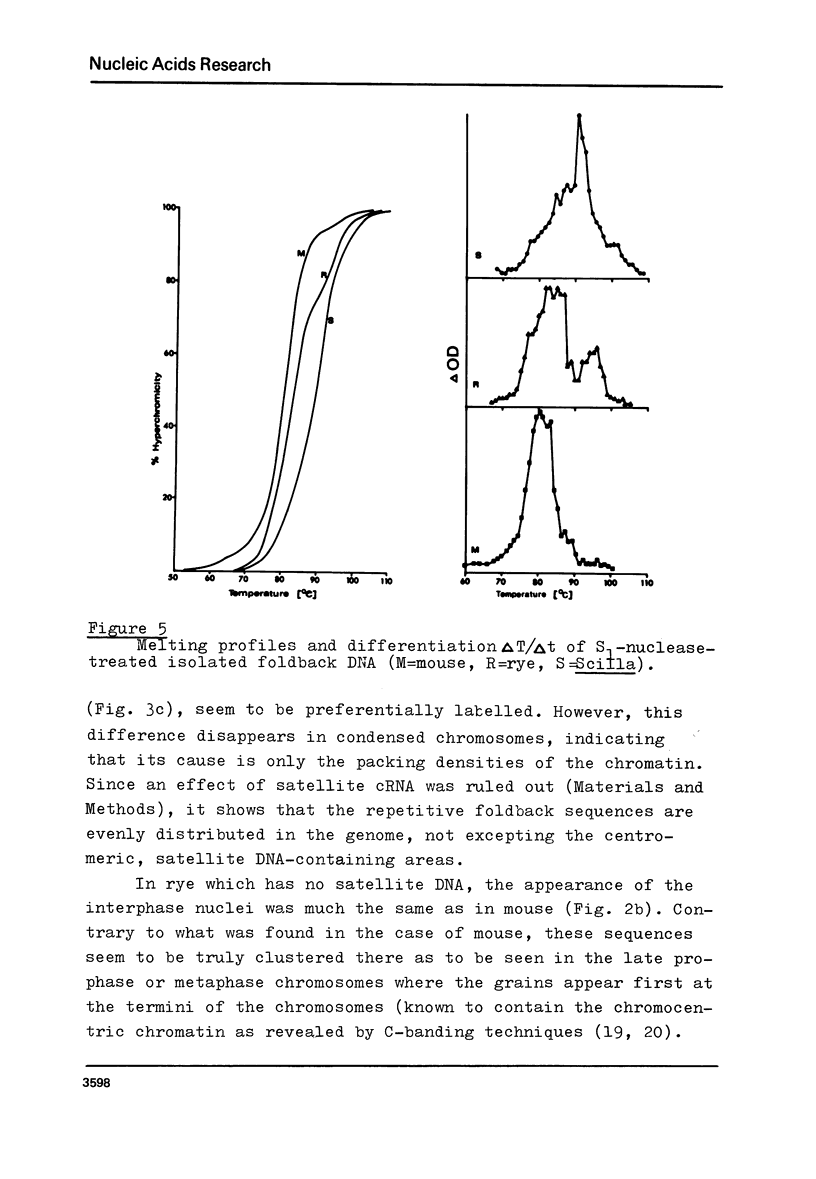
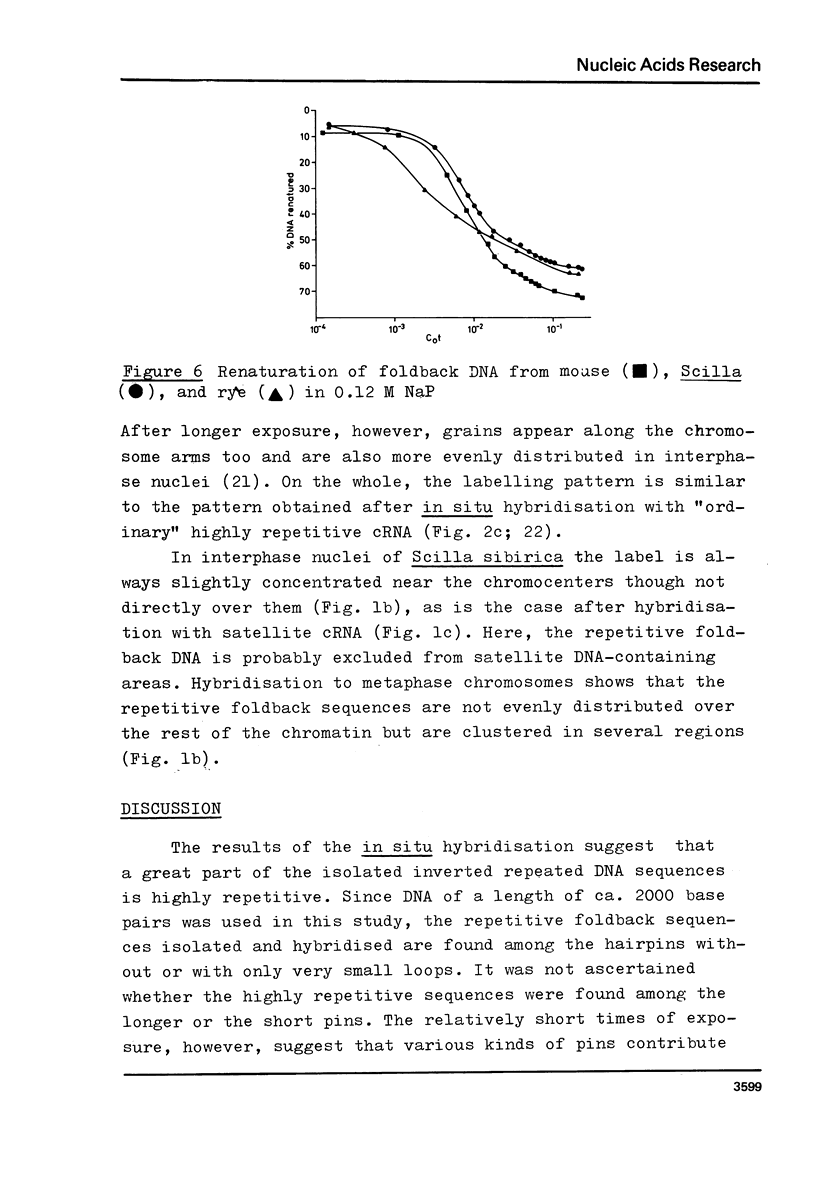
Foldback DNA, prepared from mouse and Scilla sibirica main band DNA, and from rye (Secale cereale) total DNA, was characterised by denaturation, renaturation, and electron microscopy. 3H-cRNA of this DNA was hybridised in situ to nuclei and chromosomes of the respective species. There is no universal labelling pattern among the three species. In mouse, highly repetitive foldback DNA is present in the whole chromatin including the satellite DNA-containing regions. In Scilla sibirica, on the contrary, the highly repetitive foldback sequences are excluded form the satellite DNA loci and are arranged in clusters in the remaining chromatin. In rye, there is a clear preferential labelling of the chromocenters in the interphase nuclei as well as metaphase chromosomes, indicating that highly repetitive foldback DNA is preferentially located among other highly repetitive sequences.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alonso C. Improved conditions for in situ RNA-DNA hybridization. FEBS Lett. 1973 Apr 1;31(1):85–88. doi: 10.1016/0014-5793(73)80078-x. [DOI] [PubMed] [Google Scholar]
- Bazetoux S., Jouanin L., Huguet T. Characterization of inverted repeated sequences in wheat nuclear DNA. Nucleic Acids Res. 1978 Mar;5(3):751–769. doi: 10.1093/nar/5.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A. J., Hardman N. Characterization of foldback sequences in hamster DNA using electron microsocpy. Nucleic Acids Res. 1977 Jan;4(1):247–268. doi: 10.1093/nar/4.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Palindromic base sequences and replication of eukaryote chromosome ends. Nature. 1974 Aug 9;250(5466):467–470. doi: 10.1038/250467a0. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Hearst J. E. An electron microscopic study of mouse foldback DNA. Cell. 1975 Aug;5(4):429–446. doi: 10.1016/0092-8674(75)90062-8. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Hyman R. W. A study in evolution: the DNA base sequence homology between coliphages T7 and T3. J Mol Biol. 1971 Dec 14;62(2):287–301. doi: 10.1016/0022-2836(71)90428-1. [DOI] [PubMed] [Google Scholar]
- Deininger P. L., Schmid C. W. An electron microscope study of the DNA sequence organization of the human genome. J Mol Biol. 1976 Sep 25;106(3):773–790. doi: 10.1016/0022-2836(76)90264-3. [DOI] [PubMed] [Google Scholar]
- Dott P. J., Chuang C. R., Saunders G. F. Inverted repetitive sequences in the human genome. Biochemistry. 1976 Sep 7;15(18):4120–4125. doi: 10.1021/bi00663a032. [DOI] [PubMed] [Google Scholar]
- Gill B. S., Kimber G. Giemsa C-banding and the evolution of wheat. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4086–4090. doi: 10.1073/pnas.71.10.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginelli E., Di Lernia R., Corneo G. The organization of DNA sequences in the mouse genome. Chromosoma. 1977 May 23;61(3):215–226. doi: 10.1007/BF00292806. [DOI] [PubMed] [Google Scholar]
- Hardman N., Jack P. L. Characterization of foldback sequences in Physarum polycephalum nuclear DNA using the electron microscope. Eur J Biochem. 1977 Apr 1;74(2):275–283. doi: 10.1111/j.1432-1033.1977.tb11391.x. [DOI] [PubMed] [Google Scholar]
- Jelinek W., Darnell J. E. Double-stranded regions in heterogeneous nuclear RNA from Hela cells. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2537–2541. doi: 10.1073/pnas.69.9.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin A. L., Sulimova G. E., Vanyushin B. F. Granulated hydroxyapatite: preparation and chromatographic properties. Anal Biochem. 1974 Sep;61(1):62–71. doi: 10.1016/0003-2697(74)90333-9. [DOI] [PubMed] [Google Scholar]
- Ryskov A. P., Saunders G. F., Farashyan V. R., Georgiev G. P. Double-helical regions in nuclear precursor of mRNA (pre-mRNA). Biochim Biophys Acta. 1973 Jun 8;312(1):152–164. doi: 10.1016/0005-2787(73)90060-9. [DOI] [PubMed] [Google Scholar]
- Schmid C. W., Manning J. E., Davidson N. Inverted repeat sequences in the Drosophila genome. Cell. 1975 Jun;5(2):159–172. doi: 10.1016/0092-8674(75)90024-0. [DOI] [PubMed] [Google Scholar]
- Singh L., Purdom I. F., Jones K. W. Effect of different denaturing agents on the detectability of specific DNA sequences of various base compositions by in situ hybridisation. Chromosoma. 1977 Apr 20;60(4):377–389. doi: 10.1007/BF00292860. [DOI] [PubMed] [Google Scholar]
- Spurr A. R. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969 Jan;26(1):31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- Sussenbach J. S., Kujik M. G. Initiation of adenovirus DNA replication does not occur via a hairpin mechanism. Nucleic Acids Res. 1978 Apr;5(4):1289–1295. doi: 10.1093/nar/5.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szala S., Michalska J., Paterak H., Bieniek B., Chorazy M. Inverted sequences in rat DNA. FEBS Lett. 1977 May 1;77(1):94–98. doi: 10.1016/0014-5793(77)80200-7. [DOI] [PubMed] [Google Scholar]
- Timmis J. N., Deumling B., Ingle J. Localisation of satellite DNA sequences in nuclei and chromosomes of two plants. Nature. 1975 Sep 11;257(5522):152–155. doi: 10.1038/257152a0. [DOI] [PubMed] [Google Scholar]
- Wallace B., Kass T. L. On inverted repeat sequences in chromosomal DNA. Genetics. 1976 Jan;82(1):139–140. doi: 10.1093/genetics/82.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]