Abstract
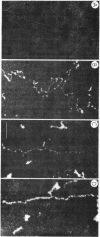
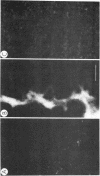
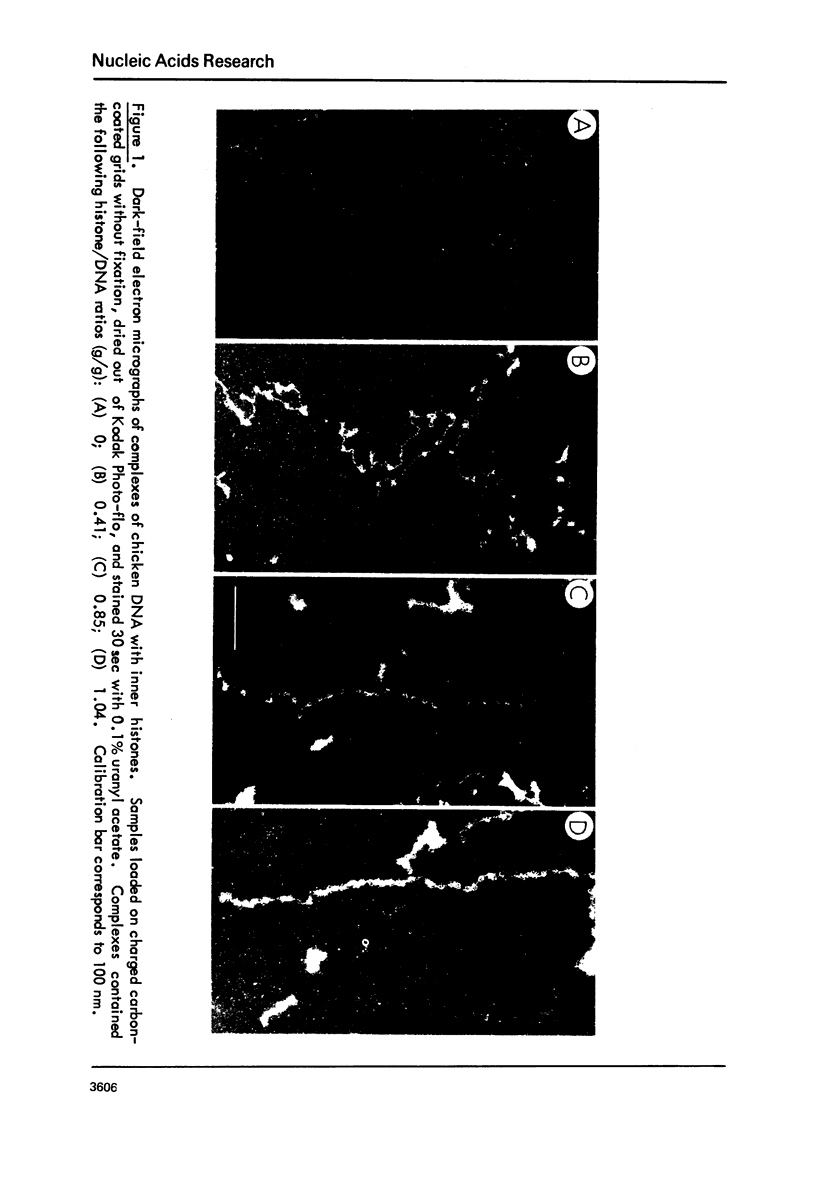
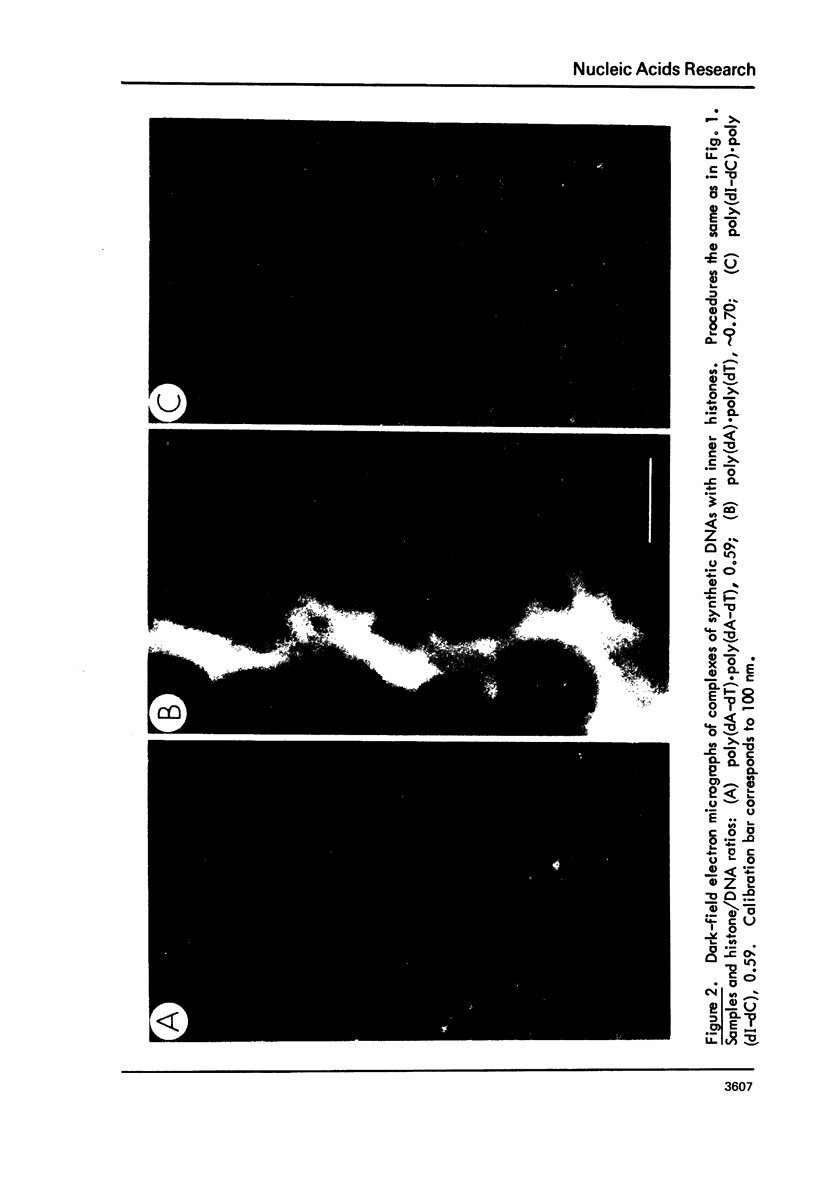
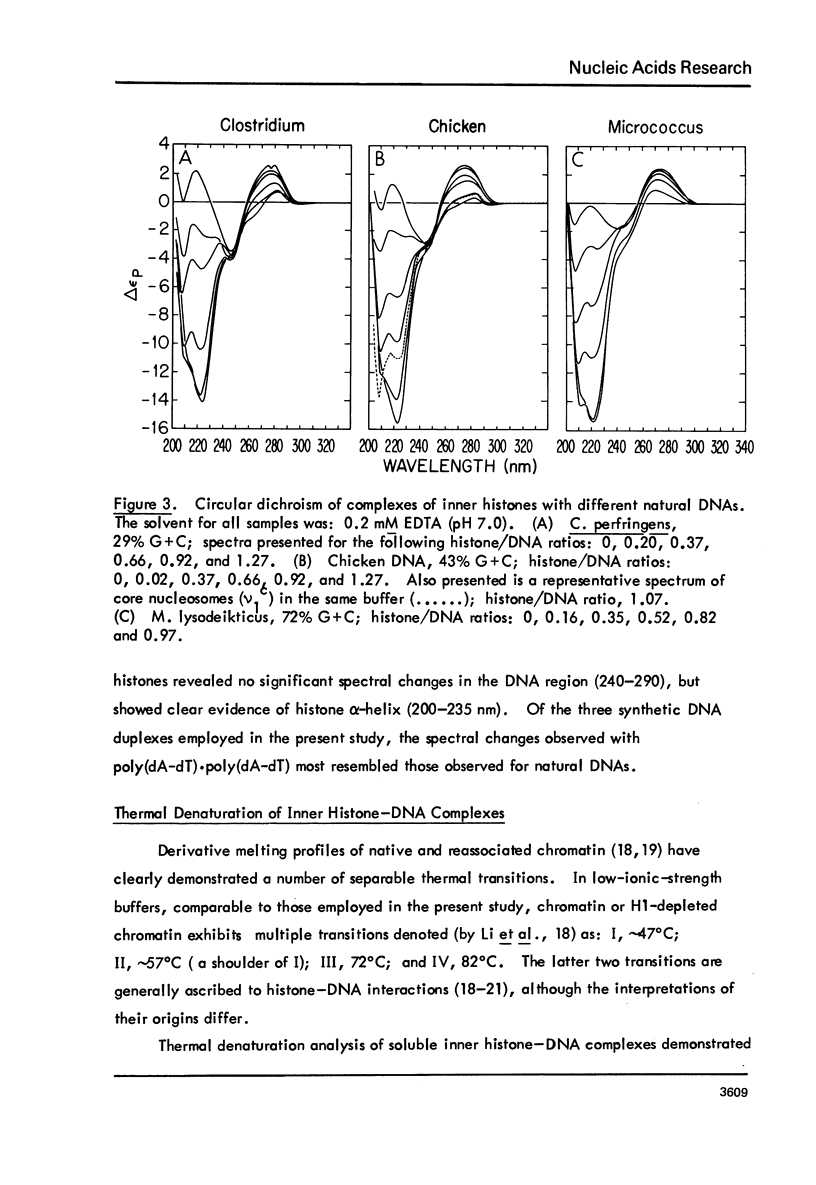
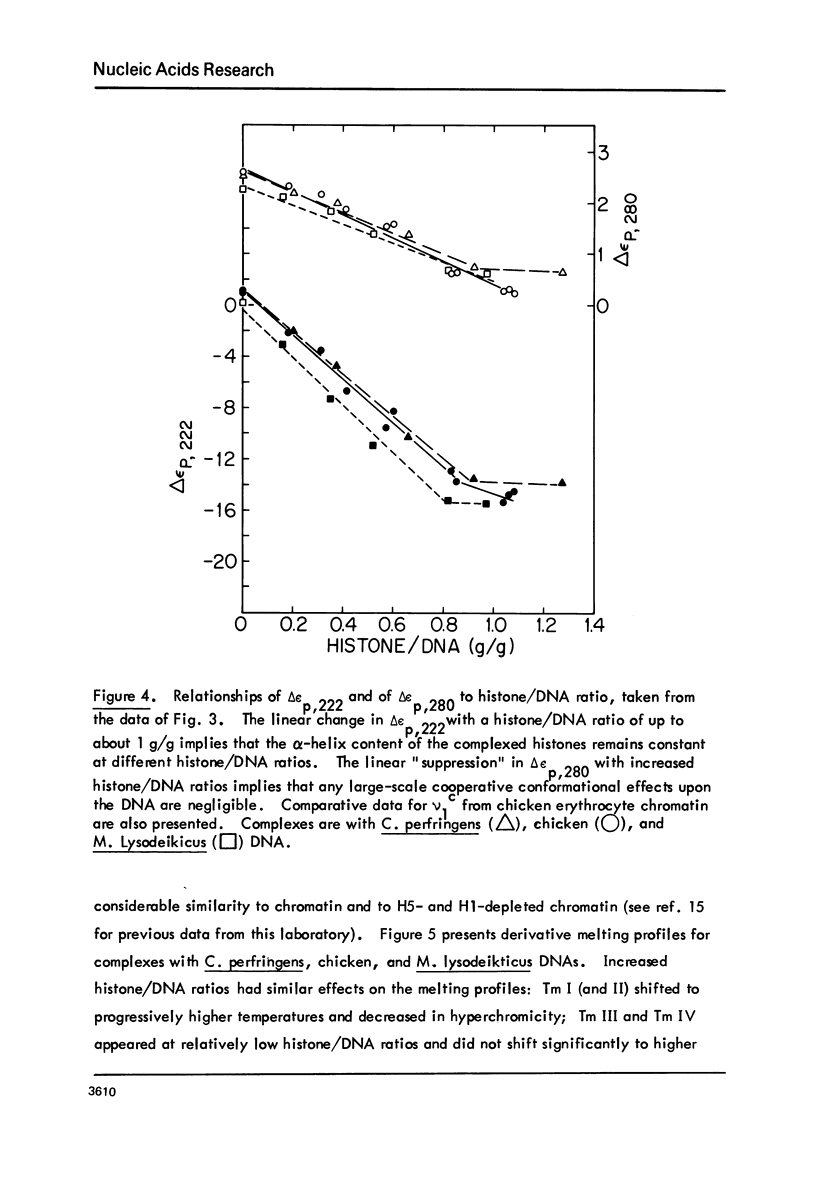
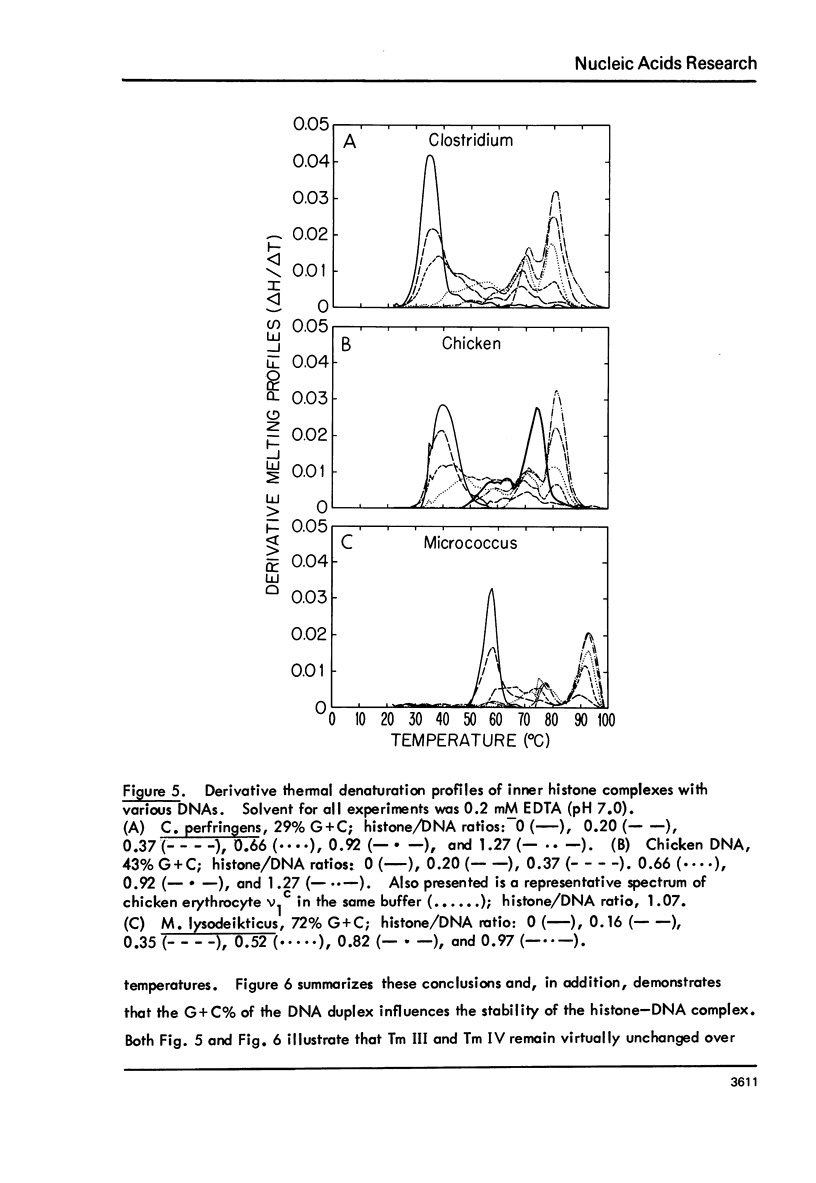
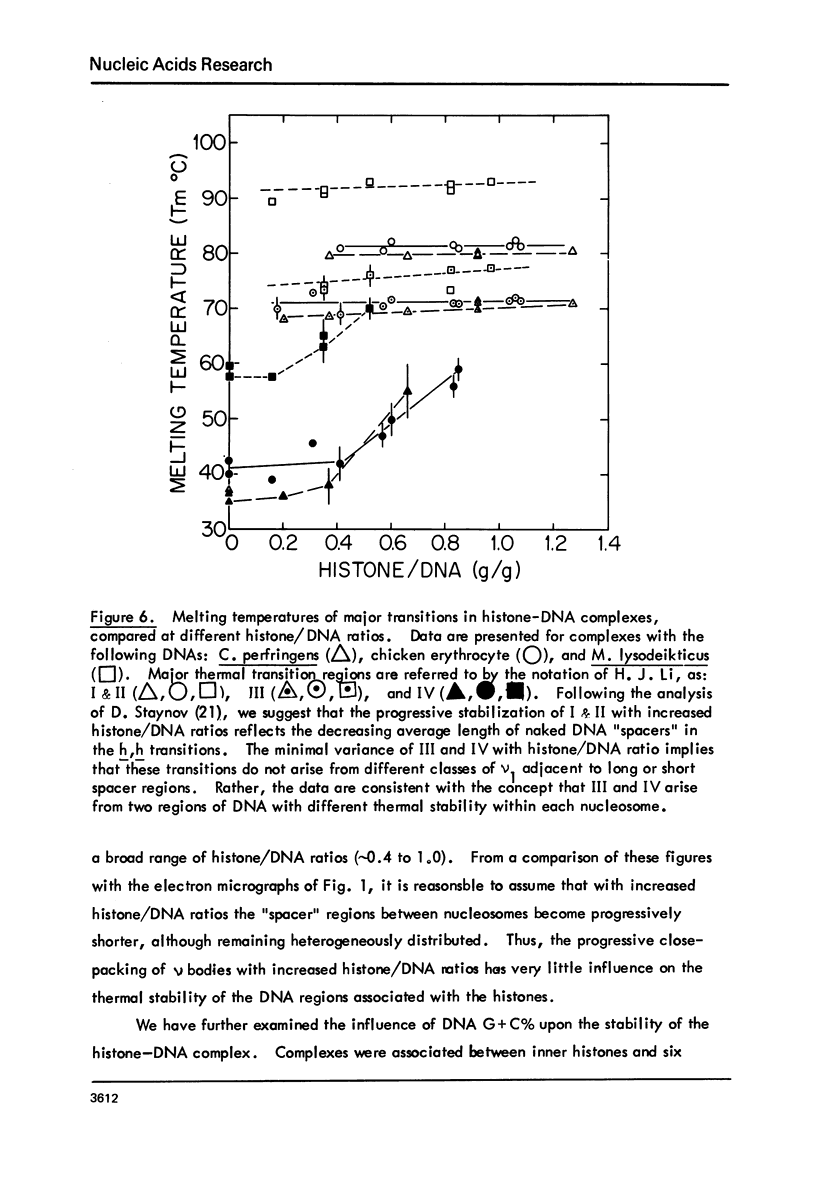
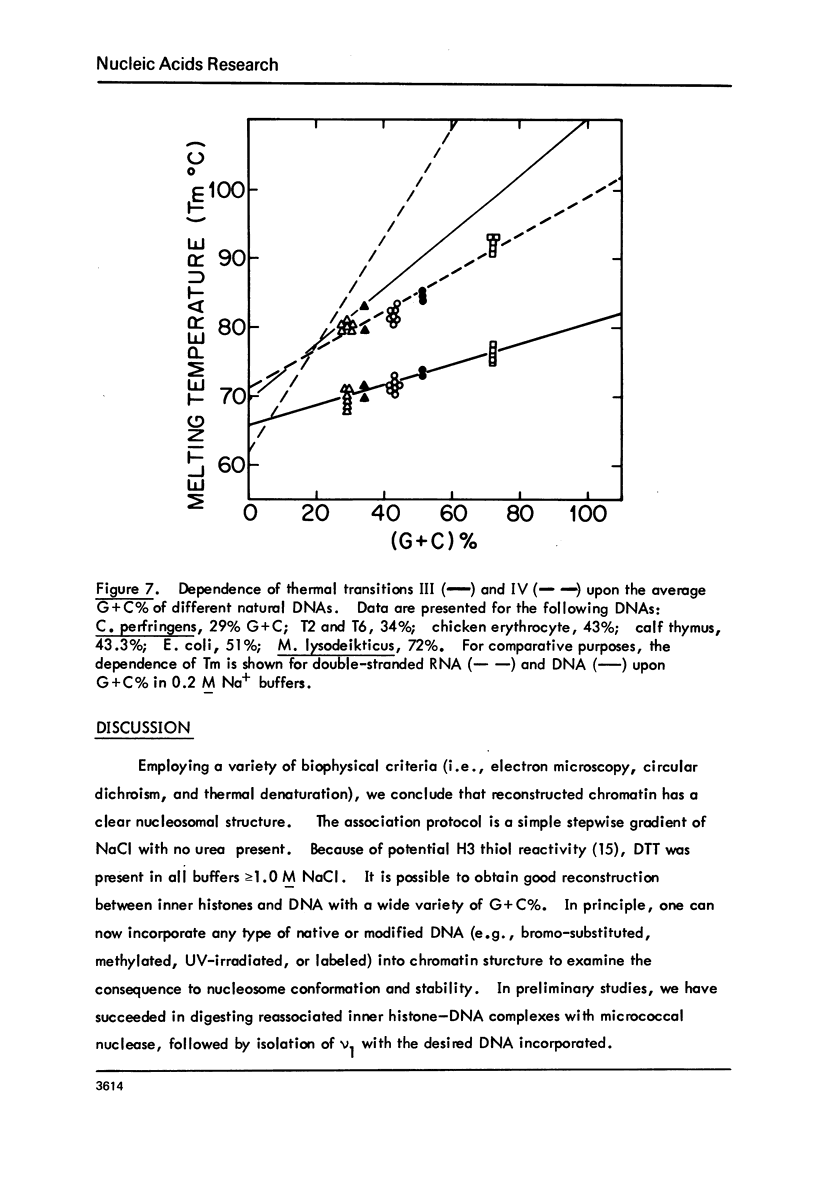
Chicken-erythrocyte inner histone tetramer has been complexed with several natural and synthetic DNA duplexes by salt-gradient dialysis at various protein/DNA ratios. The resulting complexes, in low-ionic-strength buffer, have been examined by electron microscopy, circular dichroism, and thermal denaturation. Electron microscopy reveals nucleosomes (nu bodies) randomly arranged along DNA fibers, including poly(dA-dT)-poly(dA-dT), poly(dI-dC)-poly(dI-dC), but not poly(dA)-poly(dT). Circular dichroism studies showed prominent histone alpha-helix and "suppression" of nucleic acid ellipticity (lambda less than 240 nm). Thermal denaturation experiments revealed Tm behavior comparable to that of H1- (or H5-) depleted chromatin. Tm III and Tm IV increased linearly with G + C%(natural DNAs), but were virtually independent of the histone/DNA ratio; therefore, the melting of nucleosomes along a DNA chain is insensitive to adjacent "spacer" DNA lengths. This suggests that Tm III and Tm IV arise from the melting of different domains of DNA associated with the core nu body.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brahms S., Brahms J., Van Holde K. E. Nature of conformational changes in poly[d(A-T)-d(A-T)] in the premelting region. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3453–3457. doi: 10.1073/pnas.73.10.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Felsenfeld G. Histone H3 disulfide dimers and nucleosome structure. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5519–5523. doi: 10.1073/pnas.74.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech C. L., Tinoco I., Jr Circular dichroism calculations for double-stranded polynucleotides of repeating sequence. Biopolymers. 1977 Jan;16(1):43–65. doi: 10.1002/bip.1977.360160105. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin. Nature. 1978 Jan 12;271(5641):115–122. doi: 10.1038/271115a0. [DOI] [PubMed] [Google Scholar]
- Garel A., Kovacs A. M., Champagne M., Daune M. Circular dichroism as a probe of DNA structure inside reconstituted nucleohistones. Nucleic Acids Res. 1976 Oct;3(10):2507–2519. doi: 10.1093/nar/3.10.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon S., Johnson R. S., Chan A. Relationship between protein and DNA structure in calf thymus chromatin. II. Conformational aspects. Biochemistry. 1974 Sep 10;13(19):3972–3981. doi: 10.1021/bi00716a024. [DOI] [PubMed] [Google Scholar]
- Leffak I. M., Jei Li H. Histone deoxyribonucleic acid complexes studied by thermal denaturation and circular dichroism spectroscopy. Biochemistry. 1977 Dec 27;16(26):5869–5878. doi: 10.1021/bi00645a035. [DOI] [PubMed] [Google Scholar]
- Li H. J., Chang C., Weiskopf M. Helix-coil transition in nucleoprotein-chromatin structure. Biochemistry. 1973 Apr 24;12(9):1763–1772. doi: 10.1021/bi00733a016. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Carlson R. D., Olins D. E. Visualization of chromatin substructure: upsilon bodies. J Cell Biol. 1975 Mar;64(3):528–537. doi: 10.1083/jcb.64.3.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins A. L., Carlson R. D., Wright E. B., Olins D. E. Chromatin nu bodies: isolation, subfractionation and physical characterization. Nucleic Acids Res. 1976 Dec;3(12):3271–3291. doi: 10.1093/nar/3.12.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins D. E., Bryan P. N., Harrington R. E., Hill W. E., Olins A. L. Conformational states of chromatin nu bodies induced by urea. Nucleic Acids Res. 1977 Jun;4(6):1911–1931. doi: 10.1093/nar/4.6.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Camerini-Otero R. D., Felsenfeld G. Chromatin structure as probed by nucleases and proteases: evidence for the central role of histones H3 and H4. Cell. 1976 Sep;9(1):179–193. doi: 10.1016/0092-8674(76)90063-5. [DOI] [PubMed] [Google Scholar]
- Staynov D. Z. Thermal denaturation profiles and the structure of chromatin. Nature. 1976 Dec 9;264(5586):522–525. doi: 10.1038/264522a0. [DOI] [PubMed] [Google Scholar]
- Stein A., Bina-Stein M., Simpson R. T. Crosslinked histone octamer as a model of the nucleosome core. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2780–2784. doi: 10.1073/pnas.74.7.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatchell K., Van Holde K. E. Reconstitution of chromatin core particles. Biochemistry. 1977 Nov 29;16(24):5295–5303. doi: 10.1021/bi00643a021. [DOI] [PubMed] [Google Scholar]
- Thomas G. J., Jr, Prescott B., Olins D. E. Secondary structure of histones and DNA in chromatin. Science. 1977 Jul 22;197(4301):385–388. doi: 10.1126/science.560060. [DOI] [PubMed] [Google Scholar]
- Tsai Y. H., Ansevin A. T., Hnilica L. S. Association of tissue-specific histones with deoxyribonucleic acid. Thermal denaturation of native, partially dehistonized, and reconstituted chromatins. Biochemistry. 1975 Mar 25;14(6):1257–1265. doi: 10.1021/bi00677a026. [DOI] [PubMed] [Google Scholar]
- Weischet W. O., Tatchell K., Van Holde K. E., Klump H. Thermal denaturation of nucleosomal core particles. Nucleic Acids Res. 1978 Jan;5(1):139–160. doi: 10.1093/nar/5.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C. L. Reconstitution of chromatin subunits. Science. 1977 Mar 25;195(4284):1350–1352. doi: 10.1126/science.841333. [DOI] [PubMed] [Google Scholar]