Among the most mysterious problems from both a molecular biology and medical point of view are the steps leading to the transformation of differentiated quiescent cells into oncogenic cells, which are capable of continual replication and growth. Two studies, one by Counter et al. (1) published in a recent issue of the Proceedings and a second published in this issue of the Proceedings (2), provide further evidence for a direct role(s) of telomerase in oncogenic transformation.
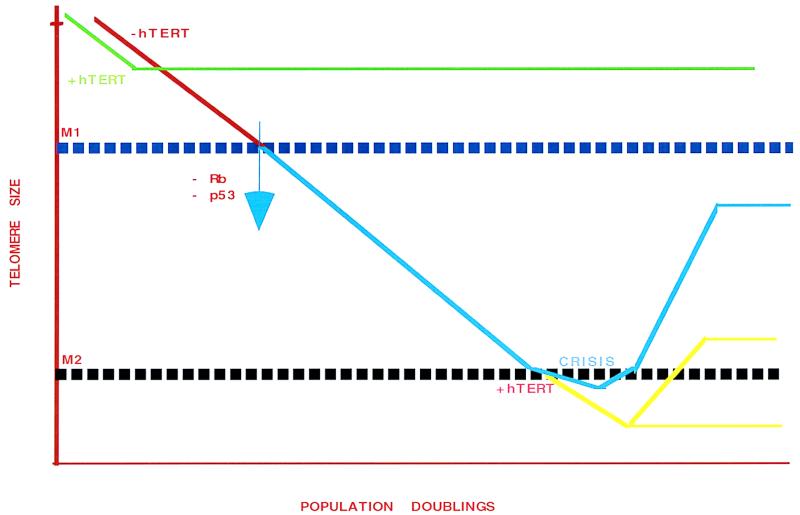
To investigate this process, the authors took advantage of a commonly used in vitro model system for defining steps in aging and in cellular immortalization in vitro (3). As first described by Hayflick (3), normal cultured cells proliferate until they reach a discrete point in which population growth ceases. This period is termed the M1 stage of aging, or replicative senescence (Fig. 1 red; ref. 3). This block, however, can be overcome by viral oncogenes. When cells are transformed with viruses that block p53 and pRB function (e.g., SV40), they continue to proliferate for an extended period of time (Fig. 1, blue). Ultimately, the cells reach a “crisis” point (M2) with a concomitant increase in both rates of death and chromosomal abnormalities (Fig. 1, blue; refs. 4–6). Only 1 × 10−7 cells survive this stringent selection. Both M1 and M2 are therefore potential suppression pathways for tumorigenesis. In the context of this commentary, M1 and senescence and M2 and crisis are used interchangeably. It is the nature of the events in M2 (specifically the role of telomerase and telomeres) that is approached in these two reports.
Figure 1.
The effect of ectopic expression of hTERT on growth and telomere size in cells cultured in vitro. As shown in red, when primary cultures are grown in vitro, telomere size decreases. Cells undergo population doublings until they reach a point, termed senescence (or M1), where populations cease to grow and telomere size is stabilized. One way to overcome senescence is expression of ectopic hTERT (green), which increases telomere size and allows continual growth. In the presence of oncogenic viruses such as SV40 virus large T antigen (and the loss of p53 or pRB functions), cells are capable of passing the senescence point (blue) and continuing to grow, with concomitant loss of telomeric sequences. This process continues until the population reaches a point called crisis (or M2) stage. At crisis, both a high rate of cell death and chromosome abnormalities have been observed. Of interest, this M2 arrest of growth appears to correlate with a particular threshold telomere length. Crisis can be overcome, however, by expression of hTERT in cells approaching M2 (yellow), with telomeres ultimately stabilizing at sizes close to or below the M2 threshold as defined in cells lacking ectopic expression of hTERT.
Telomeres, the termini of eukaryotic chromosomes, have a special problem in completing replication, termed the “end-replication” problem (7–9), which was proposed originally by Howard Cooke to be linked to the behavior of cells in M1 and M2 (10). This problem occurs as the consequence of the inability for the normal DNA replication system to overcome loss of terminal RNA primers (7, 8) and multiple exonucleolytic processes.
In most eukaryotes, the enzyme that compensates for this loss is telomerase (9). Telomerase is a unique enzyme that adds simple sequence repeats (e.g., TTAGGG in humans) onto preexisting 3′ overhangs. The core enzyme actually consists of two components: an RNA, which serves as template for the simple sequence tracts, and a catalytic subunit that acts as a reverse transcriptase on the RNA template (9, 11–13). Cloning and characterization of the human telomerase RNA (14) and the human catalytic subunit of telomerase (hTERT) (15, 16) have demonstrated that in many cell lines, hTERT, but not the telomerase RNA, is expressed concurrently with telomerase (17–19). These data suggest that hTERT, at least in some contexts, may be the limiting factor for telomerase activity.
So, what is the evidence of a relationship between telomere length, telomerase, and the M1 and M2 mortality stages? One of the primary lines of evidence was provided by the observation that telomeres of cultured somatic cells continuously erode until M1 (6). Ectopic expression of hTERT leads to cellular immortalization and a bypass of senescence (20–22), suggesting that telomere size may be causally related to senescence. If M1 is overcome by transformation with viral oncogenes, telomeres continue to decrease in size until M2 is reached, a process that may be dictated by telomere length itself.
Numerous additional studies have indicated that telomerase is involved in this in vitro system. First, telomerase activity and hTERT are absent in most primary tissue systems, while both are present in high abundance in immortalized cells (6, 15, 16). Second, whereas telomere size continually decreases during the aging process in vitro, immortalized cells reach an equilibrium, albeit at shorter-than-wild-type length. Third, continued expression of telomerase is necessary to maintain immortality (23). Finally, a loss of telomerase results in telomere shortening and subsequent inviability in yeast (24), similar to the observations in human cells.
However, the role of telomerase and hTERT as cells enter crisis has not yet been investigated, until the studies from Counter et al. (1) and Zhu et al. (2). This is a central question because ectopic expression of hTERT is likely to best mimic the alterations that occur in the survivors of crisis. The immortalizing events in crisis may in turn be a model for the steps involved in the malignant pathway in vivo.
By using a cultured HEK (human embryonic kidney) system (25), Counter et al. (1) demonstrated that cells that were virally transformed to overcome M1 and subsequently transfected with a vector containing hTERT became immortalized. Their studies indicate that telomeric tracts stabilize at sizes greater than those normally obtained during crisis. Counter et al. (1) therefore concluded that an increase in telomere size beyond an M2 telomere threshold, rather than telomerase activity per se, is responsible for immortalization (Fig. 1, yellow). In contrast, a C-terminal hemagluttinin-tagged hTERT derivative, although catalytically active (14), did not avert crisis, raising the possibility that a noncatalytic domain of hTERT may be involved in the immortalization process. However, it cannot yet be excluded that the abundance of telomerase contributes to the behavior of the tagged and untagged forms of hTERT.
Zhu et al. (2) took an analogous approach to express the hTERT in cell lines derived from primary fibroblasts. Although this study confirmed the Counter et al. (1) investigation, the report also introduces some additional puzzles. Zhu et al. (2) provided evidence for two steps after immortalization of M2 cells: (i) continual loss of telomeric sequences below known crisis thresholds, followed by (ii) the stabilization of the telomeres at a size well below the M2 threshold (Fig. 1, yellow). They postulate, therefore, that telomere size is not the deciding factor for entering crisis. Rather, an additional, previously undescribed property of telomerase may be responsible for circumventing the consequences of short telomere sizes.
Despite the discrepancies between the results shown in these two papers, a particularly intriguing result was demonstrated in both studies: transfection of the human catalytic subunit leads to immortalization of M2 cells, with the concurrent loss of chromosomal abnormalities. Both studies also imply that the most likely candidate for overcoming M2 is a mutation in the regulators of hTERT. Among the reasons for the differences in the two studies may be multiple genetic differences between regulators of telomerase in the different cell lines used in the two studies. An alternative possibility is that the M2 thresholds are changed in the presence of ectopic telomerase. Finally, the levels or activity of telomerase in the two systems are difficult to compare. Hence, the kinetics of the M2 events in the different cell lines may be modified.
In any case, the identification of the two steps by Zhu et al. (2) must be evaluated independently. Of particular importance is an understanding of the first step after immortalization by transfection with hTERT, which appears to separate telomerase activity and telomere length. In the presence of telomerase, mean telomere length continues to contract in these immortalized cells well below the normal size observed at crisis (4, 5). However, complicating these results is the fact that the conclusions are based on the statistical means of very broad distributions. A subpopulation of short telomeres in inviable cells cannot be ruled out at this point.
What is the role of telomerase in this process? First, as suggested by Zhu et al. (2), telomerase may also act as a telomeric cap. Such a cap may protect against nonhomologous end-joining factors, nucleolytic loss, or association with other end- or telomere- binding factors (e.g., TRF1) (26). The best evidence for this type of behavior of telomeres comes from investigations in the yeast Kluyueromyces lactis, in which a high level of recombinants are observed before significant selection (27).
Second, telomeric end structure may act to create anomalous single-stranded regions. For example, long single-stranded regions may be generated in the presence of ectopic telomerase. An increase in telomeric single-stranded regions has been found in several mutant strains of yeast (28–30). Such single-stranded regions may influence the interpretation of altered mobility of telomeric fragments.
Third, the stabilization of the shortest telomeres early in M2 immortalization may occur by a feedback mechanism, such as proposed in yeast and mammals (31, 32), that can “measure” deviations from average telomere length. Compensation for extensive loss may selectively induce telomerase or telomere healing of short telomeres. Further studies using specifically marked chromosomes are required to distinguish between these and other models.
Beyond the specific questions on the mechanism of telomerase activities is a far more general and basic question: To what degree does the in vitro culture system reflect the tumorigenic and oncogenic processes?
Several volumes of literature have provided support for the immortalization/oncogenesis model. First, primary somatic cells normally lack telomerase activity, whereas virtually all malignant cells contain high levels of telomerase and hTERT (6, 15, 16). Second, the population doublings that are required to reach senescence in vitro for older individuals and individuals with aging diseases are far less than their wild-type controls (33). Importantly, telomeres derived from elderly individuals tend to be shorter than those derived from younger donors (34). Finally, mutations in the catalytic subunit of the telomerase from the yeast Saccharomyces cerevisiae result in a concomitant decrease in tract size and cell viability, analogous to the in vitro crisis point in humans (24).
However, there are some emerging data that suggest that immortalization per se may not fully reflect the oncogenic process. As noted above, three recent studies (20–22) have demonstrated that transfection of primary cells with hTERT overcomes senescence. These cells do not exhibit any other manifestations of the oncogenic state, such as loss of cell-cycle control, karyotypic instability, response to serum deprivation, and the rate of tumor formation when introduced into scid mice (35, 36). These data indicate that telomerase is clearly not sufficient to overcome all of the barriers to the oncogenic state. The results may also be due to differences in “immortalization” by bypass of senescence as opposed to bypass of crisis. Further answers should arise from the continuing studies of mice disrupted in both telomerase genes (37, 38).
At present, there are three means of producing “immortalized cells” in vitro, each of which may contain unique mechanistic components: (i) ectopic expression of hTERT in presenescent cells that avoids M1; (ii) ectopic expression of hTERT of cells in crisis; and (iii) the mutagenic events that lead to immortalization during crisis. Most of these latter cells give rise to cultures that display an up-regulation of telomerase, possibly through spontaneous mutagenesis. Which, if any, of these processes are relevant to the behavior of normal and oncogenic cells remains to be determined.
The data at present suggest that telomerase and telomere length are critical components in the immortalization process. However, the rationale for the medical use of telomerase drugs depends on a strong and direct connection between telomerase activity in vivo and oncogenesis. What is truly necessary now is a manipulatable genetic system for analyzing the provocative proposals based on in vitro studies in an in vivo setting. More answers to these questions are undoubtedly around the corner.
Footnotes
References
- 1.Counter C M, Hahn W C, Wei W, Caddle S D, Beijersbergen R L, Lansdorp P M, Sedivy J M, Weinberg R A. Proc Natl Acad Sci USA. 1998;95:14723–14728. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu J, Wang H, Bishop J M, Blackburn E H. Proc Natl Acad Sci USA. 1999;96:3723–3728. doi: 10.1073/pnas.96.7.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayflick L, Moorhead P S. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 4.Shay J W, Pereira-Smith O M, Wright W E. Exp Cell Res. 1991;196:33–39. doi: 10.1016/0014-4827(91)90453-2. [DOI] [PubMed] [Google Scholar]
- 5.Hara E, Tsurui H, Shinozaki A, Nakada S, Oda K. Biochem Biophys Res Commun. 1991;179:528–534. doi: 10.1016/0006-291x(91)91403-y. [DOI] [PubMed] [Google Scholar]
- 6.Harley C. In: Telomeres. Blackburn E H, Greider C W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 247–263. [Google Scholar]
- 7.Watson J D. Nat New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 8.Olkonikov A M. J Theor Biol. 1973;41:181–190. [Google Scholar]
- 9.Greider C W. In: Telomerase. Blackburn E H, Greider C W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 35–68. [Google Scholar]
- 10.Cooke H, Smith B A. Cold Spring Harbor Symp Quant Biol. 1986;51:213–219. doi: 10.1101/sqb.1986.051.01.026. [DOI] [PubMed] [Google Scholar]
- 11.Beattie T L, Zhou W, Robinson M O, Harrington L. Curr Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- 12.Weinrich S L, Pruzan R, Ma L, Ouellette M, Tesmer V M, Holt S E, Bodnar A G, Lichtsteiner S, Kim N W, Trager J B, et al. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 13.Counter C M, Myerson M, Eaton E N, Ellisen L W, Caddle S D, Haber D A, Weinberg R A. Oncogene. 1998;16:1217–1222. doi: 10.1038/sj.onc.1201882. [DOI] [PubMed] [Google Scholar]
- 14.Feng J, Funk W D, Wang S S, Weinrich S L, Avilion A A, Chiu C-P, Adams R R, Chang E, Allsopp R C, Yu J, et al. Science. 1995;269:1236–1242. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andres W H, Lingner J, Harley C B. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 16.Myerson M, Counter C M, Eaton E N, Ellisent L W, Steiner P, Caddle S D, Ziaugra L, Beiersbergen R L, Davidoff J, Liu Q, et al. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 17.Blasco M A, Rizen M, Greider C W, Hanahan D. Nat Genet. 1996;12:200–204. doi: 10.1038/ng0296-200. [DOI] [PubMed] [Google Scholar]
- 18.Broccoli D, Young J W, de Lange T. Proc Natl Acad Sci USA. 1995;92:9082–9086. doi: 10.1073/pnas.92.20.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soder A I, Hoare S F, Muir S, Going J J, Parkinson E K, Keith W N. Oncogene. 1997;14:1013–1021. doi: 10.1038/sj.onc.1201066. [DOI] [PubMed] [Google Scholar]
- 20.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C-P, Morin G B, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 21.Vaziri H, Benchimol S. Curr Biol. 1998;8:279–282. doi: 10.1016/s0960-9822(98)70109-5. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Xie L-Y, Alllan S, Beach D, Hannon G J. Genes Dev. 1998;12:1769–1774. doi: 10.1101/gad.12.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim N W, Piatysek M A, Prowse K R, Harley C B, West M D, Ho P L, Coviello G M, Wright W E, Weinrich S L, Shay J W. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 24.Lingner J, Cech T R, Hughes T R, Lundblad V. Proc Natl Acad Sci USA. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart N, Bacchetti S. Virology. 1991;180:49–57. doi: 10.1016/0042-6822(91)90008-y. [DOI] [PubMed] [Google Scholar]
- 26.van Steensel B, de Lange T. Nature (London) 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 27.McEachern M J, Blackburn E H. Genes Dev. 1996;10:1822–1834. doi: 10.1101/gad.10.14.1822. [DOI] [PubMed] [Google Scholar]
- 28.Garvik B, Carson M, Hartwell L. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gravel S, Larrivee M, Labrecque P, Wellinger R J. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- 30.Polotnianka R M, Li J, Lustig A J. Curr Biol. 1998;8:831–834. doi: 10.1016/s0960-9822(98)70325-2. [DOI] [PubMed] [Google Scholar]
- 31.Marcand S, Gilson E, Shore D. Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- 32.van Steensel B, Smogorzewska A, de Lange T. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 33.Allsopp R C, Vaziri H, Patterson C, Goldstein S, Younglai E V, Futcher A B, Greider C W, Harley C B. Proc Natl Acad Sci USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hastie N D, Dempster M, Dunlop M G, Thompson A M, Green D K, Allshire R C. Nature (London) 1990;345:458–460. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 35.Jiang X-R, Jimenez G, Chang E, Grolkis M, Kusler B, Sage M, Beeche M, Bodnar A G, Wahl G M, Tisty T D, et al. Nat Genet. 1999;21:111–114. doi: 10.1038/5056. [DOI] [PubMed] [Google Scholar]
- 36.Morales C P, Holt S E, Ouellette M, Kaur K J, Yan Y, Wilson K S, White M A, Wright W E, Shay J W. Nat Genet. 1999;21:115–118. doi: 10.1038/5063. [DOI] [PubMed] [Google Scholar]
- 37.Blasco M A, Lee H W, Hande M P, Samper E, Landsdorp P M, DePinho R A, Greider C W. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 38.Lee H W, Blasco M A, Gottliev G J, Horner J H W, II, Greider C W, DePinho R. Nucleic Acids Res. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]