Abstract
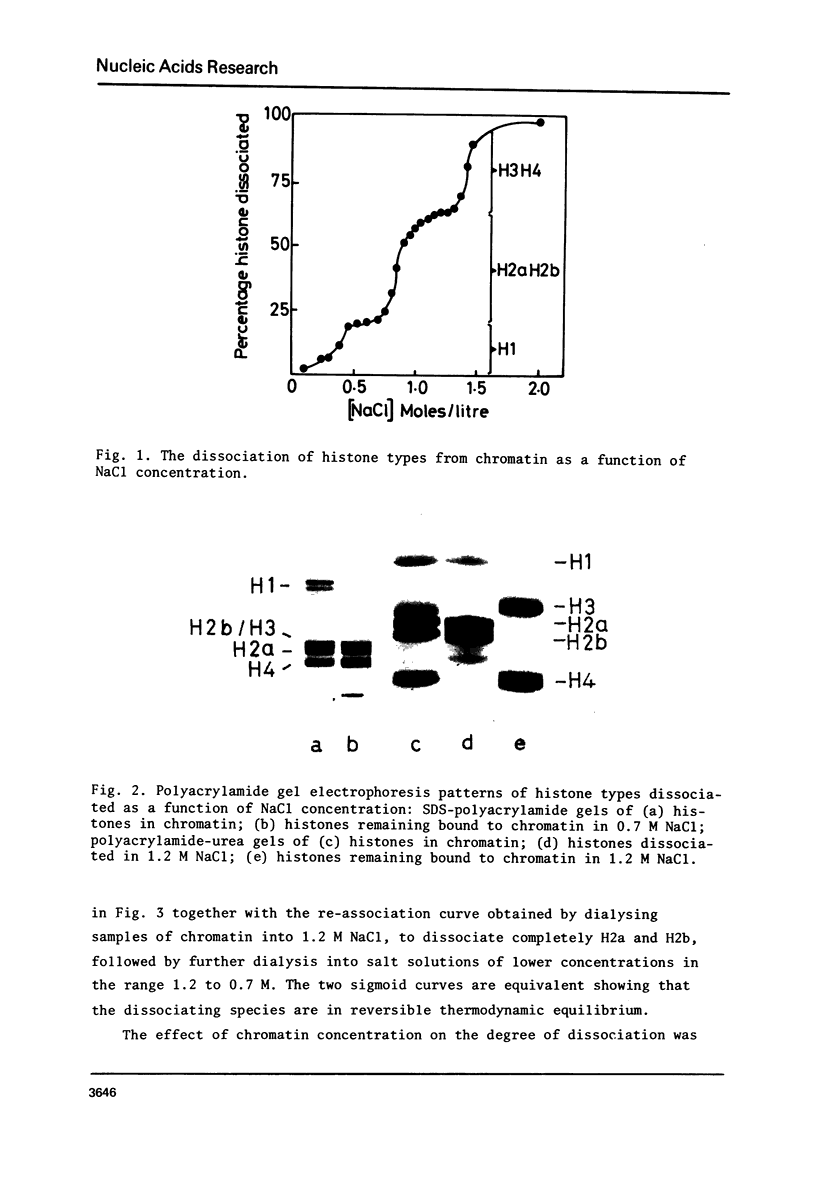
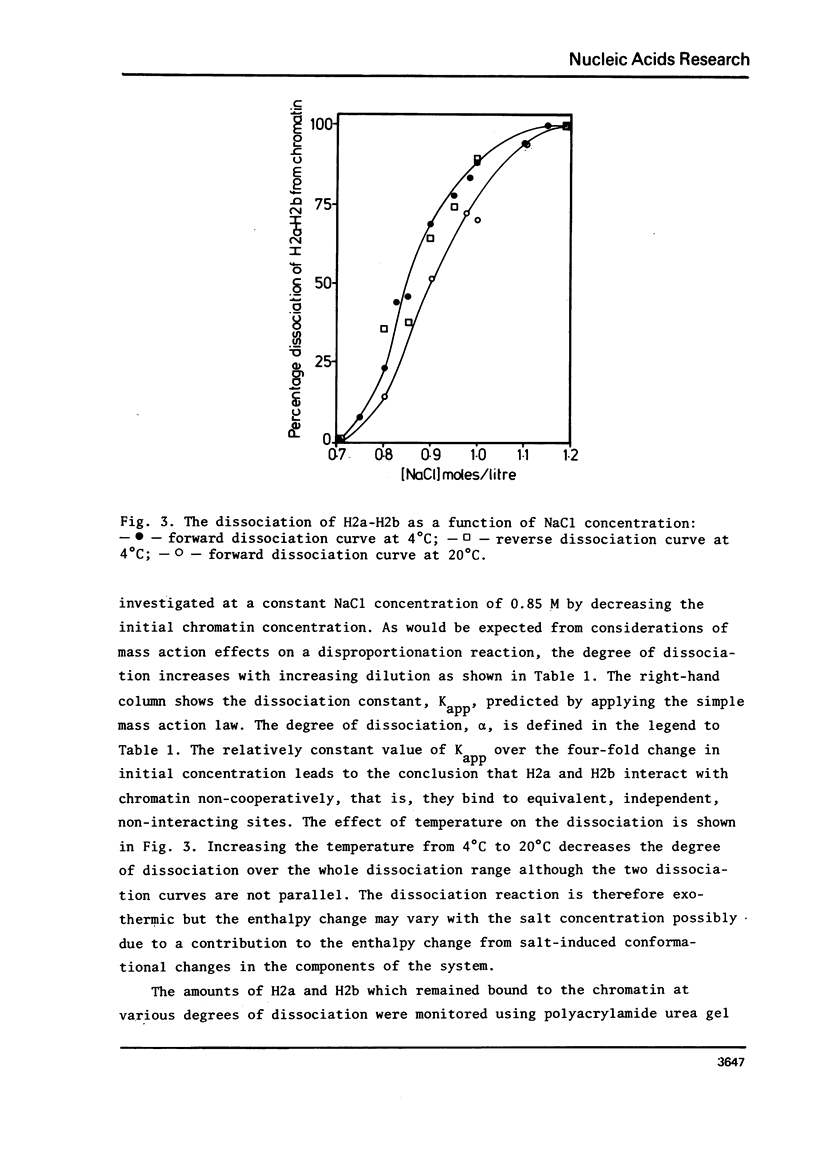
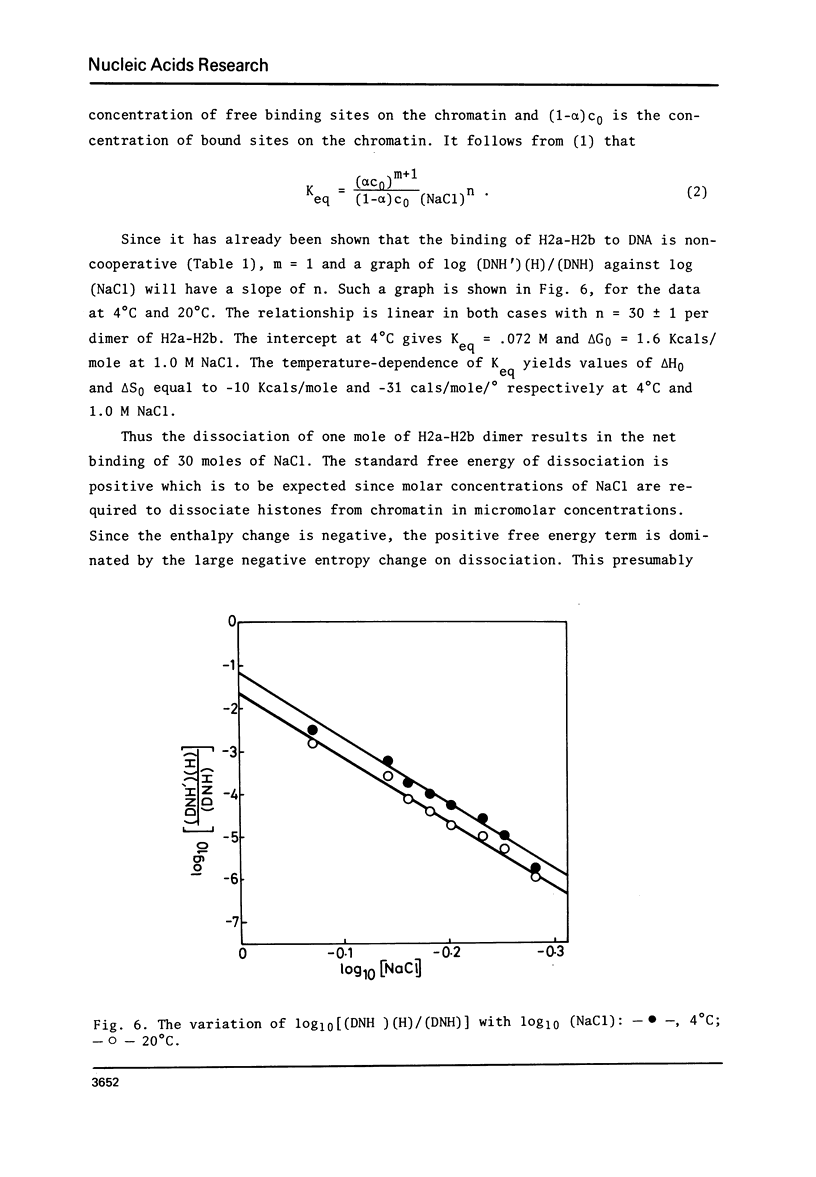
The binding of core histone proteins to DNA, measured as a function of [NaCl[ is a reversible process. Dissociation and reassociation occurs in two stages. Between 0.7 and 1.2 M NaCl H2a H2b bind non-cooperatively as an equimolar complex with deltaGo = 1.6 Kcals/mole at 4 degree C and 1.0 M NaCl. Between 1.2 and 2.0 M NaCl H3 and H4 bind cooperatively as an equimolar complex with delta Go = 7.4 Kcal/mole at 4 degree C and 1.0 M NaCl. The proper binding of H2a and H2b requires the presence of bound H3 and H4. Nuclease digestion of the H3-H4 DNA produces a tetramer of H3-H4 bound to fragments of DNA 145, 125 and 104 base pairs long. Thus an H3-H4 tetramer can protect fragments of DNA as long as those found in complete core particles and must therefore span the nucleosome core particle.
Full text
PDF
















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axel R. Cleavage of DNA in nuclei and chromatin with staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2921–2925. doi: 10.1021/bi00684a020. [DOI] [PubMed] [Google Scholar]
- Bina-Stein M., Simpson R. T. Specific folding and contraction of DNA by histones H3 and H4. Cell. 1977 Jul;11(3):609–618. doi: 10.1016/0092-8674(77)90078-2. [DOI] [PubMed] [Google Scholar]
- Burgen A. S., Roberts G. C., Feeney J. Binding of flexible ligands to macromolecules. Nature. 1975 Feb 27;253(5494):753–755. doi: 10.1038/253753a0. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Sollner-Webb B., Felsenfeld G. The organization of histones and DNA in chromatin: evidence for an arginine-rich histone kernel. Cell. 1976 Jul;8(3):333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- Elgin S. C., Weintraub H. Chromosomal proteins and chromatin structure. Annu Rev Biochem. 1975;44:725–774. doi: 10.1146/annurev.bi.44.070175.003453. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin. Nature. 1978 Jan 12;271(5641):115–122. doi: 10.1038/271115a0. [DOI] [PubMed] [Google Scholar]
- Henson P., Walker I. O. The partial dissociation of nucleohistone by salts. Circular dichroism and denaturation studies. Eur J Biochem. 1970 Nov;16(3):524–531. doi: 10.1111/j.1432-1033.1970.tb01112.x. [DOI] [PubMed] [Google Scholar]
- Hyde J. E., Walker I. O. Covalent cross-linking of histones in chromatin. FEBS Lett. 1975 Feb 1;50(2):150–154. doi: 10.1016/0014-5793(75)80477-7. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Howarth O. W., Clark V. W., Pardon J. F., Richards B. M. An investigation of the conformational and self-aggregational processes of histones using 1H and 13C nuclear magnetic resonance. Biochemistry. 1975 Oct 21;14(21):4590–4600. doi: 10.1021/bi00692a006. [DOI] [PubMed] [Google Scholar]
- Noll M., Thomas J. O., Kornberg R. D. Preparation of native chromatin and damage caused by shearing. Science. 1975 Mar 28;187(4182):1203–1206. doi: 10.1126/science.187.4182.1203. [DOI] [PubMed] [Google Scholar]
- Skidmore C., Walker I. O., Pardon J. F., Richards B. M. The structure of partially histone depleted nucleohistone. FEBS Lett. 1973 May 15;32(1):175–178. doi: 10.1016/0014-5793(73)80765-3. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Camerini-Otero R. D., Felsenfeld G. Chromatin structure as probed by nucleases and proteases: evidence for the central role of histones H3 and H4. Cell. 1976 Sep;9(1):179–193. doi: 10.1016/0092-8674(76)90063-5. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Butler P. J. Characterization of the octamer of histones free in solution. J Mol Biol. 1977 Nov;116(4):769–781. doi: 10.1016/0022-2836(77)90270-4. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Palter K., Van Lente F. Histones H2a, H2b, H3, and H4 form a tetrameric complex in solutions of high salt. Cell. 1975 Sep;6(1):85–110. doi: 10.1016/0092-8674(75)90077-x. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Worcel A., Alberts B. A model for chromatin based upon two symmetrically paired half-nucleosomes. Cell. 1976 Nov;9(3):409–417. doi: 10.1016/0092-8674(76)90085-4. [DOI] [PubMed] [Google Scholar]
- Wilhelm X., Champagne M. Dissociation de la nucléoprotéine d'érythrocytes de poulets par les sels. Eur J Biochem. 1969 Aug;10(1):102–109. [PubMed] [Google Scholar]
- Worcel A., Benyajati C. Higher order coiling of DNA in chromatin. Cell. 1977 Sep;12(1):83–100. doi: 10.1016/0092-8674(77)90187-8. [DOI] [PubMed] [Google Scholar]