Abstract
After imputation of data of the 1000 Genomes Project into a genome-wide data set of Ghanaian tuberculosis cases and controls, we identified a resistance locus on chromosome 11p13, downstream of the Wilms' tumour 1 gene. The strongest signal was obtained at SNP rs2057178 (P = 2.63 × 10−9). Replication in Gambian, Indonesian and Russian TB case-control study groups increased the significance level to P = 2.57 × 10−11.
The influence of host genetic factors on susceptibility to tuberculosis (TB) is well established by twin, linkage and candidate gene analyses1-3. Recently, in a combined genome-wide association study (GWAS) of African TB case-control groups from Ghana, The Gambia and Malawi we have identified a susceptibility locus on chromosome 18q11.24. Here, we present a novel association of a genetic locus on chromosome 11p13 with resistance to TB, obtained only after imputation of data provided by the 2010-08 release of the 1000 Genomes Project into the genome-wide Ghanaian data.
After genotyping of 1329 TB cases and 1847 controls (Affymetrix SNP Array 6.0) and quality control of SNPs, 793,964 variants were available for imputation analyses (Supplementary Note). Population stratification was low as indicated by a lambda factor (λ) of 1.03 (Supplementary Figure 1). Genotypes from the 1000 Genomes Project data set now offer African data from 78 Yoruban individuals (Nigeria), 67 Luhya in Webuye (Kenya), 24 individuals of African ancestry from Southwest USA and 5 Puerto Ricans. These genotypes were imputed into the Ghanaian dataset using the minimac software.
For association testing, the allelic dosages, which represent the expected number of copies of a distinct allele rather than the best-guess imputed genotypes of each SNP, were analysed in a logistic regression framework in order to account for imputation uncertainty. Adjustment for the population structure was performed with the mach2dat software by including the first three principal components derived from an Eigenstrat analysis of genotype data as covariates (Supplementary Note).
We chose imputed SNPs with minor allele frequencies (MAF) of > 1% and MACH RSQ values of at least 0.3 for further analyses. A MACH RSQ value is a post-imputation quality score not directly related to pairwise linkage disequilibrium measures between SNPs and indicates the correlation between true and estimated allele counts of imputed SNPs5. Applying these criteria, 10,921,004 genetic variants were successfully imputed. We selected 46 of these SNPs representing independent signals and yielding after imputation P values of < 1 × 10−5 and genotyped them using LightTyper assays in the same GWAS dataset.
Two of the 46 variants were mono-allelic in the Ghanaian study population. Eleven variants only provided after genotyping association signals of P < 1 × 10−5. The low degree of concordance between imputation with genotyping results may have several reasons. First, the data available for imputation consists of preformed and calculated haplotypes from the 1000 Genomes Project African subset under inclusion of individuals of defined ancestry. Optimal would be if the dataset would include the population currently studied. This was not the case here. Second, imputation has previously been shown to be less accurate in African populations6. Third, the imputation quality essentially depends on the size of the reference panel which is in our case, with 348 haplotypes of the African subset, suboptimal. Last, the lower the frequency of the occurrence of distinct SNPs, the lower the imputation precision.
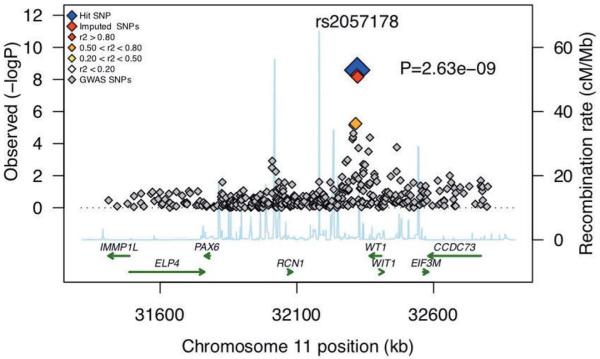
The eleven variants with P values < 1 × 10−5 were tested in a replication sample of 817 TB cases and 3805 controls, constituting a total of 7798 Ghanaian individuals (Supplementary Table 1). Variant rs2057178 on chromosome 11p13 yielded the strongest genome-wide significant association result (P value 2.63 × 10−9, odds ratio (OR) 0.77, 95% confidence interval [CI] 0.71-0.84) (Table 1). Further genotyping of variants at this locus with imputation P values of < 10−5 revealed two additional variants, rs11031728 and rs11031731, with genome-wide significant results as well (P = 5.25 × 10−9, OR = 0.77, 95% CI 0.71-0.84; P = 7.01 × 10−9, OR = 0.78, 95% CI 0.71-0.85) (Supplementary Table 2). These three variants are in strong linkage disequilibrium in the different ethnic groups represented in the HapMap Project (LD; all pairs r2 = 0.98) with each other (Supplementary Figure 2), thus making it virtually impossible to distinguish them with regard to their impact on the infection phenotype. It may be assumed that strong LD applies in other populations as well. Notably, variant rs11031728 is part of the conserved transcription factor binding site V$TCF11MAFG_01 (UCSC, HMR Conserved Transcription Factor Binding Site track, GRCh37 genome assembly). Variant rs2057178, which provided the strongest association signal (Figure 1) was investigated further after confirmation of ethnic homogeneity (Mantel-Haenszel statistic; Supplementary Table 3).
Table 1.
Meta-analysis of SNP rs2057178 in four TB case-control groups
| rs2057178 – A allele | Controls | Cases | OR (95% CI) | P value | ||
|---|---|---|---|---|---|---|
| N | Freq | N | Freq | |||
| Ghana | 5636 | 0.32 | 2127 | 0.27 | 0.77 (0.71-0.84) | 2.63E-09 |
| The Gambia | 1349 | 0.31 | 1207 | 0.27 | 0.80 (0.70-0.91) | 4.87E-04 |
| Indonesia | 983 | 0.11 | 1025 | 0.09 | 0.84 (0.68-1.03) | 0.099 |
| Russia | 5874 | 0.13 | 4441 | 0.12 | 0.91 (0.82-0.99) | 0.02 |
| Total | 13859 | 8821 | 2.57E-11 | |||
Figure 1.
Association plot of the chromosome 11 hit region with markers identified by imputation in the Ghanaian data set (red, orange and grey diamonds) and of the meta-analysis (blue diamond). Genes in the long range vicinity are given.
We genotyped SNP rs2057178 in additional TB case-control groups originating from The Gambia (1207 cases vs. 1349 controls; P = 4.9 × 10−4, OR 0.80, 95% CI 0.70-0.91), Indonesia (1025 cases vs. 983 controls; P = 9.9 × 10−2, OR 0.84, 95% CI 0.68-1.03) and Russia (4441 cases vs. 5874 controls; P = 2.0 × 10−2, OR 0.91, 95% CI 0.82-0.99). Results from the Ghanaian study group were corroborated in a meta-analysis including the results of SNP rs2057178 of the four study groups, with a combined P value of 2.57 × 10−11 (Table 1, Supplementary Figure 3, Supplementary Note). The consistent effect of rs2057178 in the study populations from West-Africa, Indonesia and Russia, that may have undergone different regional adaption and selection processes, suggests functional relevance of rs2057178 or strong linkage of rs2057178 to a causal variant yet to be identified.
Applying a fixed effect model in the Cochran Q test, the between-study heterogeneity was negligible when comparing the two African study groups (P = 0.67). A result close to significance (P = 0.062) was obtained when testing all four study groups, indicating a certain degree of inter-study heterogeneity between the two African and the two non-African groups.
SNP rs2057178, which showed the strongest association, and the other two SNPs in strong LD with it, rs11031728 and rs11031731, are located in an intergenic region 45 kb downstream of the Wilms' tumour 1 gene (WT1; MIM ID *607102). Whether the associated locus on chromosome 11 affects or even regulates WT1 expression is not clear at present. WT1 is a zinc-finger transcription factor and involved in the development of the urogenital system. Genetic variants of WT1 have been shown to be associated with the occurrence of the Wilms' tumour, but also with acute myeloid leukaemia and the Denys-Drash, Frazier and other syndromes7. WT1 also plays a role in the activation of the vitamin D receptor (VDR)8 and was, in a mouse model, found to suppress interleukin (IL)10 expression9. Both VDR and IL-10 have been claimed to be important in the pathophysiology of TB10 and genetic variation of the VDR and IL10 genes has been reported to be associated with TB susceptibility11.
Further genes telomeric of the association peak include RCN1, PAX6, ELP4 and IMMP1L and, centromerically from WT1 the genes WIT1, EIF3M and CCDC73. There is no evidence so far of an involvement of any of these genes in the phenotype arising after a M. tuberculosis infection.
More than 10 million common African variants (genotype frequencies of > 1%) have been reported in the 1000 Genomes Project and it is clear that the number of SNPs to be looked at in African groups has increased markedly. Enlargement of the number of SNPs by imputation analyses has proven useful in this study and led to the identification of a new TB locus. With the advent of affordable genome-wide sequencing technologies, more common, but also rare variants will be identified and hopefully unfold new strategies to tackle TB.
Supplementary Material
ACKNOWLEDGEMENTS
Ghana. The participation of patients and the volunteers who served as controls is gratefully acknowledged, also the contributions of field workers, nurses and physicians involved in the recruitment of participants, the staff of the Kumasi Centre for Collaborative Research in Tropical Medicine (KCCR) and the excellent assistance of Emmanuel Abbeyquaye and Lincoln Gankpala. This work was supported by the German Federal Ministry of Education and Research, Project TBornot TB (BMBF), German National Genome Research Network (NGFN1, grant number 01GS0162; NGFN2, grant number NIE-S17T20; NGFN-PLUS, grant number 01GS0811) and the BMBF Tuberculosis Research Network, grant number 01KI0780. Gambia. Sample collections were supported by MRC unit funding, European Commission framework programme awards, MRC award G0000690 (to GS) and Wellcome Trust fellowship support (to AVSH). Laboratory work in Oxford was supported by the Wellcome Trust. We thank other members of the Wellcome Trust Case Control Consortium and collaborators for previous work on the Gambian sample sets.
Indonesia. This study was supported by the Royal Netherlands Academy of Arts and Sciences (KNAW, grant number KNAW99MED01), the Netherlands Organization for Scientific Research/WOTRO (PRIOR-project) and the European Commission (grant number QLK2-CT-2003-503367). Written informed consent was obtained from all subjects, and the study was approved by the Ethical Committee of the Eijkman Institute of Molecular Biology, Jakarta, and of the Faculty of Medicine, Padjadjaran University, Hasan Sadikin Hospital, Bandung, Indonesia. We gratefully acknowledge Sangkot Marzuki (Eijkman Institute of Molecular Biology, Jakarta), Ron HH Nelwan (Faculty of Medicine, University of Indonesia, Jakarta) and Jos WM van der Meer (Department of Internal Medicine, Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands) for their continued support of the study. Russia. The participation of patients and volunteers as control individuals is gratefully acknowledged. During the course of this study SN was a Royal Society University Research Fellow and now holds the Wellcome Trust Senior Research Fellowship in Basic Biomedical Science. This study was supported by the European Union Framework Programme 7 grant 201483 (TB-EUROGEN), the Royal Society Research grant, the Wellcome Trust grant 088838/Z/09/Z and the ERC Starting grant 260477. We thank Olga Ignatyeva, Irina Kontsevaya, Svetlana Mironova, Ivan Fedorin and Nadezhda Malomanova for the recruitment of patients and controls, as well as Emma Stebbings, Liliya Kopanitsa and Arran Speirs for DNA preparation.
Footnotes
URLs. 1000 Genomes Project: http://www.1000genomes.org/;
MACH software: http://www.sph.umich.edu/csg/abecasis/MACH/download/1000G-2010-08.html;
minimac software: http://genome.sph.umich.edu/wiki/Minimac
mach2dat software: http://www.sph.umich.edu/csg/abecasis/MACH/download/mach2dat.tar.gz).
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Möller M, Hoal EG. Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis. Tuberculosis (Edinb) 2010;90:71–83. doi: 10.1016/j.tube.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Vannberg FO, Chapman SJ, Hill AV. Human genetic susceptibility to intracellular pathogens. Immunol Rev. 2011;240:105–116. doi: 10.1111/j.1600-065X.2010.00996.x. [DOI] [PubMed] [Google Scholar]
- 3.Intemann CD, et al. Autophagy gene variant IRGM -261T contributes to protection from tuberculosis caused by Mycobacterium tuberculosis but not by M. africanum strains. PLoS Pathog. 2009;5:e1000577. doi: 10.1371/journal.ppat.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thye T, et al. Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat Genet. 2010;42:739–741. doi: 10.1038/ng.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang L, Li Y, Singleton AB, Hardy JA, Abecasis G, Rosenberg NA, Scheet P. Genotype-imputation accuracy across worldwide human populations. Am J Hum Genet. 2009;84:235–250. doi: 10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huff V. Wilms' tumours: about tumour suppressor genes, an oncogene and a chameleon gene. Nat Rev Cancer. 2011;11:111–21. doi: 10.1038/nrc3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maurer U, et al. The Wilms' tumor gene product (WT1) modulates the response to 1,25-dihydroxyvitamin D3 by induction of the vitamin D receptor. J Biol Chem. 2001;276:3727–32. doi: 10.1074/jbc.M005292200. [DOI] [PubMed] [Google Scholar]
- 9.Sciesielski LK, Kirschner KM, Scholz H, Persson AB. Wilms' tumor protein Wt1 regulates the Interleukin-10 (IL-10) gene. FEBS Lett. 2010;584:4665–71. doi: 10.1016/j.febslet.2010.10.045. [DOI] [PubMed] [Google Scholar]
- 10.Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 11.Ottenhoff TH, Verreck FA, Hoeve MA, van de Vosse E. Control of human host immunity to mycobacteria. Tuberculosis. 2005;85:53–64. doi: 10.1016/j.tube.2004.09.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.