Summary
Caveolin proteins drive formation of caveolae, specialized cell-surface microdomains that influence cell signaling. Signaling proteins are proposed to use conserved caveolin-binding motifs (CBMs) to associate with caveolae via the caveolin scaffolding domain (CSD). However, structural and bioinformatic analyses argue against such direct physical interactions: In the majority of signaling proteins, the CBM is buried and inaccessible. Putative CBMs do not form a common structure for caveolin recognition, are not enriched amongst caveolin-binding proteins, and are even more common in yeast, which lack caveolae. We propose that CBM/CSD-dependent interactions are unlikely to mediate caveolar signaling, and the basis for signaling effects should therefore be reassessed.
Introduction
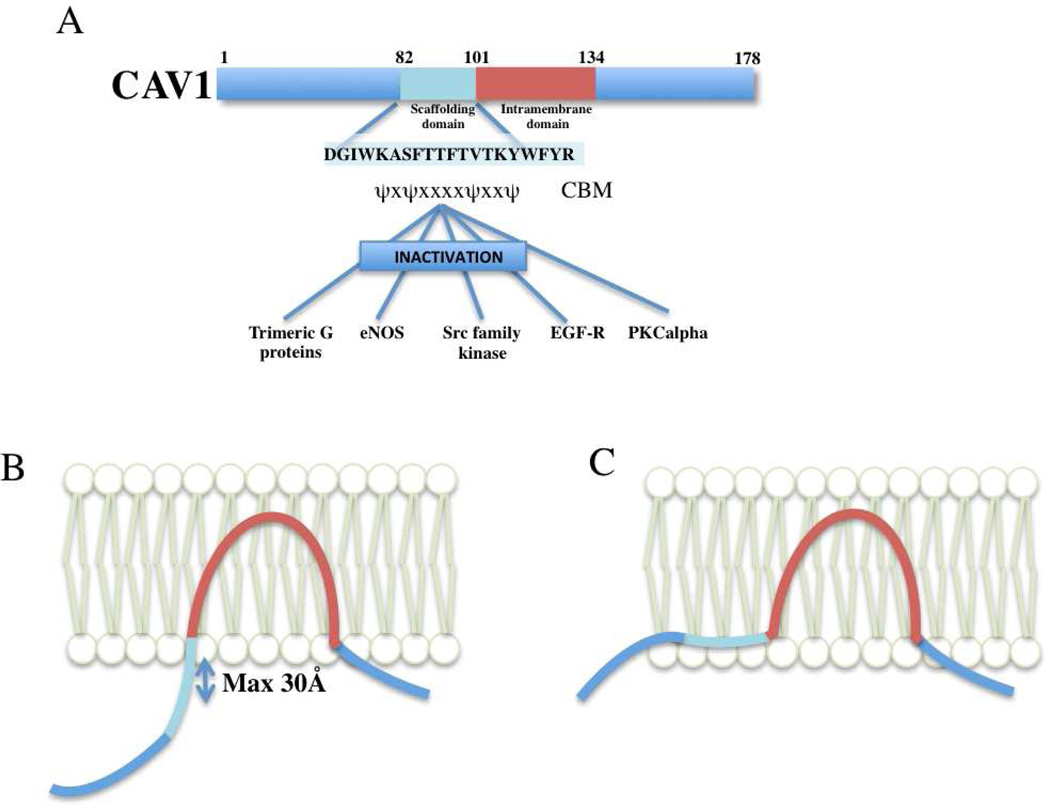
The caveolin signaling hypothesis is an enduring model for understanding spatial organization of signaling at the plasma membrane (Couet et al., 1997; Lisanti et al., 1995; Okamoto et al., 1998). The central tenet of the model is that signaling proteins can form direct protein-protein interactions with the scaffolding domain of caveolin (CSD) via a signature peptide sequence, termed the caveolin binding motif (CBM) (Couet et al., 1997; Oka et al., 1997) (Fig. 1). The characteristic CBMs were originally identified by screening of a phage display peptide library (Couet et al., 1997), and subsequently found to be present in many diverse proteins that could be immunopurified with caveolin. These consensus CBMs are hydrophobic and rich in aromatic residues (ΩxΩxxxxΩor ΩxxxxΩxxΩor the combined sequence ΩxΩxxxxΩxxΩ, where Ω is a Phe, Tyr or Trp residue and x can be any amino acid) (Table 1; Fig. S1). The caveolin interaction is generally suggested to have an inhibitory role on signaling. Thus, signaling proteins associated with the cytoplasmic face of caveolae were proposed to be held in an inactive state by the caveolin ‘brake’, prior to release from caveolae upon activation (Okamoto et al., 1998).
Figure 1.
The caveolin signalling hypothesis. (A) Schematic of the caveolin signalling hypothesis as originally proposed (Okamoto et al., 1998), with some key interacting partners highlighted. The sequence of the caveolin-1 scaffolding domain (CSD) and the consensus caveolin-binding motif (CBM) are shown. (B) and (C); Two models for caveolin association with the membrane bilayer. In model (B) the CSD is exposed and shown in an extended conformation allowing interactions with signaling proteins. However, note that the middle of the CSD is still very close to the membrane, even assuming a completely extended polypeptide conformation perpendicular to the bilayer. Model (C), in which the CSD forms part of an amphipathic cholesterol-binding in-plane helix, is an alternative model supported by a number of studies (Kirkham et al., 2008).
Table 1.
Putative Caveolin Binding Motifs (CBMs) identified in previous studiesa
| Protein | Proposed CBM(s) | Referenceb | Protein for structural analysis |
PDB ID |
Structure Reference |
Secondary structure |
Accessible surface area (%)c |
|
|---|---|---|---|---|---|---|---|---|
| Consensus CBM | ․ΩxΩxxxxΩxxΩ‥ | (Couet et al., 1997) | ||||||
| Soluble Proteins | ||||||||
| β-catenin | YTYEKLLW | (Mo et al., 2010)* | β-catenin | 2Z6G | (Xing et al., 2008) | α-helix | 36 | |
| ApoE | WELALGRFWDYLRW31 | (Yue and Mazzone, 2011) | ApoE | 1YA9 | (Hatters et al., 2005) | α-helix | 32 | |
| BTK | WAFGVLMWEIY591 | (Vargas et al., 2002) | BTK | 1K2P | (Mao et al., 2001) | α-helix | 1 | |
| Dystrophin | FHYDIKIFNQW | (Couet et al., 1997) | None | - | - | - | - | |
| eNOS | FPAAPFSGW356 | (Couet et al., 1997; Garcia-Cardena et al., 1997; Sato et al., 2004)* | eNOS | 1M9K | (Rosenfeld et al., 2002) | β-strand | 11 | |
| Gα subunits | FTFKDLHFKMF199 | (Couet et al., 1997; Li et al., 1995) | Gαi1 | 1CIP | (Coleman and Sprang, 1999) | β-hairpin | 32 | |
| Gαq | 3AH8 | (Nishimura et al., 2010) | β-hairpin | 44 | ||||
| GFPd | FAYGVQCFSRY | This study | GFP | 3OGO | (Kubala et al., 2010) | Extended coil | ||
| Heme oxygenase-1 | FLLNIELF214 | (Taira et al., 2011) | Heme oxygenase-1 | 1DVE | (Sugishima et al., 2000) | α-helix | 18 | |
| LC3B | FLYMVYASQETF119 | (Chen et al., 2010) | LC3B | 2ZJD | (Ichimura et al., 2008) | β-strand | 13 | |
| MAP kinase | YIVGFYGAF133 | (Couet et al., 1997) | MEK1 | 3PP1 | (Dong et al., 2011) | β-strand | 21 | |
| Myosin HC | WPWMKLYF836 | (Couet et al., 1997) | Myosin HC | 2MYS | (Rayment et al., 1993) | α-helix | 51 | |
| NSF | FSFNEKLF145 | (Couet et al., 1997) | NSF | 1QCS | (Yu et al., 1999) | β-hairpin | 38 | |
| Nuclear erythroid 2 p45-related factor2 |
FGDEFYSAF289 | (Li et al., 2012) | - | - | - | - | - | |
| PKCα | FSYVNPQF663 | (Oka et al., 1997) | PKCα | 3IW4 | (Wagner et al., 2009) | β-strand | 32 | |
| PPARγe | FGDFMEPKFEF370 | (Burgermeister et al., 2011)* | PPARγ | 3ETO | (Artis et al., 2009) | α-helix | 23 | |
| PTEN | FHFWVNTF278 | (Caselli et al., 2002; Xia et al., 2010)* | PTEN | 1D5R | (Lee et al., 1999) | β-strand | 7 | |
| PTP1B | FHYTTWPDF182 | (Caselli et al., 2002) | PTP1B | 2CM2 | (Ala et al., 2006) | β-strand | 28 | |
| PTP1C (SHP-1) | FVYLRQPY213 | (Caselli et al., 2002) | SHP-1 | 3PS5 | (Wang et al., 2011) | β-strand | 32 | |
| SH-PTP2 | WQYHFRYW423 | (Caselli et al., 2002) | SH-PTP2 | 3B7O | (Barr et al., 2009) | β-strand | 15 | |
| Src family kinases | WSFGILLY430 | (Couet et al., 1997) | Abl | 2G2I | (Levinson et al., 2006) | α-helix | 3 | |
| Thioredoxin reductase 1 | YHSYFWPLEW411 | (Volonte and Galbiati, 2009)* | Thioredoxin reductase 1 | 3QFA | (Fritz-Wolf et al., 2011) | β-strand | 15 | |
| Transmembrane Proteins | ||||||||
| ALK1 | WAFGLVLW406 | (Santibanez et al., 2008) | ALK1 | 3MY0 | - | α-helix | 1.8 | |
| Adiponectin Receptor R1 |
FVPWLYYSF FFPGKFDIW |
(1) (2) |
(Wang et al., 2012)* | None | - | - | - | - |
| Angiotensin | YGFLGKKFKRY | (Wyse et al., 2003) | β1AR | 2YO1 | (Warne et al., 2011) | α-helix | - | |
| Aquaporin | WIFWVGPF219 | (Couet et al., 1997) | AQP1 | 1J4N | (Sui et al., 2001) | α-helix | 22f | |
| Caveolin | FTVTKYWFY | (Couet et al., 1997) | None | - | - | - | - | |
| D1 Dopamine | FDVFVWFGW | (Kong et al., 2007)* | None | - | - | - | - | |
| Desmogleins | FCQKAYAY | (Brennan et al., 2011) | None | - | - | - | - | |
| Endothelin R | WPFDHNDFGVF | (Couet et al., 1997) | None | - | - | - | - | |
| Receptor tyrosine kinases | WSYGVTVW881 | (Couet et al., 1997; Nystrom et al., 1999; Vihanto et al., 2006)* | EGFR Ephrin A3 Insulin R |
3LZB 2QOB 1IRK |
(Fidanze et al., 2010) (Davis et al., 2008) (Hubbard et al., 1994) |
α-helix α-helix |
2 1 |
|
| IP3R3 | WKINLFMQF226 | (Sundivakkam et al., 2009) | IP3R1 | 1XZZ | (Bosanac et al., 2005) | β-strand | 32 | |
| mAcR | WTIGYWLCY | (Couet et al., 1997) | None | - | - | - | - | |
| Maxi-K channel α subunit | YNMLCFGIY1007 | (Alioua et al., 2008; Brainard et al., 2009)* | Maxi-K cytoplasmic domain | 3MT5 | (Yuan et al., 2010) | β-strand | 10 | |
| mGluR1α | FVTLIFVLY FNEAKYIAF |
(1) (2) |
(Hong et al., 2009)* | None | - | - | - | - |
| MuSK | WAYGVVLWEIF795 FSYGLPQY |
(Hezel et al., 2010) | MuSK | 1LUF | (Till et al., 2002) | α-helix | 3 | |
| Na/K ATPaseg | FCRQLFGGF93 WWFCAFPY987 |
(1) (2) |
(Cai et al., 2008) | Na/K ATPase |
3B8E | (Morth et al., 2007) | α-helix / extended coil |
37f (1) 25f (2) |
| Neu3 sialidase | YTYYIPSW | (Wang et al., 2002) | None | - | - | - | - | |
| nAChR α subunit | FSFLTGLVFY234 | (Hezel et al., 2010) | nAchR | 2BG9 | (Unwin, 2005) | α-helix | 19f | |
| P-glycoprotein | FSMFRYSNW44 | (Jodoin et al., 2003)* | P-glycoprotein | 3G61 | (Aller et al., 2009) | α-helix / extended coil | 42f | |
| Patched | YDFIAAQFKYF | (Karpen et al., 2001)* | None | - | - | - | - | |
| TLR4 | FIQSRWCIF715 | (Wang et al., 2009) | TLR2 | 1FYW | (Xu et al., 2000) | α-helix / extended coil | 29 | |
| TRPC1 | FRTSKYAMF | (Sundivakkam et al., 2009) | None | - | - | - | - | |
| β1 adrenergic receptor | FVFFNWLGY333 | (Couet et al., 1997) | β1AR | 2YO1 | (Warne et al., 2011) | α-helix | 31f | |
| Viral and other pathogen proteins | ||||||||
| Cholera toxin subunit A | YGWYRVHF132 | (Couet et al., 1997) | Cholera toxin subunit A | 1S5E | (O'Neal et al., 2004) | β-strand | 28 | |
| gp41 | WNNMTWMQW115 | (Hovanessian et al., 2004; Huang et al., 2007) | gp41 | 1QBZ | (Yang et al., 1999) | α-helix | 43 | |
| M2 channel | FFKCIYRRF54 | (Zou et al., 2009) | M2 channel | 2RLF | (Schnell and Chou, 2008) | α-helix / extended coil | 64f | |
| Matrix (M) protein | FGKSNWGLF | (Ravid et al., 2010) | None | - | - | - | - | |
| Matrix protein | FCSAEWPTF45 | (Yu et al., 2006) | Murine Leukemia Virus matrix protein | 1MN8 | (Riffel et al., 2002) | α-helix | 29 | |
| α-hemolysin | WGPYDRDSW187 | (Pany et al., 2004) | α-hemolysin | 3ANZ | (Tanaka et al., 2011) | Extended coil | 41f | |
The CBMs are derived from published studies that show association between caveolin and the identified proteins, and where direct reference is made to the caveolin binding motif (CBM) identified by phage display in Couet et al., (Couet et al., 1997). There were two motifs identified ΩxΩxxxxΩ and ΩxxxxΩxxΩ where Ω is Tyr, Phe or Trp. Couet et al., further proposed a combined CBM of ΩxΩxxxxΩxxΩ.
Asterisks (*) indicate that mutagenesis of the CBM was carried out.
The accessible surface area is expressed as an average percentage of the total possible surface area of each side-chain within the CBM sequence. Areas were calculated using the program NACCESS (http://www.bioinf.manchester.ac.uk/naccess/).
Note that the GFP sequence encompasses the cyclised Tyr that forms the chromophore.
Note the sequence in PPARα (and also PPARα,β,δ) is actually in the reverse sequence orientation to the consensus CBM.
For many transmembrane proteins “accessible surface area of CBMs” will not be an accurate indication of solvent exposure, as the sequences lie within membrane spanning regions or extracellular domains (Fig. S2).
Note that both sequences fall within the transmembrane domain.
Numerous signaling proteins have been proposed to interact with caveolin, including cytoplasmic proteins (src family kinases, trimeric G protein subunits, Ras, PPARγ, β-catenin), and single and multispan transmembrane proteins (Patched, β-adrenergic receptors (β-ARs), adiponectin receptors) (Burgermeister et al., 2011; Couet et al., 1997; Hezel et al., 2010; Ju et al., 1997; Karpen et al., 2001; Li et al., 1996; Michel et al., 1997; Mineo et al., 1997; Mineo et al., 1998; Song et al., 1996; Song et al., 1997; Toya et al., 1998; Venema et al., 1997) (Table 1; Fig. S1). The hypothesis has been extended to caveolin interactions with non-signaling proteins, including extracellular viral proteins (Benferhat et al., 2009a; Benferhat et al., 2009b; Benferhat et al., 2008; Hovanessian et al., 2004) and key autophagic regulators such as LC3 (Chen et al., 2010), and has become a paradigm for spatial regulation of signaling pathways.
Despite the elegance of the model and the wealth of literature supporting it, including indirect experimental data showing association of specific proteins with caveolin or inhibition by CSD mimetic peptides (eg. (Bucci et al., 2000)), some questions have been raised (Liu et al., 2002; Pike, 2005) and a number of crucial aspects of the model have never been systematically or rigorously addressed. For example, do the putative CBMs adopt a common structure as would be predicted by the model? Are CBMs accessible for interaction with caveolin and positioned in such a way with respect to the caveolin-containing membrane that an interaction is feasible? How common are such motifs, and are they enriched in caveolae-associated proteins? Surprisingly, a plausible molecular mechanism for the interaction of CBMs with caveolin is yet to emerge. The wealth of genomic sequence and tertiary structural information available on putative caveolin interacting proteins now means that these questions can be definitively answered. As outlined below, the answers to these questions raise major doubts about some of the founding principles on which the caveolar signaling model is based, leading us to propose that a significant reassessment of the caveolin signaling hypothesis may be needed.
Structures of putative caveolin binding proteins do not reveal a plausible caveolin binding mechanism
The putative CBM is a short, hydrophobic sequence of 8–11 amino acid residues (Table 1; Fig. S1). Two physical requirements must be met if it is to function as a bona fide caveolin interaction motif. The first requirement is that a functional CBM must either lie in a disordered region of the interacting protein (becoming ordered upon caveolin interaction), or it must form a common recognition structure for caveolin binding. The second requirement for a role of caveolin in sequestering proteins into caveolae is that the putative CBM should be exposed in the folded protein structure and accessible to the CSD.
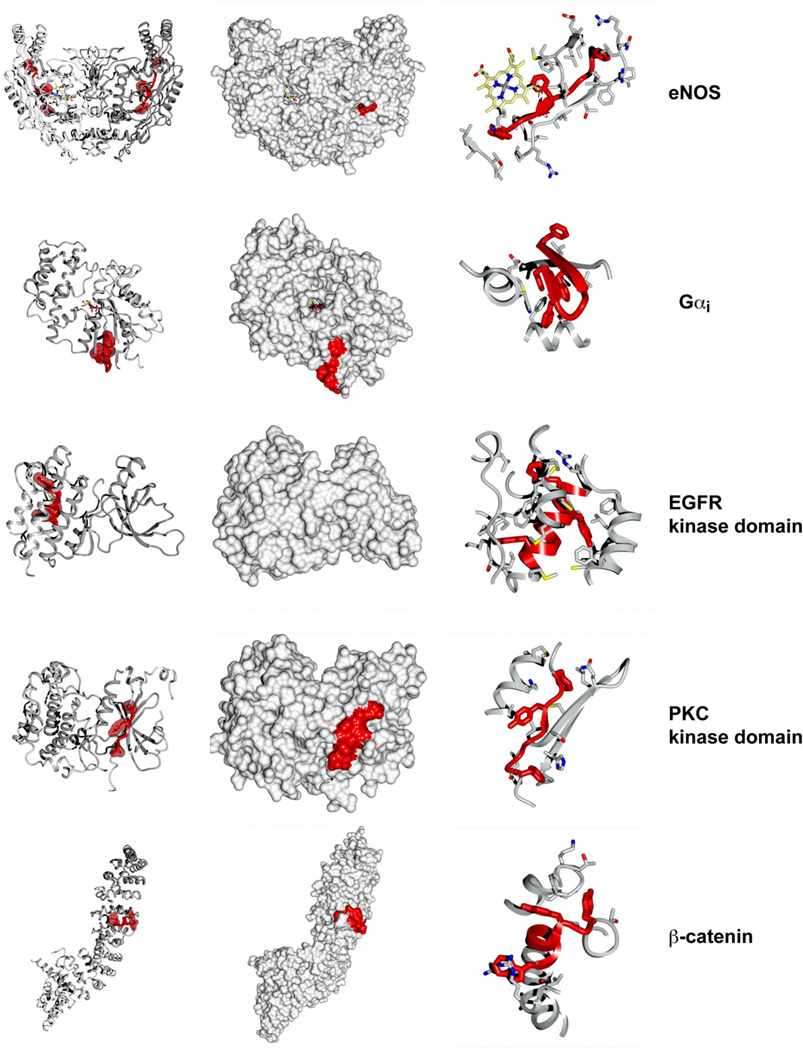
We analyzed the structures of more than 40 proteins for which caveolin interactions with specific CBMs have been described (Table 1; Fig. S2). Some specific examples are shown in greater detail in Fig. 2 and Movie S1. This clearly reveals that no single common structural motif is adopted by the putative caveolin-interacting sequences. The CBM adopts a variety of different structures within the putative caveolin binding proteins including extended structures, α-helices, β-strands and β-turns, and no consistent conformation for this peptide is observed. Even within individual protein families, including tyrosine kinases, GPCRs and protein tyrosine phosphatases, the motif adopts diverse structural orientations. For example in the EGFR and protein kinase C (PKC) kinase domains the putative CBMs are found in distinct sub-structures, forming either a central α-helix within the C-terminal lobe or a peripheral β-strand on the edge of the N-terminal lobe respectively (Fig. 2). The other major observation is that these motifs are invariably found within structured regions of the proteins, often forming essential secondary structure elements. This is in distinct contrast, for example, with the recognition of multiple sorting signals and sequence motifs during formation of the analogous clathrin coated vesicle (CCV) assembly (Owen et al., 2004; Traub, 2009). In CCVs peptide interaction motifs are always found in structurally disordered domains, and only adopt an ordered conformation upon interaction with folded domains within their partner molecule(s).
Figure 2.
Structural comparison of several examples of putative caveolin-interacting proteins. An enhanced animation of the eNOS structure is provided in Movie S1. Left panels show proteins in ribbon representation, with the CBM indicated in red. Key aromatic residues of the putative CBMs are highlighted in surface representation. In each case the key aromatic residues are tightly packed within the protein hydrophobic core. Middle panels show the same views in surface representation, with CBMs indicated in red. The right hand panel shows a close up view of the CBM and the surrounding environment. Key aromatic residues of the CBMs are shown in red stick representation, and side-chains forming direct intra-molecular contacts with these aromatic CBM residues indicated in grey stick representation. For eNOS, the core heme group is shown in yellow stick representation. All known structures of putative caveolin-interacting proteins are shown in Fig. S2, with references in Table 1. All structure images were prepared using CCP4mg (McNicholas et al., 2011).
The second crucial requirement of the model, the accessibility of the CBM to interacting proteins, is also illuminated by examination of the 3D structures. As discussed above, no common structural motif is observed for the numerous putative CBM sequences. Even more tellingly, in the large majority of cases these sequences are completely inaccessible for interaction with caveolin. Fig. 2 shows several different examples, where the CBM is not only inaccessible, but forms an essential part of the protein tertiary structure. Table 1 examines the solvent accessibility of putative CBMs based on the known crystal structures. The CBM sequence is hydrophobic and rich in aromatic side chains, and we find that in all cases the aromatic side-chains are packed within the hydrophobic core of the putative caveolin binding proteins. Focusing on just one of these, endothelial nitric oxide synthase (eNOS), for which there are numerous reports of caveolin scaffolding domain interactions (Bernatchez et al., 2011; Bucci et al., 2000; Feron et al., 1998; Garcia-Cardena et al., 1997; Hatakeyama et al., 2006; Levin et al., 2007; Zhu et al., 2004), the motif forms a key β-strand element within the hydrophobic interior of the protein. The aromatic side-chains are tightly packed in the protein core, and even more strikingly, directly contact the critical heme group within the protein’s active site (Fig. 2). It is extremely unlikely that this sequence could bind to caveolin without dramatic and detrimental conformational changes occurring. Similar observations can be made for the majority of other proteins for which structural data is available.
Could conformational changes facilitate caveolin binding?
One possibility we considered was that the CBMs could become accessible upon conformational changes in the target proteins. This also appears unlikely in view of the critical structural roles of the majority of these peptides. Invoking the hypothesis that a conformational change could lead to binding may be a reasonable explanation perhaps for a single or small number of binding events (although notably there is currently no data to support such a model). However, given the large range of different proteins from diverse structural and functional classes we have examined here, conformational change in the signaling molecules appears highly implausible as a universal explanation. Could the proteins interact with caveolin after synthesis, but before adopting a fully folded structure? We cannot rule out this possibility, but it would almost certainly give rise to a non-functional stable association with caveolin that would not be subject to the dynamic regulation required during cell signaling.
Caveolin binding motifs are not enriched in caveolae-associated proteins
This analysis raises the question of why so many proteins, particularly signaling proteins, which have been proposed to interact with caveolin possess CBMs? In fact, a systematic bioinformatics analysis of full-length coding sequences from the entire mouse genome (Carninci et al., 2005) reveals that this motif is actually present in 30% of all proteins, irrespective of localization or function (Table 2). The motif is not enriched (and is in fact less abundant) in cytoplasmic proteins and the cytoplasmic regions of transmembrane proteins that might conceivably bind caveolin at the inner leaflet of the plasma membrane. Perhaps most tellingly, the motifs show even greater prevalence in the genome of Saccharomyces cerevisae, which lacks caveolins altogether. Thus it is clear that CBM sequences are not enriched in caveolae-associated molecules, and their widespread abundance likely reflects a common requirement for hydrophobic aromatic side-chains in protein hydrophobic cores or transmembrane segments for structural stability and function.
Table 2.
Bioinformatic analysis of the abundance of consensus CBMs in mouse and yeast proteinsa
| Number of proteins |
Number containing CBM |
Overall percentage (%) |
|
|---|---|---|---|
| Mouse | |||
| Full length proteinsb | 33451 | 10076 | 30 |
| Cytoplasmic sequencesc | |||
| Soluble | 22265 | 5936 | 27 |
| Type I transmembrane | 1548 | 201 | 13 |
| Type II transmembrane | 2869 | 335 | 12 |
| Multi-pass transmembrane | 3821 | 739 | 19 |
| Total | 30503 | 7211 | 24 |
| Non-cytoplasmic sequencesc | |||
| Soluble | 2948 | 996 | 34 |
| Type I transmembrane | 1548 | 488 | 32 |
| Type II transmembrane | 2869 | 608 | 21 |
| Multi-pass transmembrane | 3821 | 773 | 20 |
| Total | 11186 | 2865 | 26 |
| Yeast | |||
| All proteins | 6736 | 2883 | 43 |
Sequences derived from CYGD database http://mips.helmholtz-muenchen.de/genre/proj/yeast/ were scanned for the presence of any of the two putative CBM sequences; ΩxΩxxxxΩ and ΩxxxxΩxxΩ or the combined consensus sequence ΩxΩxxxxΩxxΩ (Couet et al., 1997), where Ω is either Phe, Trp or Tyr.
The full set of 51135 coding sequences was reviewed, and those with annotated truncations at the N-terminus were discarded: topology with respect to the membrane cannot be accurately determined in this set.
Topology with respect to the membrane was calculated based on the presence in sequences of signal peptides and integral membrane domains using a previously published annotation pipeline (Davis et al., 2006).
In summary, it is clear from the available structural and genomic data that the proposed ΩxΩxxxxΩxxΩ CBM sequences are unlikely to represent a conserved peptide motif for direct recognition of the caveolin scaffolding domain. Another factor to consider when assessing the viability of the proposed caveolin interaction is the position of the putative CBM in the protein with respect to the membrane in which caveolin is embedded. An analogous example is the recognition of tyrosine-containing motifs by clathrin adaptors, which must be further than 7 amino acids from the membrane interface to engage with cytoplasmic proteins (Rohrer et al., 1996). This immediately raises an additional point regarding interactions with caveolin; as the maximum distance of the central-most portion of the CSD from the membrane – assuming a completely and unrealistically extended structure – is only 30 Å, corresponding to 10 amino acids (Fig. 1). This will impose severe steric constraints on any interactions with putative binding partners, which have been reported to be cytoplasmic proteins, cytoplasmic domains of transmembrane proteins, or even extracellular membrane penetrating polypeptides (eg. gp41, (Hovanessian et al., 2004)).
Implications for the caveolin signaling model
Mutations in caveolins or caveolin deficiency can clearly influence many signaling pathways as shown both in vitro and in vivo, and there is no doubting the role of caveolins in numerous cellular functions. The signaling proteins listed in Table 1 as well as many other molecules can be immunopurified in caveolin-enriched membrane fractions. However, experiments in which signaling proteins associate with caveolin as judged by immunoprecipitation must be viewed with caution given the poor solubility of caveolin-enriched domains (as discussed by (Parton and Simons, 2007)), and do not necessarily indicate a direct protein-protein interaction. A number of studies have assessed the effect of either deleting or mutating the CBM on caveolin association and in signaling assays (see Table 1), and have generally shown a disruption in caveolin interaction and function. However, the loss of an apparent interaction through mutation of the proposed CBM will be highly misleading if protein folding, trafficking, or microdomain localization are disrupted, as seems highly likely given the critical structural roles of the majority of CBM sequences. Very few reports have addressed the localization or expression of mutant signaling proteins. The mutant zebrafish β-catenin protein was found to at least localize to the nucleus similarly to the wild-type molecule (Mo et al., 2010), and the mutant Maxi-K potassium channel α subunit (Slo1) showed similar sedimentation and oligomeric properties to the WT protein in sucrose gradients (Alioua et al., 2008). In contrast the mutant EphB1 receptor tyrosine kinase was expressed at lower levels than the WT protein and was not localized to the plasma membrane (Vihanto et al., 2006). Structural integrity and correct protein folding has not been tested for any of the mutant proteins to the best of our knowledge, and should certainly be a priority in future studies.
The inhibition of signaling processes by cell permeable peptides corresponding to the caveolin scaffolding domain (CSD; amino acids 82–101 in caveolin-1) represent an additional line of evidence supporting the original caveolin scaffolding hypothesis. These studies have demonstrated a striking effect of this peptide on key signaling pathways involving proteins such as eNOS, phospholipase D (PLD), and Rac1 both in cultured cells and in tissues (Bernatchez et al., 2005; Czarny et al., 1999; Gratton et al., 2003; Kim et al., 1999; Nethe et al., 2010). In animal models administration of the caveolin-derived peptide reduced the permeability of the tumor vasculature and delayed tumor progression, an effect that was reduced in mice lacking the putative target, eNOS (Gratton et al., 2003). Conversely, a non-inhibitory version of this peptide with a single amino acid change increases basal NO release, an effect lost in tissues lacking eNOS or Caveolin-1 (Bernatchez et al., 2011). However, only a limited number of studies have attempted to directly test binding of the CSD peptide to signaling proteins, and in these cases binding was not investigated in the context of an interaction with putative CBMs (Kim et al., 1999; Nethe et al., 2010). The analyses presented here should prompt reinvestigation of the mechanisms involved in inhibition of signaling by these peptides and, more generally, the effect of loss of caveolin and/or caveolae on specific signaling pathways. Our findings certainly do not preclude the regulation of signaling pathways by caveolins through other mechanisms. These may include interactions mediated by other regions of caveolin (such as the interaction with phosphorylated caveolin-1 on tyrosine 14, (Chen et al., 2012; Place et al., 2011)), or by completely independent mechanisms including effects on lipid-based organization of the plasma membrane (Gaus et al., 2006; Hoffmann et al., 2010) or endocytosis (Cheng et al., 2010; Kirkham et al., 2005). These effects are also abrogated by mutations in the caveolin scaffolding domain (Cheng et al., 2010; Hoffmann et al., 2010).
Taken together, the findings presented here argue against a role for caveolin binding motifs in driving direct protein recruitment to caveolae. The putative CBM sequence is not enriched in proteins associated with caveolae, the motif does not adopt a common binding structure and is not exposed for caveolin binding. In most cases the CBM is part of a critical structural element, the perturbation of which is likely to lead to protein misfolding. We suggest that these considerations must be taken into account in future studies of caveolin interactions. In addition, previous work implicating caveolin as a scaffold for direct protein recruitment may need to be reassessed to reveal the actual mechanisms by which caveolins modulate specific signaling pathways.
Supplementary Material
Acknowledgements
We wish to thank Paul Curmi, Rohan Teasdale, and members of the Parton group for very helpful discussions. This work was supported by Discovery Project Grants from the Australian Research Council (ARC) to BMC and RGP (DP120101298), and MJD (DP110103384), and a Program Grant from the National Health and Medical Research Council (NHMRC) to RGP (APP1037320). RGP is supported by an NHMRC Australia Fellowship (569542). BMC is supported by an ARC Future Fellowship (FT100100027). JFH is supported by the National Institutes of Health (NIH) (GM066717).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ala PJ, Gonneville L, Hillman MC, Becker-Pasha M, Wei M, Reid BG, Klabe R, Yue EW, Wayland B, Douty B, et al. Structural basis for inhibition of protein-tyrosine phosphatase 1B by isothiazolidinone heterocyclic phosphonate mimetics. The Journal of biological chemistry. 2006;281:32784–32795. doi: 10.1074/jbc.M606873200. [DOI] [PubMed] [Google Scholar]
- Alioua A, Lu R, Kumar Y, Eghbali M, Kundu P, Toro L, Stefani E. Slo1 caveolin-binding motif, a mechanism of caveolin-1-Slo1 interaction regulating Slo1 surface expression. The Journal of biological chemistry. 2008;283:4808–4817. doi: 10.1074/jbc.M709802200. [DOI] [PubMed] [Google Scholar]
- Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, et al. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artis DR, Lin JJ, Zhang C, Wang W, Mehra U, Perreault M, Erbe D, Krupka HI, England BP, Arnold J, et al. Scaffold-based discovery of indeglitazar, a PPAR pan-active anti-diabetic agent. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:262–267. doi: 10.1073/pnas.0811325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr AJ, Ugochukwu E, Lee WH, King ON, Filippakopoulos P, Alfano I, Savitsky P, Burgess-Brown NA, Muller S, Knapp S. Large-scale structural analysis of the classical human protein tyrosine phosphatome. Cell. 2009;136:352–363. doi: 10.1016/j.cell.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benferhat R, Krust B, Rey-Cuille MA, Hovanessian AG. The caveolin-1 binding domain of HIV-1 glycoprotein gp41 (CBD1) contains several overlapping neutralizing epitopes. Vaccine. 2009a;27:3620–3630. doi: 10.1016/j.vaccine.2009.03.057. [DOI] [PubMed] [Google Scholar]
- Benferhat R, Martinon F, Krust B, Le Grand R, Hovanessian AG. The CBD1 peptide corresponding to the caveolin-1 binding domain of HIV-1 glycoprotein gp41 elicits neutralizing antibodies in cynomolgus macaques when administered with the tetanus T helper epitope. Molecular immunology. 2009b;46:705–712. doi: 10.1016/j.molimm.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Benferhat R, Sanchez-Martinez S, Nieva JL, Briand JP, Hovanessian AG. The immunogenic CBD1 peptide corresponding to the caveolin-1 binding domain in HIV-1 envelope gp41 has the capacity to penetrate the cell membrane and bind caveolin-1. Molecular immunology. 2008;45:1963–1975. doi: 10.1016/j.molimm.2007.10.036. [DOI] [PubMed] [Google Scholar]
- Bernatchez P, Sharma A, Bauer PM, Marin E, Sessa WC. A noninhibitory mutant of the caveolin-1 scaffolding domain enhances eNOS-derived NO synthesis and vasodilation in mice. The Journal of clinical investigation. 2011;121:3747–3755. doi: 10.1172/JCI44778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatchez PN, Bauer PM, Yu J, Prendergast JS, He P, Sessa WC. Dissecting the molecular control of endothelial NO synthase by caveolin-1 using cell-permeable peptides. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:761–766. doi: 10.1073/pnas.0407224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosanac I, Yamazaki H, Matsu-Ura T, Michikawa T, Mikoshiba K, Ikura M. Crystal structure of the ligand binding suppressor domain of type 1 inositol 1,4,5-trisphosphate receptor. Mol Cell. 2005;17:193–203. doi: 10.1016/j.molcel.2004.11.047. [DOI] [PubMed] [Google Scholar]
- Brainard AM, Korovkina VP, England SK. Disruption of the maxi-K-caveolin-1 interaction alters current expression in human myometrial cells. Reprod Biol Endocrinol. 2009;7:131. doi: 10.1186/1477-7827-7-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan D, Peltonen S, Dowling A, Medhat W, Green KJ, Wahl JK, 3rd, Del Galdo F, Mahoney MG. A role for caveolin-1 in desmoglein binding and desmosome dynamics. Oncogene. 2011 doi: 10.1038/onc.2011.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci M, Gratton JP, Rudic RD, Acevedo L, Roviezzo F, Cirino G, Sessa WC. In vivo delivery of the caveolin-1 scaffolding domain inhibits nitric oxide synthesis and reduces inflammation. Nature medicine. 2000;6:1362–1367. doi: 10.1038/82176. [DOI] [PubMed] [Google Scholar]
- Burgermeister E, Friedrich T, Hitkova I, Regel I, Einwachter H, Zimmermann W, Rocken C, Perren A, Wright MB, Schmid RM, et al. The Ras inhibitors caveolin-1 and docking protein-1 activate PPAR{gamma} through spatial relocalization at helix 7 of its ligand-binding domain. Mol Cell Biol. 2011 doi: 10.1128/MCB.01421-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T, Wang H, Chen Y, Liu L, Gunning WT, Quintas LE, Xie ZJ. Regulation of caveolin-1 membrane trafficking by the Na/K-ATPase. The Journal of cell biology. 2008;182:1153–1169. doi: 10.1083/jcb.200712022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Caselli A, Mazzinghi B, Camici G, Manao G, Ramponi G. Some protein tyrosine phosphatases target in part to lipid rafts and interact with caveolin-1. Biochem Biophys Res Commun. 2002;296:692–697. doi: 10.1016/s0006-291x(02)00928-2. [DOI] [PubMed] [Google Scholar]
- Chen Z, Bakhshi FR, Shajahan AN, Sharma T, Mao M, Trane A, Bernatchez P, van Nieuw Amerongen GP, Bonini MG, Skidgel RA, et al. Nitric oxide-dependent Src activation and resultant caveolin-1 phosphorylation promotes eNOS/caveolin-1 binding and eNOS inhibition. Molecular biology of the cell. 2012 doi: 10.1091/mbc.E11-09-0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZH, Lam HC, Jin Y, Kim HP, Cao J, Lee SJ, Ifedigbo E, Parameswaran H, Ryter SW, Choi AM. Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke-induced emphysema. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18880–18885. doi: 10.1073/pnas.1005574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng ZJ, Singh RD, Holicky EL, Wheatley CL, Marks DL, Pagano RE. Co-regulation of caveolar and Cdc42-dependent fluid phase endocytosis by phosphocaveolin-1. The Journal of biological chemistry. 2010;285:15119–15125. doi: 10.1074/jbc.M109.069427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman DE, Sprang SR. Structure of Gialpha1.GppNHp, autoinhibition in a galpha protein-substrate complex. The Journal of biological chemistry. 1999;274:16669–16672. doi: 10.1074/jbc.274.24.16669. [DOI] [PubMed] [Google Scholar]
- Couet J, Li S, Okamoto T, Ikezu T, Lisanti MP. Identification of peptide and protein ligands for the caveolin-scaffolding domain. Implications for the interaction of caveolin with caveolae-associated proteins. The Journal of biological chemistry. 1997;272:6525–6533. doi: 10.1074/jbc.272.10.6525. [DOI] [PubMed] [Google Scholar]
- Czarny M, Lavie Y, Fiucci G, Liscovitch M. Localization of phospholipase D in detergent-insoluble, caveolin-rich membrane domains. Modulation by caveolin-1 expression and caveolin-182-101. The Journal of biological chemistry. 1999;274:2717–2724. doi: 10.1074/jbc.274.5.2717. [DOI] [PubMed] [Google Scholar]
- Davis MJ, Zhang F, Yuan Z, Teasdale RD. MemO: a consensus approach to the annotation of a protein's membrane organization. In silico biology. 2006;6:387–399. [PubMed] [Google Scholar]
- Davis TL, Walker JR, Loppnau P, Butler-Cole C, Allali-Hassani A, Dhe-Paganon S. Autoregulation by the juxtamembrane region of the human ephrin receptor tyrosine kinase A3 (EphA3) Structure. 2008;16:873–884. doi: 10.1016/j.str.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Dong Q, Dougan DR, Gong X, Halkowycz P, Jin B, Kanouni T, O'Connell SM, Scorah N, Shi L, Wallace MB, et al. Discovery of TAK-733, a potent and selective MEK allosteric site inhibitor for the treatment of cancer. Bioorg Med Chem Lett. 2011;21:1315–1319. doi: 10.1016/j.bmcl.2011.01.071. [DOI] [PubMed] [Google Scholar]
- Feron O, Dessy C, Opel DJ, Arstall MA, Kelly RA, Michel T. Modulation of the endothelial nitric-oxide synthase-caveolin interaction in cardiac myocytes. Implications for the autonomic regulation of heart rate. The Journal of biological chemistry. 1998;273:30249–30254. doi: 10.1074/jbc.273.46.30249. [DOI] [PubMed] [Google Scholar]
- Fidanze SD, Erickson SA, Wang GT, Mantei R, Clark RF, Sorensen BK, Bamaung NY, Kovar P, Johnson EF, Swinger KK, et al. Imidazo[2,1-b]thiazoles: multitargeted inhibitors of both the insulin-like growth factor receptor and members of the epidermal growth factor family of receptor tyrosine kinases. Bioorg Med Chem Lett. 2010;20:2452–2455. doi: 10.1016/j.bmcl.2010.03.015. [DOI] [PubMed] [Google Scholar]
- Fritz-Wolf K, Kehr S, Stumpf M, Rahlfs S, Becker K. Crystal structure of the human thioredoxin reductase-thioredoxin complex. Nature communications. 2011;2:383. doi: 10.1038/ncomms1382. [DOI] [PubMed] [Google Scholar]
- Garcia-Cardena G, Martasek P, Masters BS, Skidd PM, Couet J, Li S, Lisanti MP, Sessa WC. Dissecting the interaction between nitric oxide synthase (NOS) and caveolin. Functional significance of the nos caveolin binding domain in vivo. The Journal of biological chemistry. 1997;272:25437–25440. doi: 10.1074/jbc.272.41.25437. [DOI] [PubMed] [Google Scholar]
- Gaus K, Le Lay S, Balasubramanian N, Schwartz MA. Integrin-mediated adhesion regulates membrane order. The Journal of cell biology. 2006;174:725–734. doi: 10.1083/jcb.200603034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratton JP, Lin MI, Yu J, Weiss ED, Jiang ZL, Fairchild TA, Iwakiri Y, Groszmann R, Claffey KP, Cheng YC, et al. Selective inhibition of tumor microvascular permeability by cavtratin blocks tumor progression in mice. Cancer cell. 2003;4:31–39. doi: 10.1016/s1535-6108(03)00168-5. [DOI] [PubMed] [Google Scholar]
- Hatakeyama T, Pappas PJ, Hobson RW, 2nd, Boric MP, Sessa WC, Duran WN. Endothelial nitric oxide synthase regulates microvascular hyperpermeability in vivo. The Journal of physiology. 2006;574:275–281. doi: 10.1113/jphysiol.2006.108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatters DM, Peters-Libeu CA, Weisgraber KH. Engineering conformational destabilization into mouse apolipoprotein E. A model for a unique property of human apolipoprotein E4. The Journal of biological chemistry. 2005;280:26477–26482. doi: 10.1074/jbc.M503910200. [DOI] [PubMed] [Google Scholar]
- Hezel M, de Groat WC, Galbiati F. Caveolin-3 promotes nicotinic acetylcholine receptor clustering and regulates neuromuscular junction activity. Molecular biology of the cell. 2010;21:302–310. doi: 10.1091/mbc.E09-05-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C, Berking A, Agerer F, Buntru A, Neske F, Chhatwal GS, Ohlsen K, Hauck CR. Caveolin limits membrane microdomain mobility and integrin-mediated uptake of fibronectin-binding pathogens. J Cell Sci. 2010;123:4280–4291. doi: 10.1242/jcs.064006. [DOI] [PubMed] [Google Scholar]
- Hong YH, Kim JY, Lee JH, Chae HG, Jang SS, Jeon JH, Kim CH, Kim J, Kim SJ. Agonist-induced internalization of mGluR1alpha is mediated by caveolin. J Neurochem. 2009;111:61–71. doi: 10.1111/j.1471-4159.2009.06289.x. [DOI] [PubMed] [Google Scholar]
- Hovanessian AG, Briand JP, Said EA, Svab J, Ferris S, Dali H, Muller S, Desgranges C, Krust B. The caveolin-1 binding domain of HIV-1 glycoprotein gp41 is an efficient B cell epitope vaccine candidate against virus infection. Immunity. 2004;21:617–627. doi: 10.1016/j.immuni.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Huang JH, Lu L, Lu H, Chen X, Jiang S, Chen YH. Identification of the HIV-1 gp41 core-binding motif in the scaffolding domain of caveolin-1. The Journal of biological chemistry. 2007;282:6143–6152. doi: 10.1074/jbc.M607701200. [DOI] [PubMed] [Google Scholar]
- Hubbard SR, Wei L, Ellis L, Hendrickson WA. Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature. 1994;372:746–754. doi: 10.1038/372746a0. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, Kominami E, Yamane T, Tanaka K, Komatsu M. Structural basis for sorting mechanism of p62 in selective autophagy. The Journal of biological chemistry. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- Jodoin J, Demeule M, Fenart L, Cecchelli R, Farmer S, Linton KJ, Higgins CF, Beliveau R. P-glycoprotein in blood-brain barrier endothelial cells: interaction and oligomerization with caveolins. J Neurochem. 2003;87:1010–1023. doi: 10.1046/j.1471-4159.2003.02081.x. [DOI] [PubMed] [Google Scholar]
- Ju H, Zou R, Venema VJ, Venema RC. Direct interaction of endothelial nitric-oxide synthase and caveolin-1 inhibits synthase activity. The Journal of biological chemistry. 1997;272:18522–18525. doi: 10.1074/jbc.272.30.18522. [DOI] [PubMed] [Google Scholar]
- Karpen HE, Bukowski JT, Hughes T, Gratton JP, Sessa WC, Gailani MR. The sonic hedgehog receptor patched associates with caveolin-1 in cholesterol-rich microdomains of the plasma membrane. The Journal of biological chemistry. 2001;276:19503–19511. doi: 10.1074/jbc.M010832200. [DOI] [PubMed] [Google Scholar]
- Kim JH, Han JM, Lee S, Kim Y, Lee TG, Park JB, Lee SD, Suh PG, Ryu SH. Phospholipase D1 in caveolae: regulation by protein kinase Calpha and caveolin-1. Biochemistry. 1999;38:3763–3769. doi: 10.1021/bi982478+. [DOI] [PubMed] [Google Scholar]
- Kirkham M, Fujita A, Chadda R, Nixon SJ, Kurzchalia TV, Sharma DK, Pagano RE, Hancock JF, Mayor S, Parton RG. Ultrastructural identification of uncoated caveolin-independent early endocytic vehicles. The Journal of cell biology. 2005;168:465–476. doi: 10.1083/jcb.200407078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham M, Nixon SJ, Howes MT, Abi-Rached L, Wakeham DE, Hanzal-Bayer M, Ferguson C, Hill MM, Fernandez-Rojo M, Brown DA, et al. Evolutionary analysis and molecular dissection of caveola biogenesis. J Cell Sci. 2008;121:2075–2086. doi: 10.1242/jcs.024588. [DOI] [PubMed] [Google Scholar]
- Kong MM, Hasbi A, Mattocks M, Fan T, O'Dowd BF, George SR. Regulation of D1 dopamine receptor trafficking and signaling by caveolin-1. Mol Pharmacol. 2007;72:1157–1170. doi: 10.1124/mol.107.034769. [DOI] [PubMed] [Google Scholar]
- Kubala MH, Kovtun O, Alexandrov K, Collins BM. Structural and thermodynamic analysis of the GFP:GFP-nanobody complex. Protein Sci. 2010;19:2389–2401. doi: 10.1002/pro.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, Dixon JE, Pandolfi P, Pavletich NP. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- Levin AM, Murase K, Jackson PJ, Flinspach ML, Poulos TL, Weiss GA. Double barrel shotgun scanning of the caveolin-1 scaffolding domain. ACS chemical biology. 2007;2:493–500. doi: 10.1021/cb700055t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson NM, Kuchment O, Shen K, Young MA, Koldobskiy M, Karplus M, Cole PA, Kuriyan J. A Src-like inactive conformation in the abl tyrosine kinase domain. PLoS Biol. 2006;4:e144. doi: 10.1371/journal.pbio.0040144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Couet J, Lisanti MP. Src tyrosine kinases, Galpha subunits, and H-Ras share a common membrane-anchored scaffolding protein, caveolin. Caveolin binding negatively regulates the auto-activation of Src tyrosine kinases. The Journal of biological chemistry. 1996;271:29182–29190. doi: 10.1074/jbc.271.46.29182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SW, Okamoto T, Chun MY, Sargiacomo M, Casanova JE, Hansen SH, Nishimoto I, Lisanti MP. Evidence for a regulated interaction between heterotrimeric G proteins and caveolin. J. Biol. Chem. 1995;270:15693–15701. doi: 10.1074/jbc.270.26.15693. [DOI] [PubMed] [Google Scholar]
- Li W, Liu H, Zhou JS, Cao JF, Zhou XB, Choi AM, Shen HH, Chen ZH. Caveolin-1 inhibits expression of antioxidant enzymes through direct interaction with nuclear erythroid 2 p45-related factor-2 (Nrf2) The Journal of biological chemistry. 2012 doi: 10.1074/jbc.M112.352336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisanti MP, Tang Z, Scherer PE, Kubler E, Koleske AJ, Sargiacomo M. Caveolae, transmembrane signalling and cellular transformation. Mol Membr Biol. 1995;12:121–124. doi: 10.3109/09687689509038506. [DOI] [PubMed] [Google Scholar]
- Liu P, Rudick M, Anderson RG. Multiple functions of caveolin-1. The Journal of biological chemistry. 2002;277:41295–41298. doi: 10.1074/jbc.R200020200. [DOI] [PubMed] [Google Scholar]
- Mao C, Zhou M, Uckun FM. Crystal structure of Bruton's tyrosine kinase domain suggests a novel pathway for activation and provides insights into the molecular basis of X-linked agammaglobulinemia. The Journal of biological chemistry. 2001;276:41435–41443. doi: 10.1074/jbc.M104828200. [DOI] [PubMed] [Google Scholar]
- McNicholas S, Potterton E, Wilson KS, Noble ME. Presenting your structures: the CCP4mg molecular-graphics software. Acta crystallographica. Section D, Biological crystallography. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel JB, Feron O, Sase K, Prabhakar P, Michel T. Caveolin versus calmodulin. Counterbalancing allosteric modulators of endothelial nitric oxide synthase. The Journal of biological chemistry. 1997;272:25907–25912. doi: 10.1074/jbc.272.41.25907. [DOI] [PubMed] [Google Scholar]
- Mineo C, Anderson RG, White MA. Physical association with ras enhances activation of membrane-bound raf (RafCAAX) The Journal of biological chemistry. 1997;272:10345–10348. doi: 10.1074/jbc.272.16.10345. [DOI] [PubMed] [Google Scholar]
- Mineo C, Ying YS, Chapline C, Jaken S, Anderson RG. Targeting of protein kinase Calpha to caveolae. The Journal of cell biology. 1998;141:601–610. doi: 10.1083/jcb.141.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo S, Wang L, Li Q, Li J, Li Y, Thannickal VJ, Cui Z. Caveolin-1 regulates dorsoventral patterning through direct interaction with beta-catenin in zebrafish. Dev Biol. 2010;344:210–223. doi: 10.1016/j.ydbio.2010.04.033. [DOI] [PubMed] [Google Scholar]
- Morth JP, Pedersen BP, Toustrup-Jensen MS, Sorensen TL, Petersen J, Andersen JP, Vilsen B, Nissen P. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- Nethe M, Anthony EC, Fernandez-Borja M, Dee R, Geerts D, Hensbergen PJ, Deelder AM, Schmidt G, Hordijk PL. Focal-adhesion targeting links caveolin-1 to a Rac1-degradation pathway. J Cell Sci. 2010;123:1948–1958. doi: 10.1242/jcs.062919. [DOI] [PubMed] [Google Scholar]
- Nishimura A, Kitano K, Takasaki J, Taniguchi M, Mizuno N, Tago K, Hakoshima T, Itoh H. Structural basis for the specific inhibition of heterotrimeric Gq protein by a small molecule. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13666–13671. doi: 10.1073/pnas.1003553107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystrom FH, Chen H, Cong LN, Li Y, Quon MJ. Caveolin-1 interacts with the insulin receptor and can differentially modulate insulin signaling in transfected Cos-7 cells and rat adipose cells. Mol Endocrinol. 1999;13:2013–2024. doi: 10.1210/mend.13.12.0392. [DOI] [PubMed] [Google Scholar]
- O'Neal CJ, Amaya EI, Jobling MG, Holmes RK, Hol WG. Crystal structures of an intrinsically active cholera toxin mutant yield insight into the toxin activation mechanism. Biochemistry. 2004;43:3772–3782. doi: 10.1021/bi0360152. [DOI] [PubMed] [Google Scholar]
- Oka N, Yamamoto M, Schwencke C, Kawabe J, Ebina T, Ohno S, Couet J, Lisanti MP, Ishikawa Y. Caveolin interaction with protein kinase C. Isoenzyme-dependentregulation of kinase activity by the caveolin scaffolding domain peptide. The Journal of biological chemistry. 1997;272:33416–33421. doi: 10.1074/jbc.272.52.33416. [DOI] [PubMed] [Google Scholar]
- Okamoto T, Schlegel A, Scherer PE, Lisanti MP. Caveolins, a family of scaffolding proteins for organizing "preassembled signaling complexes" at the plasma membrane. The Journal of biological chemistry. 1998;273:5419–5422. doi: 10.1074/jbc.273.10.5419. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Collins BM, Evans PR. Adaptors for clathrin coats: structure and function. Annu Rev Cell Dev Biol. 2004;20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- Pany S, Vijayvargia R, Krishnasastry MV. Caveolin-1 binding motif of alpha-hemolysin: its role in stability and pore formation. Biochem Biophys Res Commun. 2004;322:29–36. doi: 10.1016/j.bbrc.2004.07.073. [DOI] [PubMed] [Google Scholar]
- Parton RG, Simons K. The multiple faces of caveolae. Nat Rev Mol Cell Biol. 2007;8:185–194. doi: 10.1038/nrm2122. [DOI] [PubMed] [Google Scholar]
- Pike LJ. Growth factor receptors, lipid rafts and caveolae: an evolving story. Biochimica et biophysica acta. 2005;1746:260–273. doi: 10.1016/j.bbamcr.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Place AT, Chen Z, Bakhshi FR, Liu G, O'Bryan JP, Minshall RD. Cooperative role of Caveolin-1 and C-terminal Src kinase binding protein in C-terminal Src kinase-mediated negative regulation of c-Src. Mol Pharmacol. 2011;80:665–672. doi: 10.1124/mol.111.073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid D, Leser GP, Lamb RA. A role for caveolin 1 in assembly and budding of the paramyxovirus parainfluenza virus 5. J Virol. 2010;84:9749–9759. doi: 10.1128/JVI.01079-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Riffel N, Harlos K, Iourin O, Rao Z, Kingsman A, Stuart D, Fry E. Atomic resolution structure of Moloney murine leukemia virus matrix protein and its relationship to other retroviral matrix proteins. Structure. 2002;10:1627–1636. doi: 10.1016/s0969-2126(02)00896-1. [DOI] [PubMed] [Google Scholar]
- Rohrer J, Schweizer A, Russell D, Kornfeld S. The targeting of Lamp1 to lysosomes is dependent on the spacing of its cytoplasmic tail tyrosine sorting motif relative to the membrane. The Journal of cell biology. 1996;132:565–576. doi: 10.1083/jcb.132.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld RJ, Garcin ED, Panda K, Andersson G, Aberg A, Wallace AV, Morris GM, Olson AJ, Stuehr DJ, Tainer JA, et al. Conformational changes in nitric oxide synthases induced by chlorzoxazone and nitroindazoles: crystallographic and computational analyses of inhibitor potency. Biochemistry. 2002;41:13915–13925. doi: 10.1021/bi026313j. [DOI] [PubMed] [Google Scholar]
- Santibanez JF, Blanco FJ, Garrido-Martin EM, Sanz-Rodriguez F, del Pozo MA, Bernabeu C. Caveolin-1 interacts and cooperates with the transforming growth factor-beta type I receptor ALK1 in endothelial caveolae. Cardiovasc Res. 2008;77:791–799. doi: 10.1093/cvr/cvm097. [DOI] [PubMed] [Google Scholar]
- Sato Y, Sagami I, Shimizu T. Identification of caveolin-1-interacting sites in neuronal nitric-oxide synthase. Molecular mechanism for inhibition of NO formation. The Journal of biological chemistry. 2004;279:8827–8836. doi: 10.1074/jbc.M310327200. [DOI] [PubMed] [Google Scholar]
- Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song KS, Li S, Okamoto T, Quilliam LA, Sargiacomo M, Lisanti MP. Co-purification and direct interaction of Ras with caveolin, an integral membrane protein of caveolae microdomains. Detergent-free purification of caveolae microdomains. The Journal of biological chemistry. 1996;271:9690–9697. doi: 10.1074/jbc.271.16.9690. [DOI] [PubMed] [Google Scholar]
- Song KS, Sargiacomo M, Galbiati F, Parenti M, Lisanti MP. Targeting of a G alpha subunit (Gi1 alpha) and c-Src tyrosine kinase to caveolae membranes: clarifying the role of N-myristoylation. Cellular and molecular biology. 1997;43:293–303. [PubMed] [Google Scholar]
- Sugishima M, Omata Y, Kakuta Y, Sakamoto H, Noguchi M, Fukuyama K. Crystal structure of rat heme oxygenase-1 in complex with heme. FEBS Lett. 2000;471:61–66. doi: 10.1016/s0014-5793(00)01353-3. [DOI] [PubMed] [Google Scholar]
- Sui H, Han BG, Lee JK, Walian P, Jap BK. Structural basis of water-specific transport through the AQP1 water channel. Nature. 2001;414:872–878. doi: 10.1038/414872a. [DOI] [PubMed] [Google Scholar]
- Sundivakkam PC, Kwiatek AM, Sharma TT, Minshall RD, Malik AB, Tiruppathi C. Caveolin-1 scaffold domain interacts with TRPC1 and IP3R3 to regulate Ca2+ store release-induced Ca2+ entry in endothelial cells. Am J Physiol Cell Physiol. 2009;296:C403–C413. doi: 10.1152/ajpcell.00470.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira J, Sugishima M, Kida Y, Oda E, Noguchi M, Higashimoto Y. Caveolin-1 Is a Competitive Inhibitor of Heme Oxygenase-1 (HO-1) with Heme: Identification of a Minimum Sequence in Caveolin-1 for Binding to HO-1. Biochemistry. 2011;50:6824–6831. doi: 10.1021/bi200601t. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Hirano N, Kaneko J, Kamio Y, Yao M, Tanaka I. 2-Methyl-2,4-pentanediol induces spontaneous assembly of staphylococcal alpha-hemolysin into heptameric pore structure. Protein Sci. 2011;20:448–456. doi: 10.1002/pro.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till JH, Becerra M, Watty A, Lu Y, Ma Y, Neubert TA, Burden SJ, Hubbard SR. Crystal structure of the MuSK tyrosine kinase: insights into receptor autoregulation. structure. 2002;10:1187–1196. doi: 10.1016/s0969-2126(02)00814-6. [DOI] [PubMed] [Google Scholar]
- Toya Y, Schwencke C, Couet J, Lisanti MP, Ishikawa Y. Inhibition of adenylyl cyclase by caveolin peptides. Endocrinology. 1998;139:2025–2031. doi: 10.1210/endo.139.4.5957. [DOI] [PubMed] [Google Scholar]
- Traub LM. Tickets to ride: selecting cargo for clathrin-regulated internalization. Nat Rev Mol Cell Biol. 2009;10:583–596. doi: 10.1038/nrm2751. [DOI] [PubMed] [Google Scholar]
- Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Vargas L, Nore BF, Berglof A, Heinonen JE, Mattsson PT, Smith CI, Mohamed AJ. Functional interaction of caveolin-1 with Bruton's tyrosine kinase and Bmx. The Journal of biological chemistry. 2002;277:9351–9357. doi: 10.1074/jbc.M108537200. [DOI] [PubMed] [Google Scholar]
- Venema RC, Ju H, Zou R, Ryan JW, Venema VJ. Subunit interactions of endothelial nitric-oxide synthase. Comparisons to the neuronal and inducible nitric-oxide synthase isoforms. The Journal of biological chemistry. 1997;272:1276–1282. doi: 10.1074/jbc.272.2.1276. [DOI] [PubMed] [Google Scholar]
- Vihanto MM, Vindis C, Djonov V, Cerretti DP, Huynh-Do U. Caveolin-1 is required for signaling and membrane targeting of EphB1 receptor tyrosine kinase. J Cell Sci. 2006;119:2299–2309. doi: 10.1242/jcs.02946. [DOI] [PubMed] [Google Scholar]
- Volonte D, Galbiati F. Inhibition of thioredoxin reductase 1 by caveolin 1 promotes stress-induced premature senescence. EMBO reports. 2009;10:1334–1340. doi: 10.1038/embor.2009.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J, von Matt P, Sedrani R, Albert R, Cooke N, Ehrhardt C, Geiser M, Rummel G, Stark W, Strauss A, et al. Discovery of 3-(1H-indol-3-yl)-4-[2-(4-methylpiperazin-1-yl)quinazolin-4-yl]pyrrole-2,5 -dione (AEB071), a potent and selective inhibitor of protein kinase C isotypes. J Med Chem. 2009;52:6193–6196. doi: 10.1021/jm901108b. [DOI] [PubMed] [Google Scholar]
- Wang W, Liu L, Song X, Mo Y, Komma C, Bellamy HD, Zhao ZJ, Zhou GW. Crystal structure of human protein tyrosine phosphatase SHP-1 in the open conformation. J Cell Biochem. 2011;112:2062–2071. doi: 10.1002/jcb.23125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XM, Kim HP, Nakahira K, Ryter SW, Choi AM. The heme oxygenase-1/carbon monoxide pathway suppresses TLR4 signaling by regulating the interaction of TLR4 with caveolin-1. J Immunol. 2009;182:3809–3818. doi: 10.4049/jimmunol.0712437. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang X, Jasmin JF, Lau WB, Li R, Yuan Y, Yi W, Chuprun K, Lisanti MP, Koch WJ, et al. Essential role of caveolin-3 in adiponectin signalsome formation and adiponectin cardioprotection. Arteriosclerosis, thrombosis, and vascular biology. 2012;32:934–942. doi: 10.1161/ATVBAHA.111.242164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yamaguchi K, Wada T, Hata K, Zhao X, Fujimoto T, Miyagi T. A close association of the ganglioside-specific sialidase Neu3 with caveolin in membrane microdomains. The Journal of biological chemistry. 2002;277:26252–26259. doi: 10.1074/jbc.M110515200. [DOI] [PubMed] [Google Scholar]
- Warne T, Moukhametzianov R, Baker JG, Nehme R, Edwards PC, Leslie AG, Schertler GF, Tate CG. The structural basis for agonist and partial agonist action on a beta(1)-adrenergic receptor. Nature. 2011;469:241–244. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyse BD, Prior IA, Qian H, Morrow IC, Nixon S, Muncke C, Kurzchalia TV, Thomas WG, Parton RG, Hancock JF. Caveolin interacts with the angiotensin II type 1 receptor during exocytic transport but not at the plasma membrane. The Journal of biological chemistry. 2003;278:23738–23746. doi: 10.1074/jbc.M212892200. [DOI] [PubMed] [Google Scholar]
- Xia H, Khalil W, Kahm J, Jessurun J, Kleidon J, Henke CA. Pathologic caveolin-1 regulation of PTEN in idiopathic pulmonary fibrosis. Am J Pathol. 2010;176:2626–2637. doi: 10.2353/ajpath.2010.091117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Takemaru K, Liu J, Berndt JD, Zheng JJ, Moon RT, Xu W. Crystal structure of a full-length beta-catenin. Structure. 2008;16:478–487. doi: 10.1016/j.str.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Tao X, Shen B, Horng T, Medzhitov R, Manley JL, Tong L. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 2000;408:111–115. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- Yang ZN, Mueser TC, Kaufman J, Stahl SJ, Wingfield PT, Hyde CC. The crystal structure of the SIV gp41 ectodomain at 1.47 A resolution. J Struct Biol. 1999;126:131–144. doi: 10.1006/jsbi.1999.4116. [DOI] [PubMed] [Google Scholar]
- Yu RC, Jahn R, Brunger AT. NSF N-terminal domain crystal structure: models of NSF function. Mol Cell. 1999;4:97–107. doi: 10.1016/s1097-2765(00)80191-4. [DOI] [PubMed] [Google Scholar]
- Yu Z, Beer C, Koester M, Wirth M. Caveolin-1 interacts with the Gag precursor of murine leukaemia virus and modulates virus production. Virol J. 2006;3:73. doi: 10.1186/1743-422X-3-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Leonetti MD, Pico AR, Hsiung Y, MacKinnon R. Structure of the human BK channel Ca2+-activation apparatus at 3.0 A resolution. Science. 2010;329:182–186. doi: 10.1126/science.1190414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue L, Mazzone T. Endogenous adipocyte apolipoprotein E is colocalized with caveolin at the adipocyte plasma membrane. J Lipid Res. 2011;52:489–498. doi: 10.1194/jlr.M011809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Schwegler-Berry D, Castranova V, He P. Internalization of caveolin-1 scaffolding domain facilitated by Antennapedia homeodomain attenuates PAF-induced increase in microvessel permeability. American journal of physiology. Heart and circulatory physiology. 2004;286:H195–H201. doi: 10.1152/ajpheart.00667.2003. [DOI] [PubMed] [Google Scholar]
- Zou P, Wu F, Lu L, Huang JH, Chen YH. The cytoplasmic domain of influenza M2 protein interacts with caveolin-1. Arch Biochem Biophys. 2009;486:150–154. doi: 10.1016/j.abb.2009.02.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.